Abstract
NCoA6 (also referred to as NRC, ASC-2, TRBP, PRIP and RAP250) was originally isolated as a ligand-dependent nuclear receptor interacting protein. However, NCoA6 is a multifunctional coregulator or coactivator necessary for transcriptional activation of a wide spectrum of target genes. The NCoA6 gene is amplified and overexpressed in breast, colon and lung cancers. NCoA6 is a 250 kDa protein which harbors a potent N-terminal activation domain, AD1; and a second, centrally-located activation domain, AD2, which is necessary for nuclear receptor signaling. The intrinsic activation potential of NCoA6 is regulated by its C-terminal STL regulatory domain. Near AD2 is an LxxLL-1 motif which interacts with a wide spectrum of ligand-bound NRs with high-affinity. A second LxxLL motif (LxxLL-2) located towards the C-terminal region is more restricted in its NR specificity. The potential role of NCoA6 as a co-integrator is suggested by its ability to enhance transcriptional activation of a wide variety of transcription factors and from its in vivo association with a number of known cofactors including CBP/p300. NCoA6 has been shown to associate with at least three distinct coactivator complexes containing Set methyltransferases as core polypeptides. The composition of these complexes suggests that NCoA6 may play a fundamental role in transcriptional activation by modulating chromatin structure through histone methylation. Knockout studies in mice suggest that NCoA6 is an essential coactivator. NCoA6-/- embryos die between 8.5-12.5 dpc from general growth retardation coupled with developmental defects in the heart, liver, brain and placenta. NCoA6-/- MEFs grow at a reduced rate compared to WT MEFs and spontaneously undergo apoptosis, indicating the importance of NCoA6 as a prosurvival and anti-apoptotic gene. Studies with NCoA6+/- and conditional knockout mice suggest that NCoA6 is a pleiotropic coregulator involved in growth, development, wound healing and maintenance of energy homeostasis.
History
In metazoans, more than 70 different nuclear hormone receptors (NRs) have been identified (http://www.nursa.org), which comprise a superfamily. All NRs share a common modular structure comprised of an N-terminal variable "A/B" domain, a central zinc-finger containing DNA binding "C" domain (DBD), a "hinge region" referred to as the "D" region and a C-terminal "EF" domain. The C-terminal EF region (and for some receptors DEF) binds ligand and comprises the ligand binding domain (LBD), which harbors a ligand-dependent activation function referred to as AF2 [Aranda and Pascual, 2001]. A variety of small molecules such as endocrine hormones, fatty acids, cholesterol derivatives and products of lipid metabolism act as ligands for various NRs. In general, NRs are involved in growth and development, maintenance of cellular homeostasis, energy metabolism, inflammation, obesity and insulin resistance [Anghel and Wahli, 2007; Evans et al., 2004]. The mechanism that underlies these processes involves transcriptional activation or repression of NR target genes. Thus, NRs function as binary switches. In the apoform or unliganded state, some of the NRs repress transcription, while in the liganded state, NRs act as transcriptional activators. NR-mediated activation and repression has been the focus of research for the last two decades. However, the identification of NRs as sensors of cellular metabolism led to a renewed interest in identification and cloning of corepressors and coactivators for NRs.
The research regarding the mechanism of transcriptional activation by NRs evolved in the early 1990s from a simple model that depicted the association of NRs with ligands and basal factors of the transcription apparatus [Aranda and Pascual, 2001]. The current model on the mechanism of NR-mediated activation takes into account the complex macromolecular network of inter- and intramolecular interactions of chromatin remodeling factors, Mediator components and coactivator complexes. These macromolecular complexes, in concert with NRs, the basal transcription complex and RNA polymerase cause productive transcription of NR-regulated target genes. The discovery of coactivators was the result of a cumulative endeavor. A number of laboratories contributed to identification, cloning and characterization of various NR coactivators. Initial impetus in the identification of coactivators was provided by the discovery of 140 kDa and 160 kDa proteins known as ER-associated proteins (ERAPs) [Halachmi et al., 1994] and receptor-interacting proteins (RIPs) [Cavailles et al., 1994]. Thus, it became clear that the process of NR activation is orchestrated by regulators of transcription now popularly known as coactivators. In 1995, the first member of the p160 family of coactivators was cloned and identified as SRC-1 [Onate et al., 1995]. It was soon followed by cloning of TIF-2/GRIP-1 (SRC-2) [Hong et al., 1996; Voegel et al., 1996] and pCIP, ACTR, RAC-3, TRAM-1, AIB1 (SRC-3) by various groups [Anzick et al., 1997; Chen et al., 1997; Li et al., 1997; Takeshita et al., 1997; Torchia et al., 1997].
The hunt for novel coactivators for NRs continued using both yeast two-hybrid screening and biochemical purification of protein complexes associated with liganded NRs. Two similar, if not identical, multiprotein coactivator complexes referred to as TRAPs (thyroid hormone receptor interacting proteins) [Fondell et al., 1996] and DRIPs (vitamin D receptor interacting proteins) [Rachez et al., 1999] were isolated from HeLa cells using in vivo (TRAPs) and in vitro (DRIPs) biochemical approaches. The TRAP/DRIP complex is one of several related complexes which share components and are thought to modulate the activities of a variety of transcription factors including the NRs, Sp1, NF-kB (p65), VP16, E1A and p53 [Boyer et al., 1999; Ito and Roeder, 2001; Naar et al., 1999; Rachez et al., 1999]. CBP/p300 was identified as a coactivator for NRs [Chakravarti et al., 1996] and is considered a transcriptional integrator which is recruited to the NRs and other transcription factors through their association with other coactivators (e.g., the SRCs and NCoA6(NRC)) [Chan and La Thangue, 2001; Chen et al., 1997; Goodman and Smolik, 2000; Mahajan and Samuels, 2000; Tian et al., 2006; Torchia et al., 1997].
Mechanistically, coactivators are thought to act by modifying chromatin structure and/or through direct interaction with components of the TFIID basal transcription apparatus [Baek et al., 2002; McKenna and O'Malley, 2002]. Thus, liganded NRs bind members of the p160 family, which recruit CBP/p300 (and CBP bound p/CAF) to a target gene promoter. This recruitment locally modifies chromatin structure through the CBP/p300 and p/CAF histone acetyltransferase (HAT) activities. In addition, members of the p160 family associate with CARM1 (coactivator associated arginine methyltransferase 1) and PRMT1 (protein arginine methyltransferase 1), which potentiate receptor activation. The effect of CARM1 and PRMT1 on receptor activity is thought to result from chromatin changes through histone methylation [Stallcup, 2001; Strahl et al., 2001]. Thus, PRMT1 has been shown to methylate Arg-3 of histone H4, which enhances the extent of acetylation of H4 tails by CBP/p300 [Strahl et al., 2001]. The role of the DRIP/TRAP complex is less well defined, although recent studies indicate that the TRAP complex interacts with the TAFII components of the TFIID complex and affects both basal activity and stimulation of transcription by activators [Baek et al., 2002]. Chromatin immunoprecipitation assays (ChIP) indicate that p160 coactivators and CBP/p300 are recruited to NR target genes rapidly (30 min to 1 h) after ligand binding [Sharma and Fondell, 2002]. DRIP/TRAP is subsequently recruited to the receptor-bound promoter and ChIP assays show periodic cycling of p160 coactivators and TRAP220. These findings suggest that p160 coactivators and CBP/p300 modify chromatin and allow for the subsequent recruitment of the DRIP/TRAP complex, which may target the RNA Polymerase II apparatus [Sharma and Fondell, 2002].
Beyond the p160/SRC family and the DRIP/TRAP complex, nearly 250 other putative coactivators have been identified thus far (http://www.nursa.org). Coactivators which have been extensively studied include CBP, p300, p160 family members, TRAP220/DRIP205/PBP, PGC-1, NRIF3 and NCoA6(NRC) [Mahajan and Samuels, 2005]. TRAP220/DRIP205 was isolated biochemically as one of the components of the TRAP/DRIP complex and also by two-hybrid screening as PBP using PPARγ as a bait [Zhu et al., 1997]. PGC-1 was isolated as a coactivator for PPARγ involved in thermogenesis in brown adipose tissue [Puigserver et al., 1998]. NRIF3 acts as a receptor selective coactivator for the TRs and the RXRs and interaction with these NRs occurs through a novel LxxIL motif [Li et al., 1999; Li et al., 2001; Li et al., 2005]. NRC (Nuclear Receptor Coregulator) [Mahajan and Samuels, 2000], also referred to as ASC-2 [Lee et al., 1999], RAP250 [Caira et al., 2000], TRBP [Ko et al., 2000] and PRIP [Zhu et al., 2000], was cloned and characterized as a coactivator simultaneously by a number of laboratories [Mahajan and Samuels, 2005]. The NCBI has annotated human NRC and the other identical factors (ASC-2, RAP250, PRIP and TRBP) as NCoA6 (nuclear receptor coactivator 6) (NCBI Accession # NM_014071). AIB3 (Amplified in Breast Cancer 3) is a N-terminal variant of NCoA6 (NCBI Accession # 208277), which was first identified as one of the genes on chromosome 20 that was overexpressed in BT-474 breast cancer cells [Guan et al., 1996]. However, in some papers NCoA6 has been inadvertently referred to as AIB3. In the short time following its discovery, NCoA6 has emerged as an important coactivator not only for NRs, but also for a number of other well known transcription factors such as c-Fos, c-Jun, CREB, NF-kB, ATF-2, heat shock factors, E2F-1, SRF, Rb, p53 and Stat2 [Goo et al., 2004; Hong et al., 2004a; Hong et al., 2004b; Ko et al., 2000; Kong et al., 2003; Lee et al., 1999; Lee et al., 2000; Mahajan et al., 2007; Mahajan et al., 2002; Mahajan and Samuels, 2000; Mahajan and Samuels, 2005]. The biological importance of NCoA6 as an essential, non-redundant coactivator for embryonic growth and viability is evident from recent biochemical and genetic studies involving NCoA6 knockout mice [Antonson et al., 2003; Kuang et al., 2002; Mahajan et al., 2004; Zhu et al., 2003]. A wealth of information has accumulated on NCoA6 over the past 7 years thanks to contributions from a number of laboratories. In this review, we summarize this published information and highlight the recent advances in our understanding of how NCoA6 functions as a transcriptional coregulator. For brevity, we refer to NRC, ASC-2, RAP250, TRBP and PRIP as NCoA6, as designated by NCBI. However, the original nomenclature has been retained while citing a specific laboratory’s work on NCoA6.
Cloning, domain structure, functional organization and expression of the nuclear receptor coregulator/coactivator, NCoA6
Cloning
NCoA6 was cloned as NRC, ASC-2, RAP250, TRBP and PRIP [Caira et al., 2000; Ko et al., 2000; Lee et al., 1999; Mahajan and Samuels, 2000; Zhu et al., 2000] by various groups. It is interesting to note that all the groups cloned NCoA6 by yeast two-hybrid cDNA screening using various NRs as baits. ASC-2 was isolated from a Xenopus cDNA library using RXR as a bait [Lee et al., 1999], RAP250 from a mouse embryo cDNA library using PPARα as a bait [Caira et al., 2000], TRBP was cloned from a rat GC cell library using TR as a bait [Ko et al., 2000], while PRIP was cloned from a mouse liver cDNA library using PPARγ as a bait [Zhu et al., 2000]. A second isoform of human NCoA6 has been identified and is referred to as AIB3 (Amplified in Breast Cancer 3). AIB3 is believed to be a 2001 amino acid protein, which differs from NCoA6 at their N-termini, where the first 88 amino acids of NCoA6 are replaced by 26 different amino acids in AIB3 (NCBI Accession # AF208277). The physical isolation of AIB3 cDNA and detailed studies may reveal some functional differences between the two. Our laboratory cloned NRC [Mahajan and Samuels, 2000] from a GH4C1-derived cDNA library using full length TR as a bait. Both structural and functional studies carried out in detail in our laboratory and others demonstrated that NCoA6 functions as a coactivator for NRs.
Protein domain structure and functional organization of NCoA6
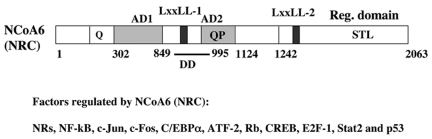
Human NCoA6 is a ∼250 kDa protein containing 2063 amino acids, while mouse NCoA6(PRIP) is a protein of 2068 amino acids [Zhu et al., 2000]. The rat genome database has recently annotated a sequence for rat NCoA6(NRC) mRNA with a predicted protein of 1844 amino acids. The identity among human, mouse and rat NCoA6(NRC) is over 90% at the amino acid level. Structure-function analysis of human NCoA6(NRC) in yeast and in mammalian cells [Mahajan and Samuels, 2000] has revealed a modular structure for NCoA6(NRC) (Figure 1) consisting of: 1) a potent activation domain (AD1), 2) a second Glu-, Pro-rich activation domain (AD2) involved in NR signaling, 3) LxxLL-1, which plays an essential role for ligand-dependent interactions with a wide variety of NRs and LxxLL-2, which is more limited in its NR interactions, 4) a dimerization region and 5) an inhibitory region at the C-terminus rich in Ser, Thr, and Leu. AD1 of NCoA6(NRC) is localized towards the N-terminus of NCoA6(NRC) and is highly Glu- and Pro-rich and moderately high in Gly, Asn and Ser amino acids. AD1, encompassing 481 amino acid residues (amino acids 302 -783), is a potent activator in mammalian cells and is functionally as potent in activity as the Herpes Simplex VP16 activation domain. Although it is generally believed that Glu-rich sequences are potential activating motifs in eukaryotes, the first 302 amino acids of NCoA6(NRC) flanking the AD1 and containing a Glu stretch (34 residues) does not function as an autonomous activation domain when expressed in mammalian cells as a Gal4-DBD fusion [Mahajan and Samuels, 2000]. Interestingly, AD1 was not found to be necessary for NR activation [Mahajan et al., 2007]. AD2, containing 184 residues (amino acids 940-1124), is juxtaposed to the LxxLL-1 region, which is very rich in Glu and Pro amino acids. This Glu/Pro rich region is necessary for AD2 activity [Mahajan and Samuels, 2000]. Interestingly, the AD2 domain was found to be essential for NR signaling and AF2-dependent enhancement of NRs by NCoA6(NRC) [Mahajan et al., 2007]. The C-terminal end of NCoA6(NRC) (∼600 amino acids) contains a region we designated as STL, which is rich in Ser, Thr and Leu. The STL region is inhibitory in the context of full length NCoA6(NRC) and its deletion enhances the intrinsic activation potential of AD1 and AD2 in NCoA6(NRC) [Mahajan and Samuels, 2000]. Thus, the role of the STL domain may be to control the transcriptional output of NCoA6(NRC) through inter- and intramolecular interactions. The STL region contains 9 predicted protein kinase A phosphorylation sites in two clusters and 14 predicted casein kinase II phosphorylation sites in four clusters. Thus, the function of the STL region may potentially be modulated by phosphorylation-dephosphorylation, which could alter the activity of NCoA6(NRC). Although rich in Leu, the C-terminal region of NCoA6(NRC) does not contain any apparent leucine zipper-like motifs. However, the high content of Leu, Ile and Val may facilitate certain hydrophobic-driven interactions of NCoA6(NRC) with other proteins to regulate the activation potential of NCoA6(NRC) in vivo. At this juncture, experimental evidence for any posttranslational modification in the C-terminus is lacking.
Figure 1. Domain organization of NCoA6(NRC).
NCoA6(NRC) contains two activation domains (AD1 and AD2) and two LxxLL receptor interacting motifs (LxxLL-1 and LxxLL-2). LxxLL-1 interacts with a wide variety of NRs, while LxxLL-2 is more restricted in NR recognition. The C-terminal region of NCoA6(NRC) is rich in Ser, Thr and Leu (STL), and appears to function to repress the activation domains of the protein. NCoA6(NRC) forms homodimers through a dimerization domain (DD), which is located near LxxLL-1. Factors that are known to be regulated by NCoA6(NRC) are also listed.
LxxLL modules as NR interaction motifs (NR boxes) have been identified in most of the coactivators. p160 family members contain multiple LxxLL modules, all of which interact with a variety of NRs, albeit with some differences in specificity [McInerney et al., 1998]. Barring one initial report with NCoA6(ASC-2), which suggested that NRs interact through a region of NCoA6(ASC-2) (amino acids 586-860) that did not contain any LxxLL motif [Lee et al., 1999], all other studies confirmed that of the two LxxLL motifs in NCoA6, LxxLL-1 is necessary for interaction with a wide spectrum of NRs including TRs, RXRs, RARs, GR, ERs, VDR and the PPARs [Caira et al., 2000; Ko et al., 2000; Mahajan and Samuels, 2000]. Mutation of LVNLL (LxxLL-1) to AVNAA or to AVNAL or to LVAAA abrogated the association of NCoA6 with NRs in vitro [Caira et al., 2000; Ko et al., 2000; Mahajan and Samuels, 2000] and in vivo [Mahajan and Samuels, 2000]. Through the use of combinatorial peptide libraries, LxxLL modules have been characterized as Class 1, 2 or 3 depending on the nature of the amino acid residue at position -1 or -2 of LxxLL (LxxLL=+1+2+3+4+5, respectively) [Chang et al., 1999]. The Class 2 LxxLL module with a Pro at -2 has been shown to interact with various NRs. The LxxLL-1 region of NCoA6(NRC) (LTSPLLVNLLQSDIS) resembles the Class 2 LxxLL module and contains a Pro at the -2 position. Members of the p160 family of coactivators contain multiple Class I LxxLL modules (SRLxxLL), all of which associate with a variety of NRs, albeit with some difference in specificity. In contrast, NCoA6(NRC) contains a single functional LxxLL-1 module that associates with a variety of NRs in vitro and in vivo with high affinity. Interestingly, the Pro at the -2 position found in the NCoA6(NRC) LxxLL-1 motif is also found in the LxxLL (PILTSLL) of TRAP220/DRIP205/PBP and RIP140 LxxLL (PILYYML) motif. The NCoA6(NRC) LxxLL-1 module and surrounding amino acids is very hydrophobic and interacts with virtually all liganded NRs with high affinity. Helix 12 of the NR AF2 domain is critical in the formation of an interface which contacts the NCoA6(NRC) LxxLL-1 module. Point mutations in helix 12 of cTRα (L398R), or deletion of helix 12, abolish the association of the NR with NCoA6(NRC) in vitro and in vivo [Mahajan and Samuels, 2000]. Similarly, deletion of helix 12 (last 12 amino acids) of PPARγ resulted in the abolition of the interaction with NCoA6(PRIP) [Zhu et al., 2000].
In yeast two-hybrid assays, full-length NCoA6(NRC) interacts with ligand-bound NRs with similar or higher affinity than full-length SRC-1 containing multiple functional LxxLL motifs. The NCoA6(NRC) LxxLL-1 motif appears to display broad specificity, yet high affinity towards a large spectrum of NRs [Mahajan and Samuels, 2000]. The Ser at the -3 position (Ser884) of LxxLL-1 in NCoA6(TRBP) has also been shown to be important for binding with the LBDs of NRs, particularly for ERβ, TR and RXR [Ko et al., 2002]. Although phosphorylation of Ser 884 by MAPK in vitro reduced its interaction with NRs, it is not known if such regulation by phosphorylation occurs in vivo [Ko et al., 2002]. In another study, the LxxLL-1 module was shown to associate more avidly to mouse ERβ1 than to ERβ2, consistent with the idea that estradiol-induced mouse ERβ1 is a more potent transcriptional activator than mouse ERβ2 [Zhao et al., 2005]. Studies with a ∼300 amino acid region from rat NCoA6(NRC) containing LxxLL-1 and AD2 indicated that LxxLL-1 enhances both ligand-dependent activity and the intrinsic basal activity of AD2 [Mahajan and Samuels, 2000]. Thus, the LxxLL-1 module of NCoA6(NRC) embedded in AD2 influences not only the association of NCoA6(NRC) with NRs, but also the activation potential of AD2. As opposed to AD1, the AD2 region has now been shown to be necessary for transcriptional activation by NRs [Mahajan et al., 2007].
NCoA6(NRC) contains a second LxxLL motif towards the C-terminus (EAPTSLSQLLDNSGA) designated as LxxLL-2, which does not contain a conserved hydrophobic amino acid residue at the -1 position [Mahajan and Samuels, 2000]. LxxLL-2 does not interact with TR, RAR, RXR and GR, although it interacts with ERα with ∼10-fold lower affinity compared with LxxLL-1 [Mahajan and Samuels, 2000]. It has also been reported that NCoA6(ASC-2) can associate with LXR through LxxLL-2 [Lee et al., 2001; Toresson et al., 2004]. Interestingly, LxxLL-2 does not resemble any of the known LxxLL motifs identified thus far in various coregulators or from combinatorial phage peptide libraries [Chang et al., 1999].
Homodimerization region in NCoA6
NRs activate transcription from target genes preferably as homodimers (ER, GR, PR) or heterodimers (TR, RAR, PPAR, VDR, LXR, FXR) with RXR [Aranda and Pascual, 2001]. p160 family coactivators contain multiple LxxLL motifs; thus a single molecule of p160 coactivator, via its LxxLL motifs, interacts directly with various NR dimers [Nolte et al., 1998]. Similarly, TRAP220, as a monomer with two LxxLL motifs, one of which primarily interacts with RXR, while the other interacts preferentially with VDR or TR [Ito and Roeder, 2001], has the potential to bind NR/RXR heterodimers [Ito and Roeder, 2001]. In contrast, NCoA6 contains a single essential LxxLL-1 motif that is involved in interaction with most of the NRs. An interesting question relates to how NCoA6 with a single LxxLL-1 motif binds to and activates NR dimers? Recent studies suggest the formation of NCoA6(NRC) homodimers, which have been shown to occur using both yeast two-hybrid and in vitro binding assays [Mahajan and Samuels, 2000]. The homodimerization region (DD) (Figure 1) of NCoA6(NRC) has been mapped to 146 amino acids (amino acid 849-995 of human NCoA6(NRC) and the corresponding homologous sequence in rat NCoA6(NRC) [Mahajan et al., 2007; Mahajan and Samuels, 2000]. Embedded in this region is the LxxLL-1 motif, which is involved in NR interaction, but not homodimerization of NCoA6(NRC), as suggested by mutational analysis and dimerization data. Thus, a homodimer of NCoA6(NRC) with two LxxLL-1 motifs has the potential of binding NR homo- and hetero-dimers with high affinity [Mahajan et al., 2007; Mahajan and Samuels, 2000].
Conformational change in NCoA6
It is well known that ligand binding induces a conformational change in NRs which renders them transcriptionally active. For example, the TR-LBD undergoes a conformational change upon ligand binding which is necessary for binding of the LBD with LxxLL modules of coactivators [Aranda and Pascual, 2001]. It is an interesting paradigm to study the regulation of activity of transcription factors and other proteins through conformational transitions which may render the molecules active, inactive, stable, unstable or induce complex formation or signal transduction. Thus, the activation properties of a coactivator could be modulated or fine tuned through conformational alteration if the activation domain of the coactivator molecule is buried or unexposed when not engaged with ligand-bound NR. Whether ligand-bound NR leads to a conformational transition of the coactivator from a relatively inactive to active form has been studied in NCoA6(NRC). LexA-NRC (NRC containing AD1 and AD2 activation domains) when expressed in yeast was not found to be constitutively active, however, 100 fold activation was detected when RXR without any heterologous activation domain was co-expressed in the presence of its ligand. Thus, yeast two-hybrid assays were used to provide evidence that association with ligand-bound NR leads to a change in NCoA6(NRC) which exposes its activation domains. Similar evidence for a conformational change in NCoA6(NRC) by liganded NR was also provided by experiments in mammalian cells. When a region of NCoA6(NRC) containing LxxLL-AD2 was expressed in mammalian cells, it was found to be only moderately active when expressed alone. However, its activity is markedly enhanced when co-expressed with the liganded TR-LBD [Mahajan and Samuels, 2000]. It is already known that the LBD of liganded TR binds a single LxxLL motif of a coactivator through the hydrophobic groove of the liganded-LBD [Nolte et al., 1998], with the enhanced activation suggesting a conformational change in NCoA6(NRC) leading to enhanced AD2 activity. Similarly, PGC-1 has been shown to undergo a conformational change upon NR binding [Puigserver et al., 1999]. Thus, like NRs which undergo a conformational change when they bind their cognate ligands, the activity of certain coactivators appears to be masked until they associate with ligand-bound NR.
NCoA6 mRNAs, isoforms, and expression pattern
Analysis of the gene organization of human NCoA6(RAP250) on chromosome 20q11 predicted a single gene that spans ∼111 kb and consists of 15 exons and 14 introns [Antonson et al., 2004]. Thus, the various sized NCoA6 mRNAs identified in different tissues likely represent alternative splicing products of the NCoA6 gene. The sequence of NCoA6(RAP250) gene promoter indicates that the promoter is TATA-less and contains 4 predicted binding sites for Sp1, and sites for C/EBP and Myc/Max [Antonson et al., 2004], suggesting that the gene might be modulated by growth regulatory signals. The mouse gene is localized on chromosome 2 (H2) with 13 exons [Zhu et al., 2000], while rat NCoA6 resides on chromosome 3 (q42).
NCoA6 is a 250 kDa protein expressed from an mRNA of ∼8-9 kb in various human tissues which include spleen, thymus, testis, ovary, peripheral blood leukocytes and brain [Mahajan and Samuels, 2000]. NCoA6 mRNAs of 6.8 kb, 4.5 kb and 3.6 kb of varying abundance were also detected in these tissues [Mahajan and Samuels, 2000]. Interestingly, the 3.6 kb transcript was the predominant NCoA6 mRNA species detected in heart and skeletal muscle, while the 4.5 kb transcript was the major form found in testes [Mahajan and Samuels, 2000]. Mouse NCoA6(PRIP) (∼8-9 kb mRNAs) encodes a protein of 2068 amino acids [Zhu et al., 2000]. The 4.5 kb mRNA species found in human testis was identified as an alternatively spliced form of human NCoA6(RAP250) encoding 1070 amino acids (1-971 and 1965-2063) [Caira et al., 2000]. This isoform lacks LxxLL-2 and a large component (amino acids 972-1964) of the C-terminus. A partial cDNA (rNRC.1) encoding the LxxLL-1 and AD2 followed by a stop codon and a poly A tail was isolated from a rat pituitary GH4C1 cell cDNA library [Mahajan and Samuels, 2000]. The presence of rNRC.1 suggests an mRNA lacking LxxLL-2 and the entire C-terminus containing the STL region, and likely represents one of the shorter NCoA6(NRC) isoforms expressed in the pituitary. Since the C-terminal STL region appears to harbor an inhibitory domain, shorter isoforms lacking this region may have elevated intrinsic activity in vivo. The rat genome database has recently annotated a sequence for rat NCoA6 mRNA that encodes a predicted protein of 1844 amino acids. However, the assembled rat NCoA6 protein lacking parts of AD2 and the C-terminal STL region is presently hypothetical.
The identity among human, mouse and rat NCoA6 is over 90% at the amino acid level. The distribution of NCoA6 protein in mice has been studied by immunochemistry using antibody against NCoA6(RAP250) and AIB3/NCoA6. These studies indicate that relatively higher levels of NCoA6 are present in cells of endocrine target tissues, including testicular sertoli cells, follicular granulosa cells and epithelial cells of the prostate, uterus, mammary gland, as well as kidney tubules [Zhang et al., 2003]. NCoA6 protein expression was also detected in thyroid and parathyroid cells and the pancreatic islets of langerhans. Although medium to low expression was reported in a variety of tissues, the relatively higher levels of NCoA6 observed in reproductive and many types of endocrine tissues suggests that NCoA6 supports NR functions in reproduction and regulation of the endocrine system [Zhang et al., 2003]. It should be noted that in these studies expression has been studied using antibodies that do not distinguish between NCoA6 and AIB3 isoforms.
Dominant negative forms of NCoA6 and NCoA6-siRNA
One of the ways to study the nature of a coactivator molecule is by expressing its dominant negative form and studying its effects. Alternatively, the biological role of a coactivator can also be assessed by using a siRNA to extinguish expression of the coactivator in vivo. A region of NCoA6 containing an NR-interaction domain has been successfully used as a dominant negative inhibitor. Since a broad spectrum of NRs bind LxxLL-1 of NCoA6, the ectopic expression of a region of NCoA6 containing LxxLL-1 blocks ligand-dependent activation by NRs. These dominant negative constructs work very well in cell culture studies [Mahajan et al., 2007]. A peptide with 80 amino acids (DN1) of NCoA6(ASC-2) containing LxxLL-1 when expressed in transgenic mice was found to block activation by NRs [Kim et al., 2002]. Transgenic expression of another region (DN2) (amino acids 1431-1511) containing the LxxLL-2 region of NCoA6(ASC-2) was found to block the activity of LXRs in vivo [Kim et al., 2003]. An unexpected result of the transgenic expression of DN1 or DN2 was that these factors were reported to selectively block the activity of endogenous NCoA6(ASC-2), but not other coactivators. Since DN1 containing LxxLL-1 and DN2 containing LxxLL-2 would be expected to compete with the binding of other coactivators for liganded NRs, the mechanism(s) by which these LxxLL regions specifically block the activity of NCoA6(ASC-2), but not other coactivators, is unclear and requires further study. However, these transgenic studies have provided clues to the probable biological role of NCoA6(NRC) in vivo. The livers from the DN2 transgenic mice showed similar changes as seen in livers from LXRα-/- mice, suggesting that NCoA6(NRC) plays a role in cholesterol and lipid metabolism in the liver, presumably through regulation of LXR [Kim et al., 2003]. More recently, a number of laboratories have used specific siRNAs against mouse and human NCoA6 in cell culture studies to attenuate the expression of NCoA6 in vivo to study transcription factors that are regulated by NCoA6. For example, p53 and Sox9 were found to be regulated by NCoA6(NRC), as suggested by siRNA studies [Mahajan et al., 2007]. siRNA against NCoA6(ASC-2) was used in MCF-7 cells to demonstrate the E2-dependent induction of pS2 gene expression [Ju et al., 2006].
AIB3/NCoA6 is amplified and overexpressed in various cancers
Human NCoA6 is localized on chromosome 20 (20q11). The AIB3 gene was initially mapped to the 20q11 segment of chromosome 20 during a search for amplified and overexpressed genes mapping to chromosome 20q in breast cancer [Guan et al., 1996]. Subsequently, using FISH analysis, the NCoA6(ASC-2) copy number was found to be increased to a moderate level (4-6 copies) in 14 out of 335 (4.2%) cases of breast cancer and to a high level (>6 copies) in 15 out of 335 (4.5%) cases of breast cancer [Lee et al., 1999]. In addition, NCoA6(ASC-2) mRNA was also detected in 11 different breast cancer cell lines, with the highest expression in BT-474 cells [Lee et al., 1999]. The NCoA6(ASC-2) gene was also found to be amplified in lung and colon cancers. The high level of NCoA6 expression in breast cancer cell lines does not correlate with the level of ER expression. It should be noted that the probes used to assess gene amplification or mRNA expression do not differentiate between AIB3 and NCoA6(ASC-2) mRNAs. Therefore, it is unclear whether it is AIB3 or NCoA6 (or both) which is overexpressed in these tumors. Amplification and overexpression of NCoA6/AIB3 in various cancers underscores its functional importance (discussed later) as a prosurvival gene necessary for normal growth and development.
NCoA6 interacts with and forms complexes with a variety of transcription factors
Central to the mechanistic basis of ligand-dependent transcriptional activation by NRs and coactivator function, is the identification of factors which interact with coactivators and form components of a functional coactivator protein complex(es). Recently, a number of laboratories have identified proteins that interact with NCoA6 employing yeast two-hybrid screens or biochemical purifications (Figure 2 and Figure 3). Since NCoA6 is a large protein with multiple domains, it is likely that distinct regions of NCoA6 might associate with factors involved in transcription, such as transcriptional activators, components of the basal transcription apparatus, the DRIP/TRAP complex or chromatin remodeling factors. Thus, like CBP, NCoA6 may act as a transcriptional co-integrator through its interaction with a variety of transcription factors, including the NRs. Consistent with this model, NCoA6 plays a role in activating not only NRs, but other factors (see Figure 1) such as c-Fos, c-Jun, NF-kB, ATF-2, CREB, heat shock factors, SRF, Rb, p53 and Stat2 [Goo et al., 2004; Hong et al., 2004a; Hong et al., 2004b; Ko et al., 2000; Lee et al., 1999; Lee et al., 2000; Mahajan et al., 2007; Mahajan et al., 2002; Mahajan and Samuels, 2000]. The remainder of this section reviews the factors that have been identified to interact with and play a role in the action of NCoA6(NRC).
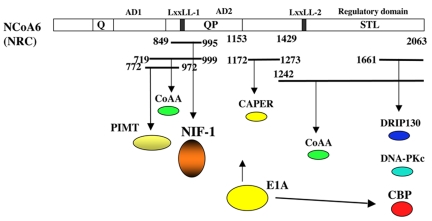
Figure 2. NRC associates with a number of cofactors in vitro and in vivo that have been characterized.
Schematic of NCoA6(NRC) to indicate the regions of NRC(NCoA6) which have been reported to associate with the indicated factors such as CAPER, CBP/p300, CoAA, DNA-PKc, DRIP130, NIF-1 and PIMT in vitro and/or in vivo. E1A was shown to block NRC activity (Mahajan and Samuels, 2000), although its direct interaction with NRC has not been checked. EIA is known to associate with CBP.
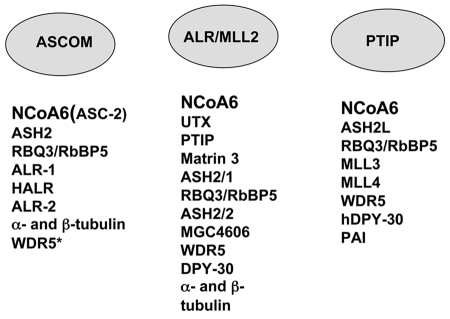
Figure 3. NCoA6(NRC) is a component of three nuclear protein complexes.
Three protein complexes referred to as ASCOM (Goo et al., 2003), ALR (Issaeva et al., 2007) and PTIP (Cho et al., 2007) have been purified and the polypeptides that represent each complex are listed. The 3 complexes share NCoA6 and several other polypeptides. Recently, WDR5 has also been shown to be present in ASCOM (Lee et al., 2006).
CBP/p300
NCoA6 has no apparent histone acetyltransferase (HAT) domain or HAT activity, although this has not been rigorously examined. However, NCoA6 could modulate chromatin structure by recruiting factors with HAT activity to the promoter (e.g., CBP/p300). Consistent with this notion is the finding that NCoA6(NRC) forms a high affinity complex with CBP (Figure 2) in vivo [Mahajan and Samuels, 2000]. Expression of NCoA6(NRC) in 293 cells followed by extraction, affinity adsorption and Western blotting indicated that a significant amount of CBP in the cell is associated with NCoA6(NRC). Association of NCoA6(NRC) with CBP involves the C-terminal region of NCoA6(NRC), which includes AD2. p300 could not be detected in the same Western blot, suggesting that NCoA6(NRC) might preferentially associate with CBP in vivo [Mahajan and Samuels, 2000]. This preferential association may in part reflect the non-redundant physiological functions of CBP and p300. Although NCoA6(NRC) associates with CBP in mammalian cells, no direct association between full length NCoA6(NRC) and full length CBP was identified in yeast [Tian et al., 2006]. This suggests that the association of NCoA6(NRC) with CBP in mammalian cells may be mediated through another factor. In contrast with these findings, another study reported that NCoA6(ASC-2) associated with various regions of CBP in yeast and by in vitro binding, although full length CBP was not examined [Lee et al., 2001]. Other in vitro binding studies indicated that the C-terminal region of p300 (amino acids 1661-2414) can associate with the C-terminal region of NCoA6(TRBP) [Ko et al., 2000]. Whether association of NCoA6 with CBP/p300 is essential for ligand-dependent activation by NRs or other transcription factors activated by NCoA6 has not been directly addressed using CBP-/- cells. However, ligand-dependent activation of NRs by NCoA6(NRC) was completely blocked by expression of E1A, which is known to inactivate CBP/p300 [Mahajan and Samuels, 2000]. Earlier interaction studies indicated a direct interaction between NR and CBP [Chakravarti et al., 1996]. However, it is now believed that CBP is a component of a large coactivator complex containing a primary coactivator that directly interacts with NRs. NCoA6(NRC) or SRC-1 was shown to primarily bind to TRβ2-LBD, while CBP was found to associate with the N-terminal AB domain of TRβ2 [Tian et al., 2006]? . Thus, in a concerted manner, binding of both NCoA6(NRC) and CBP may lead to a maximum transcription potential of certain NRs. These results support the notion that NCoA6(NRC) may act as a coactivator by recruiting a complex containing CBP/p300 and associated factors to ligand-bound NRs on gene promoters.
DNA repair components and methyl transferase-containing protein complexes: PTIP, ASCOM, ALR/MLL2 complex
Some recent studies have hinted that NCoA6 may be involved in DNA repair. At this juncture, most of the clues come from interaction studies. For example, the NCoA6(TRBP) C-terminal region (amino acids 1237-2063) has been shown to associate in vitro with some components of the DNA-dependent protein kinase complex [Ko et al., 2000]. The components included the catalytic subunit (DNA-PKc) (Figure 2), regulatory subunits (Ku70 and Ku80), poly(ADP-ribose)-polymerase (PARP), NuMA (p200) and DNA topoisomerase I. DNA-PK in vitro phosphorylates NCoA6(TRBP) containing amino acids 714-999 in a DNA-dependent manner and amino acids 1237-2063 in a DNA-independent manner [Ko et al., 2000]. In addition, NCoA6(TRBP) activates DNA-PK in vitro which has a phosphorylation pattern that differs from its DNA-mediated activation form. Although the role of DNA-PK in recombination and DNA repair has been very well characterized [Nishino and Morikawa, 2002], the involvement of DNA-PK in transcription is less clear. However, studies of NCoA6(TRBP) activity in cells deficient in DNA-PK indicate a marked reduction in NCoA6(TRBP)-mediated enhancement of GR activity, suggesting a role for DNA-PK in transcriptional activation [Ko and Chin, 2003]. A protein called PTIP (Pax transactivation domain-interacting protein), believed to be involved in DNA damage response upon ionizing radiation, associates with a Set1-like histone methyltransferase component consisting of ∼9 polypetides, including NCoA6 [Cho et al., 2007]. NCoA6 was also shown to localize to NucE (nucleosome of pS2 promoter containing ER binding site) containing TopoIIβ-mediated double strand breaks [Ju et al., 2006].
A steady state ∼2 MDa complex referred to as ASCOM, consisting of ∼8 proteins was isolated from HeLa nuclei using antibody against NCoA6(ASC-2) [Goo et al., 2003]. In addition to NCoA6(ASC-2), the complex includes the Trithorax-related proteins ALL-1/MLL, ALR-1(MLL2), ALR-2, HALR (MLL-3), ASH2, the 66 kDa retinoblastoma binding protein, RBQ3 and 48 kDa α- and β-tubulin proteins (Figure 4). WDR5 was recently added to the list of polypetides that are present in ASCOM [Lee et al., 2006]. Given the composition and the nature of proteins associated with NCoA6(ASC-2), the ASCOM complex is clearly distinct from other known coactivator complexes identified for NRs and other transcription factors. How ASCOM acts to regulate transcription is not known, although recombinant ALR-1 and HALR exhibit weak histone H3 lysine 4 methylation activities in vitro. Using ChIP assays, ASH2, ALR and NCoA6(ASC-2) were reported to be recruited to the RARβ and the p21WAF1 gene promoters which are activated by liganded RAR, and this recruitment was inhibited when the DN1 dominant negative peptide containing LxxLL-1 was expressed [Goo et al., 2003]. DN1 did not affect the recruitment of TRAP220 or SRC-1 to the p21WAF1 promoter. This is surprising since NCoA6(ASC-2), TRAP220 and all the p160 coactivators are thought to use the same interaction surface of liganded-RAR. Although the mechanism of such specific inhibition is unclear [Goo et al., 2003], a possible explanation is that the DN1 LxxLL-1 region blocks dimerization of NCoA6, thus preventing its binding to NR dimers [Mahajan and Samuels, 2000].
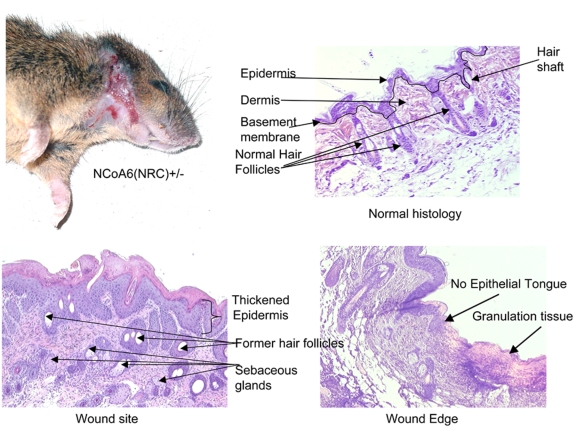
Figure 4. NCoA6(NRC)+/- mice exhibit a wound healing defect.
Shown is a representative mouse with skin lesions in the neck and grooming area. The skin from the wound site, edge of the wound and a normal skin area was processed for histology and stained with hematoxylin-eosin. The skin lesions show an increase in sebaceous glands, a lack of hair follicles and no keratinocyte migration (Epithelial Tongue). Reproduced with permission from Mahajan et al. (Mahajan et al., 2004) by the American Society of Microbiology (Mol. Cell. Biol.).
Recently, NCoA6 was reported to be a component of multiprotein ALR [Issaeva et al., 2007] and PTIP [Cho et al., 2007] complexes (Figure 3). PTIP complex contained ASH2L, RbBP5 (RBQ3), WDR5, hDPY-30, MLL3, MLL4, PA1 and NCoA6. In the ALR complex, 12 distinct polypeptides were identified as UTX, PTIP, Matrin 3, ASH2/1, RBQ3, ASH2/2, MGC4606, WDR5, DPY30, α- and β-tubulin and NCoA6 [Issaeva et al., 2007]. Interestingly, all the complexes containing NCoA6 contain Set proteins as core components, which harbor methyltransferase activity involved in histone, H3K4 methylation of chromatin. However, NCoA6 was not found to be present in partially purified ER-MLL2 complex [Mo et al., 2006]. The composition of these complexes is similar but not identical, invoking the possibility that association of NCoA6 with these complexes may be regulated in a cell type- or target gene-specific manner. It is unclear whether NCoA6 acts only through these complexes to activate NRs and other transcription factors such as c-Fos, c-Jun, CREB and NF-kB.
Although NCoA6 was shown to be a component of the ASCOM, ALR and PTIP complexes, CBP/p300 and a number of other NRC-interacting factors referred to as NIF-1 [Mahajan et al., 2002], CoAA [Iwasaki et al., 2001], CAPER [Jung et al., 2002] and PIMT [Zhu et al., 2001] have been identified as NCoA6-interacting proteins which may play important roles in the action of NCoA6. However, as of now, none of these factors have been shown to be a component of any known coactivator complex.
NIF-1 and NIF-2
NRC-interacting factor-1 (NIF-1) was isolated in a yeast two-hybrid screen as an NRC-interacting factor (Figure 2) [Mahajan et al., 2002]. NIF-1 is a zinc finger protein which is highly conserved across avian, mouse, rat, and human species. Human NIF-1 is a 1342 amino acid nuclear protein which contains a number of highly conserved domains including six zinc fingers, an N-terminal acidic rich region, an LxxLL and a C-terminal leucine zipper-like motif. The LxxLL motif of NIF-1 does not directly interact with Type I or Type II NRs [Mahajan et al., 2002; Mahajan and Samuels, 2005]. Human NIF-2 was identified as an alternatively spliced isoform that is identical to NIF-1, but lacks the first three zinc fingers [Mahajan et al., 2002]. NCoA6(NRC) associates with NIF-1 through a region containing 146 amino acids (amino acids 849-995) of NCoA6(NRC) referred to as the NIF-interaction domain (NIF-ID). NIF-1 interacts with NCoA6(NRC) through a region containing zinc finger 6. Although the NIF-ID of NCoA6(NRC) contains LxxLL-1, this motif is not directly involved in interaction with NIF-1 [Mahajan et al., 2002]. It is interesting that although the NIF-ID and the NR interaction region of NCoA6(NRC) are in close physical proximity (Figure 2), associations of NIF-1 and NRs with NCoA6(NRC) are not mutually exclusive.
Zinc fingers 1, 2 and 3 in NIF-1 form a unique structure referred to as BED fingers [Aravind, 2000], a motif conserved in Drosophila BEAF and DREF [Hart et al., 1999] and several other eukaryotic transcription factors such as Sp1 [Ishii and Laemmli, 2003]. BED finger proteins are thought to function as either activators or repressors by modifying local chromatin structure upon binding to GC-rich sequences [Aravind, 2000]. Thus, NIF-1 may modulate chromatin structure as a part of a coactivator complex and facilitate the activation potential of NRs and other factors. Another possibility is that NIF-1 may also bind insulator DNA sequences directly and facilitate the process of transcription. Interestingly, NIF-2 lacks the BED fingers, but retains the ability to bind NCoA6(NRC) through zinc finger 6. This raises the possibility that NIF-2 may be a naturally-occurring dominant negative form of NIF-1, although this has not been verified experimentally. Both NIF-1 and NIF-2 are ubiquitously expressed in a number of human tissues, although NIF-1 mRNA levels are expressed at much higher levels than NIF-2 [Mahajan et al., 2002]. NIF-1 contains a number of potential phosphorylation sites for various kinases which may play a role in modulating the function of NIF-1. Although NIF-1 does not directly interact with ligand-bound NRs, it markedly enhances ligand-dependent activation [Mahajan et al., 2002]. In addition to NRs, NIF-1 enhances the transcriptional activity of c-Fos, c-Jun and p53 [Mahajan et al., 2007; Mahajan et al., 2002]. Thus, NIF-1 displays an activation profile somewhat similar to NCoA6(NRC) and interacts with NCoA6(NRC) in vivo. Since NIF-1 does not directly bind to NRs, its action is likely through its association with a primary coactivator such as NCoA6(NRC). Factors like NIF-1, which do not directly bind transcriptional activators but modulate the activity of coactivators which directly bind transcriptional activators (NRs and other transcription factors), have been referred to as co-transducers [Mahajan et al., 2002; Mahajan and Samuels, 2005].
RNA binding proteins, CAPER, CoAA and PIMT
CAPER, PIMT and CoAA were identified in yeast two-hybrid screens to interact with NCoA6 (Figure 2). In contrast with NIF-1, CAPER, CoAA and PIMT contain RNA recognition motifs (RRMs). Although the extent of activation of NRs by PIMT, CAPER and CoAA is only moderate (1.8-3 fold), this does not exclude important roles for these factors since many established coactivators only moderately enhance activation in transfection assays. CAPER (Coactivator protein for AP-1 and ER receptor) was isolated from a mouse liver cDNA library using the C-terminal region (amino acids 1172-1729) of NCoA6(ASC-2) as bait [Jung et al., 2002]. CAPER is identical to the nuclear autoantigens HCC1.3 and HCC1.4 reported in hepatocarcinoma [Imai et al., 1993]. HCC1.3 and HCC1.4 are identical except for 6 additional amino acids in HCC1.4. Mouse CAPER (HCC1.3) was reported to enhance activation of ERα and c-Jun. CAPER contains three RRMs and associates with c-Jun and the ligand-bound ER-LBD region via RRM3, while the C-terminus of CAPER was shown to bind NCoA6(ASC-2) [Jung et al., 2002]. CAPER did not appear to enhance activation of other NRs such as RARs, RXRs, TR and GR. The basis for ER specificity was not clarified, but the specificity implies that CAPER may not enhance ER function through association with NCoA6(ASC-2), which interacts with and enhances activation of NRs including RARs, RXRs, TR and GR.
Another RRM-containing factor referred to as “Coactivator activator” (CoAA) is a 669 amino acid protein isolated from a GC cell cDNA library in a yeast two-hybrid screen using the C-terminal region (amino acids 1641-2063) of NCoA6(TRBP) as bait [Iwasaki et al., 2001]. CoAA contains two RRMs near its N-terminus, while its C-terminal region interacts with NCoA6(TRBP). Whether CoAA directly interacts with various NRs has not been examined. Expression of CoAA enhances activation by a number of transcription factors including NF-kB, CREB, AP-1, TR, ER and GR. CoAA was also shown to associate with DNA-PK regulatory subunit Ku86 and PARP from GH3 cells, suggesting that CoAA may exist as part of a DNA-PK/NCoA6 complex in vivo.
PIMT (PRIP-interacting methyltransferase) was also isolated in a yeast two-hybrid screen from a human liver cDNA library with the bait containing the C-terminal region (amino acids 773-2067) of NCoA6(PRIP) [Zhu et al., 2001]. PIMT is a ubiquitously expressed putative RNA methyltransferase which contains an invariant GXXGXXI motif near its N-terminus found in K-homology motifs of many RNA binding proteins. Expression of PIMT enhances the activity of PPARγ and RXR, which is further enhanced by expression of NCoA6(PRIP). The putative methyltransferase activity of PIMT does not appear to be involved in its role as an activator since deletion of the methyltransferase domain does not affect its activating function [Zhu et al., 2001]. Expression of PIMT enhances activation by PPARγ and RXR. Like NCoA6(NRC), PIMT homodimerizes in vitro and may also form homo-oligomers [Zhu et al., 2001]. PIMT does not appear to methylate histones, but binds CBP/p300 and PBP(DRIP205/TRAP220) [Misra et al., 2002]. CAPER, CoAA, PIMT, and NIF-1 were not identified as components of the ASCOM complex [Goo et al., 2003]. DRIP130, a component of the DRIP/TRAP mediator complex, was reported to associate with the C-terminal region of NCoA6(TRBP) in vitro (amino acids 1237-2063) [Ko et al., 2000]. Whether DRIP130 and NCoA6 synergistically activate NRs and/or other transcription factors has not been explored.
Role of NCoA6 in systemic biology and physiology: NCoA6 is an essential coactivator and a pleiotropic modulator affecting growth, development, reproduction, apoptosis and wound healing
Most of the coactivators that have been characterized in detail are complex in structure, containing multiple domains. Although mechanistically, coactivator molecules are primarily involved in orchestrating transcription, their role in biological and physiological functions has become known by knocking out their genes in mice. CBP [Goodman and Smolik, 2000; Tanaka et al., 1997], p300 [Yao et al., 1998], SRC-1 [Xu et al., 1998], SRC-2/GRIP1/TIF-2 [Gehin et al., 2002], SRC-3 [Xu et al., 2000] (p160/SRC family), TRAP220/PBP [Crawford et al., 2002; Ito et al., 2000; Landles et al., 2003], RIP140 [White et al., 2000] and (NCoA6) NRC/AIB3/PRIP/RAP250 [Antonson et al., 2003; Kuang et al., 2002; Mahajan et al., 2004; Zhu et al., 2003] have been deleted in mice by homologous recombination to gain insight into the biological role(s) of these coactivators. Based on these studies, coactivator molecules can be grouped into non-essential and essential coactivators. RIP140 and p160 family members (SRC-1, SRC-2 and SRC-3), when knocked out, are not essential for embryonic growth and development since mice carrying homozygous deletion of either SRC-1, SRC-2, SRC-3 or RIP140 are viable [White et al., 2000]. Although members of the p160 family of coactivators can mediate different functions [Gehin et al., 2002; Mark et al., 2004], the gene deletion studies suggest redundancy in function of the p160 coactivator family. On the other hand, CBP, p300, TRAP220/PBP and NCoA6 null mutant embryos do not survive beyond 12.5 dpc since these coactivators are essential for embryonic growth and development.
Transgenic animal model systems using dominant negative peptides
As an alternative to gene targeting, transgenic mouse models have been generated to address the role of NCoA6(ASC-2) in NR signaling using a dominant negative approach [Kim et al., 2002; Kim et al., 2003]. For example, DN1 peptide (residues 849-929) contains the LxxLL-1 motif [Kim et al., 2002] and functions as a dominant negative inhibitor, while DN2 peptide contains the LxxLL-2 motif. NCoA6(ASC-2) associates with the majority of the NRs through LxxLL-1, while LxxLL-2 specifically associates with LXRs. Several transgenic mice lines, each expressing either DN1 or DN2, were produced [Kim et al., 2003]. DN1 transgenic lines displayed pathological abnormalities in many different organs that included heart, pituitary, adrenal glands, brain, spleen, liver and lung. In particular, eyes with microphthalmia and posterior lanticonus with cataract were scored out [Kim et al., 2002]. On a high fat cholesterol diet, DN2 transgenic lines displayed rapid accumulation of large amounts of cholesterol and downregulation of known lipid metabolizing target genes of LXR [Kim et al., 2003]. Both DN1 and DN2 caused repression of some NR-mediated target gene expression, which was found to be completely reversed by co-transfection of wild-type NCoA6(ASC-2), but not by co-expression of other coactivators such as TRAP220 or SRC-1. These results were interpreted to indicate that DN1 and DN2 transgenic expression blocks specific interaction of endogenous NCoA6(ASC-2) with nuclear receptors. Recent studies on conditional knockout of NCoA6(PRIP) in liver [Sarkar et al., 2007], however, suggest that phenotypes observed in DN1 transgenic mice may not be simply ASC-2-dependent. It is likely that a broad spectrum of Type I and Type II endogenous receptors are inhibited by DN1(LxxLL-1 peptide). Thus, the phenotypes may result not just from inhibition of NCoA6(ASC-2)-NR interaction, but also from inactivation of a number of receptors that bind DN1 in vivo. Thus, the mechanism of DN1-mediated phenotypes in transgenic mice needs a broader interpretation.
Deletion of both NCoA6 genes is embryonic lethal and leads to apoptosis
The NCoA6 gene has been knocked out in mice in a mixed C57BL/6-129S6 (C57/129) genetic background [Antonson et al., 2003; Kuang et al., 2002; Mahajan et al., 2004; Zhu et al., 2003]. These studies confirmed that NCoA6 null mutation is embryonic lethal and mutant embryos die in utero between 8.5-12.5 dpc, while C57/129 NCoA6+/- mutant mice appear normal and grow similar to wild-type mice. The cause of lethality of NCoA6-/- embryos is attributed to placental dysfunction and a number of developmental defects involving the heart, liver and brain [Antonson et al., 2003; Kuang et al., 2002; Zhu et al., 2003]. The role of NCoA6(NRC) in growth is particularly striking since NCoA6(NRC)-/- embryos do not survive beyond 12.5 dpc, are growth retarded and half the size of NCoA6(NRC)+/- or NCoA6(NRC)+/+ embryos [Mahajan et al., 2004]. The decreased growth could result from a disruption in cell cycle progression, increased apoptosis, or both. Interestingly, NCoA6(NRC)-/- mouse embryo fibroblasts (MEFs) derived from 12.5 dpc NCoA6(NRC)-/- embryos also exhibit growth retardation in culture compared to NCoA6(NRC)+/+ MEFs. In addition, NCoA6(NRC)-/- MEFs undergo apoptosis as they enter into the late log phase of growth [Mahajan et al., 2004]. NCoA6(NRC)+/+ MEFs do not undergo apoptosis under the same growth conditions, while NCoA6(NRC)+/- cells exhibit reduced apoptosis. Knockdown of NCoA6(NRC) in wild-type NCoA6(NRC)+/+ MEFs using RNAi leads to a similar extent of apoptosis as found with NCoA6(NRC)-/- MEFs [Mahajan et al., 2004]. Apoptosis of the NCoA6(NRC)-/- cells seems caspase-mediated since zVAD-fmk, a pan-caspase inhibitor, blocks the apoptogenic response in NCoA6(NRC)-/- MEFs [Mahajan et al., 2004]. However, which caspase(s) is involved and what triggers apoptosis in these cells remain to be identified. These findings suggest that NCoA6(NRC) is involved in regulating antiapoptotic and/or prosurvival genes necessary for cell growth and development. Interestingly, growth retardation has also been observed in MEFs derived from Trap220-/- embryos [Ito et al., 2000].
A number of laboratories have used NCoA6-/- MEFs to show that activation by RXR, RAR, TR and PPAR is compromised in NCoA6-/- MEF cells as opposed to wild-type cells [Antonson et al., 2003; Kuang et al., 2002; Qi et al., 2003; Zhu et al., 2003]. Although different laboratories have reported differences in the extent of activation by various NRs in NCoA6-/- MEFs, the overall conclusion is that NCoA6 functions as a coactivator for RXR, RAR, TR and PPAR. Also, NCoA6(PRIP)-/- MEF cells in culture were reported to be refractory to PPARγ-mediated adipogenesis and fail to express adipogenic markers like aP2. Thus, much like PBP/TRAP220, this study concluded that NCoA6(PRIP) is required for the adipogenic program [Qi et al., 2003].
Growth and reproductive phenotypes of NCoA6+/- 129S6 newborn and adult mice
Although NCoA6(NRC)+/- mice in the C57/129 mixed genetic background appear outwardly normal and grow and reproduce similar to NCoA6(NRC)+/+ mice, the penetrance of a specific phenotype is frequently dependent on the genetic background of the mouse strain. Since the ES cells used for homologous recombination are derived from the 129S6 strain, the effect of NCoA6(NRC) heterozygosity was examined in 129S6 mice generated in our laboratory [Mahajan et al., 2004]. The NCoA6(NRC)+/- mice in 129S6 and C57/129 mixed genetic background exhibit a number of interesting phenotypes, primarily involving growth, reproduction and wound healing [Mahajan et al., 2004].
Growth retardation phenotype
NCoA6(NRC)+/- 129S6 mice exhibit a neonatal growth phenotype; the newborn pups are ∼10-15% smaller than their wild-type litter mates [Mahajan et al., 2004]. However, after weaning, the NCoA6(NRC)+/- mice exhibit “catch-up” growth, and within 2 months these mice become similar in size to their NCoA6(NRC)+/+ littermates. Although similar growth retardation has also been noted among NCoA6(NRC)+/- neonates in the C57/129 background, it is much less profound, and is found in less than 5% of neonates. Interestingly, ∼3% of the NCoA6(NRC)+/- 129S6 newborn pups are extremely growth stunted, weighing 70% less than their wild-type littermates. These mice often die before weaning. However, a small number of these mice survive and exhibit a similar “catch-up” growth, as described earlier [Mahajan et al., 2004]. Interestingly, the growth phenotype found in NCoA6(NRC)+/- 129S6 mice resembles that found with TIF-2-/- (SRC-2) mice, which also exhibit transient postnatal growth retardation [Gehin et al., 2002]. However, this phenotype seems different from pCIP-/- (SRC-3) newborn mice that are also uniformly smaller in size, but their weight deficit persists through adulthood. This is thought to reflect a role for pCIP-/- (SRC-3) in mediating effects through the IGF-1 pathway [Liao et al., 2002; Xu et al., 2000]. The molecular mechanisms underlying growth retardation in NCoA6(NRC)+/- 129S6 mice have not yet been defined, but likely reflect an effect of NCoA6(NRC) on one or more transcription factors which play an important role in growth (e.g., AP-1).
Male and female hypofertility, and mammary glands
Although NCoA6(NRC)+/- C57/129 mice exhibit no reproductive phenotype, the fertility of both sexes of NCoA6(NRC)+/- mice in 129S6 background is compromised [Mahajan et al., 2004]. Both male and female mice are hypofertile (average litter size of 3 pups) and ∼20% of NCoA6(NRC)+/- females are sterile. These infertile females appear to mate based on the formation of vaginal plugs and exhibit normal estrus cycles, suggesting problems in oogenesis, implantation of fertilized ova, or a defect in placental function, since no embryos were detected between 8.5-12.5 dpc. The number of newborn NCoA6(NRC)+/- pups obtained from crosses between NCoA6(NRC)+/- 129S6 hypofertile males and females is far less than expected based on Mendelian distribution [Mahajan et al., 2004]. This appears to result from a significant number of NCoA6(NRC)+/- embryos dying in utero. Those NCoA6(NRC)+/- 129S6 female mice which are hypofertile exhibit a progressive decline in fertility as they age and their newborn pups have a high rate of neonatal mortality. In another study, NCoA6(PRIP)-deficient mammary glands [Qi et al., 2004] during pregnancy exhibited decreased alveolar density. It was reported that the null mammary glands could not produce enough milk to nurse all the pups during lactation. The results suggested that NCoA6(PRIP) is required for efficient ductal branching of mammary glands in response to estrogen. The reproductive phenotypes in NCoA6(NRC)+/- 129S6 mice are similar to those found for TIF-2-/- (SRC-2) mice [Gehin et al., 2002]. TIF-2-/- males exhibit hypofertility resulting from age-dependent testicular degeneration and defective sperm, while female hypofertility is due to placental hypoplasia and the need for maternal TIF-2 in the decidua stromal cells of the placenta [Gehin et al., 2002]. Although the reproductive phenotype of NCoA6(NRC)+/- 129S6 mice resemble TIF-2-/- mice, the precise mechanism(s) underlying male and female hypofertility in NCoA6(NRC)+/- 129S6 mice needs further clarification. Whether TIF-2 and NCoA6 are needed together requires further study. It is of interest that LXRα/β-/- male mice become completely sterile within 6 months [Volle et al., 2004]. Since NCoA6 acts as a coactivator for LXR [Kim et al., 2003], the progressive loss of fertility in NCoA6(NRC)+/- male mice may reflect this effect of NCoA6(NRC) on the LXRs in the testes.
Recently, mice have been generated with Set domain mutations in MLL3 to elucidate the physiological role of ASCOM (coactivator complex that associates with NCoA6(ASC-2) [Lee et al., 2006]. MLL3Δ/Δ mutant mice phenotypes are the result of the deletion of the Set catalytic domain involved in methylation. MLL3Δ/Δ mutation leads to partial embryonic lethality. MLL3Δ/Δ mice [Lee et al., 2006] are viable and share some similar phenotypes with NCoA6(NRC)+/- isogenic mice [Mahajan et al., 2004]. MLL3Δ/Δ mice were stunted in their overall growth, weighing 30-40% less at birth and females exhibited a range of reproductive phenotypes from infertility to hypofertility, while males were generally hypofertile. Interestingly, MEFs generated from MLL3Δ/Δ strain doubled at approximately half the rate of wild-type MEFs [Lee et al., 2006], much like NCoA6(NRC)-/- MEFs [Mahajan et al., 2004].
NCoA6+/- mice exhibit a wound healing phenotype resulting from a defect in keratinocyte migration
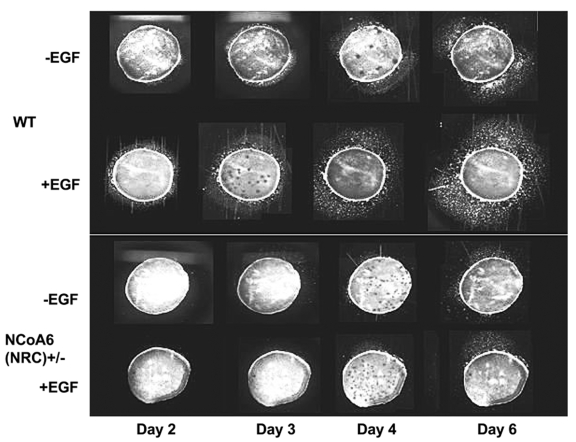
In general, NCoA6+/- C57/129 mice appear normal, therefore these mice have not been followed through adulthood in some laboratories. In our laboratory, we have followed the postnatal growth of NCoA6(NRC)+/- mice. As they grow older, these mice exhibit a wound healing phenotype [Mahajan et al., 2004]. The mice spontaneously develop skin lesions or ulcers around the neck, ears, snout and facial area (Figure 4). These regions correspond to the areas where mice groom and likely scratch themselves. This occurs in approximately 25% of both male and female NCoA6(NRC)+/- C57/129, as well as NCoA6(NRC)+/- 129S6 mice. These spontaneous lesions develop as early as 4-6 months of age in the NCoA6(NRC)+/- C57/129 mice and somewhat earlier in the 129S6 background [Mahajan et al., 2004]. Histopathology of the affected and normal regions of the skin show thickening of the epidermis, increased sebaceous glands and a reduction or total loss of hair follicles. In addition, there is no leading edge of keratinocyte migration (Epithelial Tongue), which is seen in normal wound healing (Figure 4). This lack of keratinocyte migration was reproduced using ex vivo skin explant cultures from two day old C57/129 NCoA6(NRC)+/- mice [Mahajan et al., 2004]. While NCoA6(NRC)+/+ keratinocytes showed robust migration when incubated with epidermal growth factor (EGF), keratinocytes from the NCoA6(NRC)+/- explants showed little or no response to EGF (Figure 5).
Figure 5. Lack of keratinocyte migration in explants from NCoA6(NRC)+/- mice.
Keratinocyte migration was determined from skin explants of 2 day old wild-type (left) and NCoA6(NRC)+/- mice (right). Explants from NCoA6(NRC)+/- mice exhibit a delay in the initiation of and a diminished capacity for migration, as well as a lack of response to EGF. Reproduced with permission from Mahajan et al. (Mahajan et al., 2004) by the American Society of Microbiology (Mol. Cell. Biol.).
Interestingly, the wound healing phenotype identified in the NCoA6(NRC)+/- mice is similar to that observed in mice where c-Myc is targeted to the basal layer of mouse epidermis and hair follicles using a keratin K14 promoter expression vector [Waikel et al., 2001]. Although mice heterozygous for expression of K14/c-Myc are viable and have no phenotype at birth, like the NCoA6(NRC)+/- mice as they age, they develop chronic skin lesions in the facial, ear and neck grooming areas. The mechanism for development of the c-Myc-mediated chronic wounds was suggested to be secondary to repression of integrins (e.g., α6 and β1) [Waikel et al., 2001]. These integrins are thought to play an important role in keratinocyte migration and in self-renewal of epidermal stem cells, leading to their gradual depletion over time.
A similar phenotype found in mice heterozygous for expression of K14/c-Myc and in the NCoA6(NRC)+/- mice suggests that NCoA6(NRC) may play a role in regulating the genes targeted by overexpressed c-Myc. For example, NCoA6(NRC) is a potent coregulator of c-Jun activity [Ko et al., 2000; Lee et al., 2000; Mahajan et al., 2002], which plays an important role in regulating genes involved in wound healing (integrins, laminins, and extracellular matrix genes in the epithelium) [Angel et al., 2001]. In addition, c-Jun has recently been shown to stimulate expression of heparin-binding-epidermal growth factor (HB-EGF) in the basal layer of wounded epithelium which, in turn, stimulates signaling by EGFR to regulate keratinocyte migration through regulation of focal adhesion kinase [Li et al., 2003]. Since NCoA6 is known to play an important role in c-Jun signaling [Ko et al., 2000; Lee et al., 2000; Mahajan et al., 2002], the impaired wound healing phenotype of NCoA6(NRC)+/- mice can be explained, at least in part, by a reduction in c-Jun activity. In addition to c-Jun, other transcription factors and signaling molecules (e.g., NF-kB and the PPARs) involved in wound healing are known to be regulated by NCoA6 [Ko et al., 2000; Lee et al., 1999; Mahajan et al., 2002; Mahajan and Samuels, 2000; Zhu et al., 2000; Zhu et al., 2003]. Both PPARα and PPARβ have been shown to be important in wound healing [Michalik and Wahli, 2007; Tan et al., 2001]. The precise mechanism(s) by which a reduction of NCoA6(NRC) in the skin of NCoA6(NRC)+/- mice leads to chronic skin wounds needs to be further defined. However, its role as a coactivator for c-Jun and the PPARs, and its role as a prosurvival antiapoptotic factor, suggests that NCoA6 acts through regulation of one or more of the above factors and functional pathways involved in the wound healing process.
NCoA6+/- mice exhibit decreased insulin secretion
In the pancreas, NCoA6 is expressed in the endocrine cells of islets of langerhans and thus the role of NCoA6(ASC-2) in insulin secretion from pancreatic β-cells has been assessed in heterozygous mice and NCoA6(ASC-2) overexpressed in cells [Yeom et al., 2006]. Overexpressed NCoA6(ASC-2) increased glucose-elicited insulin secretion, whereas insulin secretion was decreased in islets from NCoA6(ASC-2)+/- mice. Primary rat islets ectopically expressing DN1 or DN2 (dominant negative fragments from NCoA6(ASC-2) that contain NR boxes 1 and 2, respectively) exhibited decreased insulin secretion. Islet mass and number were also found to be decreased in heterozygous mice, which is believed to be due to increased apoptosis and decreased proliferation of NCoA6+/- islets [Yeom et al., 2006].
NCoA6+/- mice exhibit accelerated polyoma middle-T antigen-induced mammary tumorigenesis
In one of the studies, AIB3(NCoA6)+/- were crossed with mice carrying polyoma middle-T antigen (pyMT) and these mice were directly compared with transgenic pyMT mice for mammary tumorigenesis [Zhang et al., 2004]. Mammary tumor development in AIB3+/- pyMT female and male mice was substantially accelerated as compared with that in WT/pyMT mice. Tumor formation in nude mice that received premalignant NCoA6/AIB3+/- pyMT mammary tissue was much faster than in nude mice that received transplants from premalignant WT/pyMT mammary tissue. This tumor acceleration was reported as being due to increased cell proliferation and ductal hyperplasia and mammary intraepithelial neoplasia. The mechanism of pyMT-induced tumorigenesis was believed to be through a partial impairment of activated PPARγ/RXR [Zhang et al., 2004].
Phenotypes from conditional knockouts
NCoA6 null mutation is embryonic lethal, which poses a challenge in deciphering the in vivo functions of NCoA6 in adult mice. To overcome this hurdle, some laboratories have recently produced conditional knockout mice for NCoA6 to study the role of NCoA6 in select tissues of adult mice. In NCoA6(PRIP)-deficient mammary glands [Qi et al., 2004], the elongation of ducts during puberty was not affected, but the number of ductal branches was decreased and during pregnancy the null mammary glands exhibited decreased alveolar density. The lactating NCoA6(PRIP)-deficient glands contained scant lobuloalveoli with many adipocytes, while the wild-type glands lacked adipocytes. It was reported that the null mammary glands could not produce enough milk to nurse all the pups during lactation. The results suggested that NCoA6(PRIP) contributes to efficient ductal branching of mammary glands in response to estrogen. A slight increase in apoptosis was found in the terminal buds from NCoA6(PRIP)-deficient glands, raising the possibility that abnormal apoptosis contributes to the impaired ductal branching [Qi et al., 2004].
NCoA6(PRIP) liver conditional knockout mice have been generated to study the in vivo role of NCoA6 as a coactivator for PPARα and CAR [Sarkar et al., 2007]. NCoA6(ASC-2) had been earlier shown to interact with CAR and enhance the transcriptional activation by CAR in transfection assays [Choi et al., 2005]. Similarly, NCoA6 was shown to activate PPARs in in vitro studies [Caira et al., 2000; Zhu et al., 2000]. The in vivo data obtained from conditional knockout studies show that NCoA6(PRIP) deficiency in liver fails to alter acetaminophen-induced hepatotoxicity in mice pretreated with CAR ligands phenobarbital (PB) or TCPOBOP. No differences in the upregulation of cytochrome P450 gene expression (CYP1A2, CYP2B10, CYP3A11 and CYP2E1) were seen following PB or TCPOBOP treatment between NCoA6(PRIP)+/+ or liver NCoA6(PRIP)-/- conditional knockout mice. CAR mRNA and protein levels between these mice were found to be similar, suggesting that NCoA6(PRIP) does not alter hepatic CAR expression [Sarkar et al., 2007]. These in vivo findings were somewhat contradictory to earlier studies in transgenic mice expressing DN1, a fragment of NCoA6(ASC-2) containing the LxxLL-1 motif [Kim et al., 2002]; these mice failed to show acetaminophen-induced hepatic necrosis. Similarly, RAP250/NCoA6(PRIP) has been shown to activate PPARs with in vitro studies [Caira et al., 2000; Zhu et al., 2000]. However, in conditional knockout mice, the degree of PPARα ligand-mediated peroxisome proliferation in liver cells was essentially similar to that seen in wild-type liver cells, implying that in liver, NCoA6 is not involved for PPARα-mediated proliferation. Also, the PPARα-specific target genes fatty acyl-CoA oxidase, enoyl-CoA hydratase/L-3 hydoxyacyl-CoA hydrogenase (L-bifunctional enzyme; L-PBE), peroxisomal thiolase (PTL) and CYP4A1 increased markedly in both PRIP+/+ mice and in mice lacking hepatic PRIP, following treatment with the PPARα ligand, Wy-14643. Also, PPARα mRNA levels remained essentially similar in control and Wy-14,643-treated mice [Sarkar et al., 2007]. Based on these results, this study concluded that NCoA6 is redundant in liver in activating PPARα- and CAR-mediated gene expression, while PBP/TRAP220 was found to be essential for PPARα and CAR activation [Sarkar et al., 2007].
Conclusions and future perspectives
Although NCoA6 was initially cloned as coactivator protein for NRs, its detailed characterization by a number of laboratories has uncovered its potential to enhance the activity of a wide variety of other transcription factors including c-Fos, c-Jun, CREB, NF-kB, ATF-2, heat shock factors, E2F-1, SRF, and Rb, p53 and Sox9 [Goo et al., 2004; Hong et al., 2004a; Hong et al., 2004b; Ko et al., 2000; Kong et al., 2003; Lee et al., 1999; Lee et al., 2000; Mahajan et al., 2007; Mahajan et al., 2002; Mahajan and Samuels, 2000; Mahajan and Samuels, 2005]. Thus, like CBP/p300, NCoA6 may function as a transcriptional co-integrator of NRs and other important transcription factors involved in growth, proliferation, cytokine signaling, metabolism, the immune response and apoptosis. Given the wide spectrum of factors that are regulated by NCoA6, an interplay of signaling pathways is expected to be affected as a result of NCoA6 expression. It would be of significant interest to determine if these factors associate directly with NCoA6 or whether they interact with other components of NCoA6 protein complexes. In this regard, many of the factors described above have been shown to interact with CBP/p300, suggesting that NCoA6 may function to enhance the activity of such factors through its interaction with CBP/p300, which occurs in vivo. Although NCoA6 seems to be devoid of HAT activity, its ability to associate with a large number of transcription factors and signaling molecules strongly suggests that NCoA6 plays a fundamental role in various coactivator complexes. It is already known that NCoA6 associates with methyltransferases, and therefore, is involved in modifying nucleosome architecture through histone methylation. ALR/MLL2-knockdown affects cell adhesion-related processes and suppresses cell growth and migration capacity of cells [Issaeva et al., 2007]. Genes regulated by ALR include ECM-degrading enzyme, heparanase, uPA/PL, BH-protocadherin, L1CAM, ALCAM and Keratin17, consistent with decreased expression of these molecules. ALR knockdown cells display reduced growth kinetics, higher levels of intrinsic apoptosis and impaired anchorage-independent growth in vitro [Issaeva et al., 2007]. Interestingly, these ALR knockdown phenotypes show similarities with the heterozygous NCoA6(NRC) mice, which show defects in wound healing; and growth retardation, slower cell proliferation and enhanced apoptosis in NRC null embryos and null MEF cells.
In addition to conformational alterations in NCoA6 mediated by liganded NRs, and possibly other transcription factors, posttranslational modifications that may regulate NCoA6 activity (e.g., phosphorylation of the C-terminal STL inhibitory region) could play an important role in modulating the intrinsic activity of NCoA6. Such phosphorylation of NCoA6 by PKA and other kinases may serve to communicate and integrate regulatory events mediated by NRs or other transcription factors and the signaling cascades generated by cell surface receptors. NCoA6 has been shown to interact with a number of factors in vitro and in vivo including CBP/p300, DNA-PKc, DRIP130, NIF-1, CAPER, CoAA and PIMT. In addition, NCoA6 is a component of ASCOM, ALR and PTIP complexes. This raises the possibility that NCoA6 may be a component of distinct transcriptional regulatory complexes, as has been found for the various “Mediator” (DRIP/TRAP) complexes, which have somewhat different protein compositions. In addition, a systematic analysis of protein complexes of the yeast proteome indicates that many proteins are shared by distinct protein complexes which otherwise have very different compositions. Given the complexity of the intermolecular network of NCoA6 and its multiple functions, it is also likely that NCoA6 is a component of different regulatory complexes in the cell.
Unlike the p160/SRC family of coactivators, NCoA6 is a single copy gene and has no apparent closely related genes in the human genome. Since NCoA6 appears to modulate the activity of many transcription factors, it is not surprising that deletion of both NCoA6 alleles is embryonic lethal. Thus, like CBP, p300 or TRAP220, NCoA6 appears to be an essential gene involved in the regulation of growth, development and cell survival. Although full length NCoA6 has been studied in detail, various sized mRNAs are expressed in different tissues, which likely reflect alternative splicing of the NCoA6 gene. Most of these isoforms, some of which are tissue-specific, have not been well characterized. Depending on the composition of the protein, such isoforms may mediate specific effects or selectively interact with specific protein complexes in the cell. Thus, some of the described isoforms lack the C-terminal STL region which interacts with CBP/p300, DRIP130 and DNA-PKc, while retaining the interaction domain for NIF-1, PIMT and CoAA. In addition, such an isoform would lack LxxLL-2, which exhibits preference for the LXRs. Furthermore, an NCoA6 isoform lacking the inhibitory C-terminal STL region, but which retains LxxLL-1 and AD2, might be expected to be a more active isoform of NCoA6. AIB3 is a distinct isoform of NCoA6 with a unique N-terminus. Studies on AIB3 may reveal some interesting functional difference with NCoA6. Thus, although NCoA6 is a single copy gene, tissue-specific alternative splicing of the gene may generate isoforms which may exhibit differences in intrinsic activity, and/or lead to specific or restricted target tissue response(s) compared with full length NCoA6.
In summary, NCoA6 (NRC/ASC-2/RAP250/TRBP/PRIP) is an essential coregulator of NRs and other transcription factors and exhibits pleiotropic effects on a wide variety of processes [Mahajan and Samuels, 2005]. Recent studies of NCoA6+/- mice indicate that haploid expression of NCoA6 leads to a variety of phenotypes, indicating that NCoA6+/- mice may prove useful in deciphering the role(s) of NCoA6 in various processes in vivo. These studies, and studies on the various NCoA6 isoforms and NCoA6 complexes, should lead to a more comprehensive understanding of the contribution(s) of NCoA6 in mediating effects of the NRs and other factors involved in controlling growth, development, apoptosis, wound healing, reproduction and metabolic regulation. Due to embryonic lethality, the role of NCoA6 in various cell types and in the adult mice has not been fully explored. It is likely that NCoA6 is regulated in a cell and tissue type-specific manner, thus potentially making NCoA6 a very important molecule in the regulation of NR activities. NCoA6 may emerge as a specific coactivator for a specific set of NRs due to the fact that NCoA6 displays NR-specific interaction through LxxLL-2. Already, some data has accumulated regarding the role of NCoA6 in vivo; however, future studies should be focused on conditional knockouts of NCoA6 in various tissues to gain insights into tissue-specific functions of NCoA6. NCoA6 may be involved in regulating fat metabolism and energy homeostasis because of its interactions with PPARs, LXRs and TRs. An interesting question relates to why does NCoA6 associate itself with at least 3 different known protein complexes, such as ASCOM, ALR and PTIP? Thus, researchers interested in mechanism of transcription should address the role of NCoA6 in proteomics. Soon after p160 coactivators were discovered, various groups followed the kinetics of their appearance and disappearance. Some models proposed ordered recruitment, while some argued for cyclical recruitment. Studies on NCoA6 clearly reflect that it is not a core molecule of one particular coactivator complex. Instead, NCoA6 dynamically associates itself with at least 3 different coactivator complexes (ASCOM, ALR and PTIP). Thus, these studies raise the possibility that NCoA6, through its associations, and much like CBP, may be a co-integrator regulating transcription and cell signaling. The role of NCoA6 in cholesterol metabolism and efflux should be of general interest to biologists. Thus, NCoA6 opens possibilities for pharmacological interventions to clinical researchers interested in wound healing, cardiovascular diseases, obesity and insulin resistance.
Acknowledgements:
This work was supported in part by a grant from the National Institutes of Health (NIH; DK16636) (H.H.S.) and a grant from the Entertainment Industry Foundation (EIF) (H.H.S.). Due to space limitation, we have cited review articles where appropriate. We apologize to the many investigators whose original references have not been cited.
Abbreviations
- ACTR
activator of transcription of nuclear receptors
- AD
activation domain
- AF
activation function
- AIB3
amplified in breast cancer 3
- Akt1
acute transforming retrovirus thymoma protein kinase 1
- AP-1
activator protein 1
- AR
androgen receptor
- ASC-2
activating signal cointegrator-2
- ASCOM
ASC-2 complex
- ATF-2
Activating transcription factor-2
- CAPER
Coactivator protein for AP-1 and ER receptor
- CAR
constitutive androstane receptor
- CARM1
coactivator-associated arginine (R) methyltransferase-1
- CBP
cAMP response element binding protein (CREB)-binding protein
- c-Fos
cellular homologue of the vfos oncogene
- ChIP
chromatin immunoprecipitation
- c-Jun
cellular homologue of an oncogene in the avian sarcoma virus 17
- CoAA
coactivator activator
- CREB
cAMP response element-binding protein
- DBD
DNA-binding domain
- DD
dimerization domain
- DNA-PKc
DNA-dependent protein kinase catalytic subunit
- DRIP
vitamin D receptor-interacting protein
- E1A
adenovirus early expressed protein 1
- E2F-1
transcription factor that binds Rb
- EGF
epidermal growth factor
- ER
estrogen receptor
- GR
glucocorticoid receptor
- GRIP
GR-interacting protein
- HAT
histone acetyltransferase
- LBD
ligand-binding domain
- LXR
liver x receptor
- MAPK
mitogen-activated protein kinase
- MEF
mouse embryo fibroblast
- MR
mineralocorticoid receptor
- NCoA-1
nuclear receptor coactivator-1
- NCoA6
nuclear receptor coactivator 6
- NF-kB
nuclear factor kB
- NIF-1
NRC interacting factor-1
- NIF-2
NRC interacting factor-2
- NR
nuclear receptor
- NRC
nuclear receptor coactivator/coregulator
- NRIF3
nuclear receptor interacting factor 3
- p/CAF
p300/CBP-associated factor
- p/CIP
p300/CBP/co-integrator-associated protein
- p160
160 kDa proteins of the SRC family
- PBP
PPAR-binding protein
- PGC-1
PPAR coactivator-1
- PIMT
PRIP-interacting methyltransferase
- PKA
protein kinase A
- Pol II
RNA polymerase II
- PPAR
peroxisome proliferator-activated receptor
- PR
progesterone receptor
- PRIP
PPAR-interacting protein
- PTEN
phosphatase and tensin homologue deleted from chromosome 10
- PXR
pregnane x receptor
- RAC3
receptor associated coactivator
- RAP250
250 kDa receptor associated protein
- RAR
retinoic acid receptor
- Rb
retinoblastoma gene product
- RIP
receptor-interacting protein
- RRM
RNA recognition motif
- RXR
retinoid x receptor
- Sp1
SV40 virus promoter-specific transcription factor
- SRC
steroid receptor coactivator
- SRF
serum response factor
- STL
serine, threonine, leucine
- TIF
transcriptional intermediary factor
- TR
thyroid hormone receptor
- TRAM-1
thyroid receptor activator molecule-1
- TRAP
TR-associated protein
- VP16
Herpes Simplex Virus protein 16
References
- Angel P., Szabowski A., Schorpp-Kistner M. Function and regulation of AP-1 subunits in skin physiology and pathology. Oncogene. 2001;20:2413–23. doi: 10.1038/sj.onc.1204380. [DOI] [PubMed] [Google Scholar]
- Anghel S. I., Wahli W. Fat poetry: a kingdom for PPARgamma. Cell Res. 2007;17:486–511. doi: 10.1038/cr.2007.48. [DOI] [PubMed] [Google Scholar]
- Antonson P., Schuster G. U., Wang L., Rozell B., Holter E., Flodby P., Treuter E., Holmgren L., Gustafsson J. A. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Mol Cell Biol. 2003;23:1260–8. doi: 10.1128/MCB.23.4.1260-1268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonson P., Al-Beidh F., Matthews J., Gustafsson J. A. The human RAP250 gene: genomic structure and promoter analysis. Gene. 2004;327:233–8. doi: 10.1016/j.gene.2003.11.022. [DOI] [PubMed] [Google Scholar]
- Anzick S. L., Kononen J., Walker R. L., Azorsa D. O., Tanner M. M., Guan X. Y., Sauter G., Kallioniemi O. P., Trent J. M., Meltzer P. S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–8. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Aranda A., Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- Aravind L. The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem Sci. 2000;25:421–3. doi: 10.1016/s0968-0004(00)01620-0. [DOI] [PubMed] [Google Scholar]
- Baek H. J., Malik S., Qin J., Roeder R. G. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol Cell Biol. 2002;22:2842–52. doi: 10.1128/MCB.22.8.2842-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer T. G., Martin M. E., Lees E., Ricciardi R. P., Berk A. J. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–9. doi: 10.1038/20466. [DOI] [PubMed] [Google Scholar]
- Caira F., Antonson P., Pelto-Huikko M., Treuter E., Gustafsson J. A. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J Biol Chem. 2000;275:5308–17. doi: 10.1074/jbc.275.8.5308. [DOI] [PubMed] [Google Scholar]
- Cavailles V., Dauvois S., Danielian P. S., Parker M. G. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci U S A. 1994;91:10009–13. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D., LaMorte V. J., Nelson M. C., Nakajima T., Schulman I. G., Juguilon H., Montminy M., Evans R. M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- Chang C., Norris J. D., Gron H., Paige L. A., Hamilton P. T., Kenan D. J., Fowlkes D., McDonnell D. P. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors α and β. Mol Cell Biol. 1999;19:8226–39. doi: 10.1128/mcb.19.12.8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan H. M., La Thangue N. B. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–73. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- Chen H., Lin R. J., Schiltz R. L., Chakravarti D., Nash A., Nagy L., Privalsky M. L., Nakatani Y., Evans R. M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–80. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Choi E., Lee S., Yeom S. Y., Kim G. H., Lee J. W., Kim S. W. Characterization of activating signal cointegrator-2 as a novel transcriptional coactivator of the xenobiotic nuclear receptor constitutive androstane receptor. Mol Endocrinol. 2005;19:1711–9. doi: 10.1210/me.2005-0066. [DOI] [PubMed] [Google Scholar]
- Cho Y. W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G. R., Copeland T. D., Kalkum M., Ge K. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford S. E., Qi C., Misra P., Stellmach V., Rao M. S., Engel J. D., Zhu Y., Reddy J. K. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J Biol Chem. 2002;277:3585–92. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- Evans R. M., Barish G. D., Wang Y. X. PPARs and the complex journey to obesity. Nat Med. 2004;10:355–61. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- Fondell J. D., Ge H., Roeder R. G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci U S A. 1996;93:8329–33. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehin M., Mark M., Dennefeld C., Dierich A., Gronemeyer H., Chambon P. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol Cell Biol. 2002;22:5923–37. doi: 10.1128/MCB.22.16.5923-5937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo Y. H., Sohn Y. C., Kim D. H., Kim S. W., Kang M. J., Jung D. J., Kwak E., Barlev N. A., Berger S. L., Chow V. T., Roeder R. G., Azorsa D. O., Meltzer P. S., Suh P. G., Song E. J., Lee K. J., Lee Y. C., Lee J. W. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–9. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman R. H., Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–77. [PubMed] [Google Scholar]
- Goo Y. H., Na S. Y., Zhang H., Xu J., Hong S., Cheong J., Lee S. K., Lee J. W. Interactions between activating signal cointegrator-2 and the tumor suppressor retinoblastoma in androgen receptor transactivation. J Biol Chem. 2004;279:7131–5. doi: 10.1074/jbc.M312563200. [DOI] [PubMed] [Google Scholar]
- Guan X. Y., Xu J., Anzick S. L., Zhang H., Trent J. M., Meltzer P. S. Hybrid selection of transcribed sequences from microdissected DNA: isolation of genes within amplified region at 20q11-q13.2 in breast cancer. Cancer Res. 1996;56:3446–50. [PubMed] [Google Scholar]
- Halachmi S., Marden E., Martin G., MacKay H., Abbondanza C., Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–8. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- Hart C. M., Cuvier O., Laemmli U. K. Evidence for an antagonistic relationship between the boundary element-associated factor BEAF and the transcription factor DREF. Chromosoma. 1999;108:375–83. doi: 10.1007/s004120050389. [DOI] [PubMed] [Google Scholar]
- Hong S., Choi H. M., Park M. J., Kim Y. H., Choi Y. H., Kim H. H., Choi Y. H., Cheong J. Activation and interaction of ATF2 with the coactivator ASC-2 are responsive for granulocytic differentiation by retinoic acid. J Biol Chem. 2004a;279:16996–7003. doi: 10.1074/jbc.M311752200. [DOI] [PubMed] [Google Scholar]
- Hong S., Kim S. H., Heo M. A., Choi Y. H., Park M. J., Yoo M. A., Kim H. D., Kang H. S., Cheong J. Coactivator ASC-2 mediates heat shock factor 1-mediated transactivation dependent on heat shock. FEBS Lett. 2004b;559:165–70. doi: 10.1016/S0014-5793(04)00028-6. [DOI] [PubMed] [Google Scholar]
- Hong H., Kohli K., Trivedi A., Johnson D. L., Stallcup M. R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci U S A. 1996;93:4948–52. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H., Chan E. K., Kiyosawa K., Fu X. D., Tan E. M. Novel nuclear autoantigen with splicing factor motifs identified with antibody from hepatocellular carcinoma. J Clin Invest. 1993;92:2419–26. doi: 10.1172/JCI116848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Laemmli U. K. Structural and dynamic functions establish chromatin domains. Mol Cell. 2003;11:237–48. doi: 10.1016/s1097-2765(03)00010-8. [DOI] [PubMed] [Google Scholar]
- Issaeva I., Zonis Y., Rozovskaia T., Orlovsky K., Croce C. M., Nakamura T., Mazo A., Eisenbach L., Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Yuan C. X., Okano H. J., Darnell R. B., Roeder R. G. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell. 2000;5:683–93. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- Ito M., Roeder R. G. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab. 2001;12:127–34. doi: 10.1016/s1043-2760(00)00355-6. [DOI] [PubMed] [Google Scholar]
- Iwasaki T., Chin W. W., Ko L. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM) J Biol Chem. 2001;276:33375–83. doi: 10.1074/jbc.M101517200. [DOI] [PubMed] [Google Scholar]
- Ju B. G., Lunyak V. V., Perissi V., Garcia-Bassets I., Rose D. W., Glass C. K., Rosenfeld M. G. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- Jung D. J., Na S. Y., Na D. S., Lee J. W. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J Biol Chem. 2002;277:1229–34. doi: 10.1074/jbc.M110417200. [DOI] [PubMed] [Google Scholar]
- Kim S. W., Park K., Kwak E., Choi E., Lee S., Ham J., Kang H., Kim J. M., Hwang S. Y., Kong Y. Y., Lee K., Lee J. W. Activating signal cointegrator 2 required for liver lipid metabolism mediated by liver X receptors in mice. Mol Cell Biol. 2003;23:3583–92. doi: 10.1128/MCB.23.10.3583-3592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. W., Cheong C., Sohn Y. C., Goo Y. H., Oh W. J., Park J. H., Joe S. Y., Kang H. S., Kim D. K., Kee C., Lee J. W., Lee H. W. Multiple developmental defects derived from impaired recruitment of ASC-2 to nuclear receptors in mice: implication for posterior lenticonus with cataract. Mol Cell Biol. 2002;22:8409–14. doi: 10.1128/MCB.22.24.8409-8414.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong H. J., Yu H. J., Hong S., Park M. J., Choi Y. H., An W. G., Lee J. W., Cheong J. Interaction and functional cooperation of the cancer-amplified transcriptional coactivator activating signal cointegrator-2 and E2F-1 in cell proliferation. Mol Cancer Res. 2003;1:948–58. [PubMed] [Google Scholar]
- Ko L., Chin W. W. Nuclear receptor coactivator thyroid hormone receptor-binding protein (TRBP) interacts with and stimulates its associated DNA-dependent protein kinase. J Biol Chem. 2003;278:11471–9. doi: 10.1074/jbc.M209723200. [DOI] [PubMed] [Google Scholar]
- Ko L., Cardona G. R., Iwasaki T., Bramlett K. S., Burris T. P., Chin W. W. Ser-884 adjacent to the LXXLL motif of coactivator TRBP defines selectivity for ERs and TRs. Mol Endocrinol. 2002;16:128–40. doi: 10.1210/mend.16.1.0755. [DOI] [PubMed] [Google Scholar]
- Ko L., Cardona G. R., Chin W. W. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc Natl Acad Sci U S A. 2000;97:6212–7. doi: 10.1073/pnas.97.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S. Q., Liao L., Zhang H., Pereira F. A., Yuan Y., DeMayo F. J., Ko L., Xu J. Deletion of the cancer-amplified coactivator AIB3 results in defective placentation and embryonic lethality. J Biol Chem. 2002;277:45356–60. doi: 10.1074/jbc.C200509200. [DOI] [PubMed] [Google Scholar]
- Landles C., Chalk S., Steel J. H., Rosewell I., Spencer-Dene B., Lalani el N., Parker M. G. The thyroid hormone receptor-associated protein TRAP220 is required at distinct embryonic stages in placental, cardiac, and hepatic development. Mol Endocrinol. 2003;17:2418–35. doi: 10.1210/me.2003-0097. [DOI] [PubMed] [Google Scholar]
- Lee S. K., Anzick S. L., Choi J. E., Bubendorf L., Guan X. Y., Jung Y. K., Kallioniemi O. P., Kononen J., Trent J. M., Azorsa D., Jhun B. H., Cheong J. H., Lee Y. C., Meltzer P. S., Lee J. W. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J Biol Chem. 1999;274:34283–93. doi: 10.1074/jbc.274.48.34283. [DOI] [PubMed] [Google Scholar]
- Lee S. K., Na S. Y., Jung S. Y., Choi J. E., Jhun B. H., Cheong J., Meltzer P. S., Lee Y. C., Lee J. W. Activating protein-1, nuclear factor-kappaB, and serum response factor as novel target molecules of the cancer-amplified transcription coactivator ASC-2. Mol Endocrinol. 2000;14:915–25. doi: 10.1210/mend.14.6.0471. [DOI] [PubMed] [Google Scholar]
- Lee S., Lee D. K., Dou Y., Lee J., Lee B., Kwak E., Kong Y. Y., Lee S. K., Roeder R. G., Lee J. W. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc Natl Acad Sci U S A. 2006;103:15392–7. doi: 10.1073/pnas.0607313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. K., Jung S. Y., Kim Y. S., Na S. Y., Lee Y. C., Lee J. W. Two distinct nuclear receptor-interaction domains and CREB-binding protein-dependent transactivation function of activating signal cointegrator-2. Mol Endocrinol. 2001;15:241–54. doi: 10.1210/mend.15.2.0595. [DOI] [PubMed] [Google Scholar]
- Liao L., Kuang S. Q., Yuan Y., Gonzalez S. M., O'Malley B. W., Xu J. Molecular structure and biological function of the cancer-amplified nuclear receptor coactivator SRC-3/AIB1. J Steroid Biochem Mol Biol. 2002;83:3–14. doi: 10.1016/s0960-0760(02)00254-6. [DOI] [PubMed] [Google Scholar]
- Li G., Gustafson-Brown C., Hanks S. K., Nason K., Arbeit J. M., Pogliano K., Wisdom R. M., Johnson R. S. c-Jun is essential for organization of the epidermal leading edge. Dev Cell. 2003;4:865–77. doi: 10.1016/s1534-5807(03)00159-x. [DOI] [PubMed] [Google Scholar]
- Li D., Wang F., Samuels H. H. Domain structure of the NRIF3 family of coregulators suggests potential dual roles in transcriptional regulation. Mol Cell Biol. 2001;21:8371–84. doi: 10.1128/MCB.21.24.8371-8384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. M., Muktar A., Samuels H. H. Functional consequences of interactions between thyroid hormone receptors and retinoid X receptor. Current Opinion in Endocrinology & Diabetes. 2005;12:356–362. [Google Scholar]
- Li D., Desai-Yajnik V., Lo E., Schapira M., Abagyan R., Samuels H. H. NRIF3 is a novel coactivator mediating functional specificity of nuclear hormone receptors. Mol Cell Biol. 1999;19:7191–202. doi: 10.1128/mcb.19.10.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Gomes P. J., Chen J. D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci U S A. 1997;94:8479–84. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan M. A., Samuels H. H. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol Cell Biol. 2000;20:5048–63. doi: 10.1128/mcb.20.14.5048-5063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan M. A., Murray A., Samuels H. H. NRC-interacting factor 1 is a novel cotransducer that interacts with and regulates the activity of the nuclear hormone receptor coactivator NRC. Mol Cell Biol. 2002;22:6883–94. doi: 10.1128/MCB.22.19.6883-6894.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan M. A., Samuels H. H. Nuclear hormone receptor coregulator: role in hormone action, metabolism, growth, and development. Endocr Rev. 2005;26:583–97. doi: 10.1210/er.2004-0012. [DOI] [PubMed] [Google Scholar]
- Mahajan M. A., Murray A., Levy D., Samuels H. H. Nuclear receptor coregulator (NRC): mapping of the dimerization domain, activation of p53 and STAT-2, and identification of the activation domain AD2 necessary for nuclear receptor signaling. Mol Endocrinol. 2007;21:1822–34. doi: 10.1210/me.2005-0529. [DOI] [PubMed] [Google Scholar]
- Mahajan M. A., Das S., Zhu H., Tomic-Canic M., Samuels H. H. The nuclear hormone receptor coactivator NRC is a pleiotropic modulator affecting growth, development, apoptosis, reproduction, and wound repair. Mol Cell Biol. 2004;24:4994–5004. doi: 10.1128/MCB.24.11.4994-5004.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark M., Yoshida-Komiya H., Gehin M., Liao L., Tsai M. J., O'Malley B. W., Chambon P., Xu J. Partially redundant functions of SRC-1 and TIF2 in postnatal survival and male reproduction. Proc Natl Acad Sci U S A. 2004;101:4453–8. doi: 10.1073/pnas.0400234101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney E. M., Rose D. W., Flynn S. E., Westin S., Mullen T. M., Krones A., Inostroza J., Torchia J., Nolte R. T., Assa-Munt N., Milburn M. V., Glass C. K., Rosenfeld M. G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–68. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna N. J., O'Malley B. W. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–74. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Michalik L., Wahli W. Peroxisome proliferator-activated receptors (PPARs) in skin health, repair and disease. Biochim Biophys Acta. 2007;1771:991–8. doi: 10.1016/j.bbalip.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Misra P., Qi C., Yu S., Shah S. H., Cao W. Q., Rao M. S., Thimmapaya B., Zhu Y., Reddy J. K. Interaction of PIMT with transcriptional coactivators CBP, p300, and PBP differential role in transcriptional regulation. J Biol Chem. 2002;277:20011–9. doi: 10.1074/jbc.M201739200. [DOI] [PubMed] [Google Scholar]
- Mo R., Rao S. M., Zhu Y. J. Identification of the MLL2 complex as a coactivator for estrogen receptor α. J Biol Chem. 2006;281:15714–20. doi: 10.1074/jbc.M513245200. [DOI] [PubMed] [Google Scholar]
- Naar A. M., Beaurang P. A., Zhou S., Abraham S., Solomon W., Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–32. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- Nishino T., Morikawa K. Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors. Oncogene. 2002;21:9022–32. doi: 10.1038/sj.onc.1206135. [DOI] [PubMed] [Google Scholar]
- Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., Milburn M. V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–43. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- Onate S. A., Tsai S. Y., Tsai M. J., O'Malley B. W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–7. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–39. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Adelmant G., Wu Z., Fan M., Xu J., O'Malley B., Spiegelman B. M. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–71. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- Qi C., Kashireddy P., Zhu Y. T., Rao S. M., Zhu Y. J. Null mutation of peroxisome proliferator-activated receptor-interacting protein in mammary glands causes defective mammopoiesis. J Biol Chem. 2004;279:33696–701. doi: 10.1074/jbc.M401266200. [DOI] [PubMed] [Google Scholar]
- Qi C., Surapureddi S., Zhu Y. J., Yu S., Kashireddy P., Rao M. S., Reddy J. K. Transcriptional coactivator PRIP, the peroxisome proliferator-activated receptor γ (PPARgamma)-interacting protein, is required for PPARgamma-mediated adipogenesis. J Biol Chem. 2003;278:25281–4. doi: 10.1074/jbc.C300175200. [DOI] [PubMed] [Google Scholar]
- Rachez C., Lemon B. D., Suldan Z., Bromleigh V., Gamble M., Naar A. M., Erdjument-Bromage H., Tempst P., Freedman L. P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–8. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- Sarkar J., Qi C., Guo D., Ahmed M. R., Jia Y., Usuda N., Viswakarma N., Rao M. S., Reddy J. K. Transcription coactivator PRIP, the peroxisome proliferator-activated receptor (PPAR)-interacting protein, is redundant for the function of nuclear receptors PParalpha and CAR, the constitutive androstane receptor, in mouse liver. Gene Expr. 2007;13:255–69. doi: 10.3727/000000006780666948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D., Fondell J. D. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc Natl Acad Sci U S A. 2002;99:7934–9. doi: 10.1073/pnas.122004799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallcup M. R. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene. 2001;20:3014–20. doi: 10.1038/sj.onc.1204325. [DOI] [PubMed] [Google Scholar]
- Strahl B. D., Briggs S. D., Brame C. J., Caldwell J. A., Koh S. S., Ma H., Cook R. G., Shabanowitz J., Hunt D. F., Stallcup M. R., Allis C. D. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Takeshita A., Cardona G. R., Koibuchi N., Suen C. S., Chin W. W. TRAM-1, A novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–34. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Naruse I., Maekawa T., Masuya H., Shiroishi T., Ishii S. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci U S A. 1997;94:10215–20. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan N. S., Michalik L., Noy N., Yasmin R., Pacot C., Heim M., Fluhmann B., Desvergne B., Wahli W. Critical roles of PPAR β/δ in keratinocyte response to inflammation. Genes Dev. 2001;15:3263–77. doi: 10.1101/gad.207501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Mahajan M. A., Wong C. T., Habeos I., Samuels H. H. The N-Terminal A/B domain of the thyroid hormone receptor-beta2 isoform influences ligand-dependent recruitment of coactivators to the ligand-binding domain. Mol Endocrinol. 2006;20:2036–51. doi: 10.1210/me.2005-0437. [DOI] [PubMed] [Google Scholar]
- Torchia J., Rose D. W., Inostroza J., Kamei Y., Westin S., Glass C. K., Rosenfeld M. G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–84. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Toresson G., Schuster G. U., Steffensen K. R., Bengtsson M., Ljunggren J., Dahlman-Wright K., Gustafsson J. A. Purification of functional full-length liver X receptor β produced in Escherichia coli. Protein Expr Purif. 2004;35:190–8. doi: 10.1016/j.pep.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Voegel J. J., Heine M. J., Zechel C., Chambon P., Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. Embo J. 1996;15:3667–75. [PMC free article] [PubMed] [Google Scholar]
- Volle D. H., Dechelotte P., Colombier M., Veyssiere G., Mangelsdorf D. J., Benhamed M., Lobaccaro J. M. A. Testicular function in LXR-deficient mice. Keystone Symposia-Nuclear Receptors: Orphan Brothers, Abstract 337. 2004.
- Waikel R. L., Kawachi Y., Waikel P. A., Wang X. J., Roop D. R. Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet. 2001;28:165–8. doi: 10.1038/88889. [DOI] [PubMed] [Google Scholar]
- White R., Leonardsson G., Rosewell I., Ann Jacobs M., Milligan S., Parker M. The nuclear receptor co-repressor nrip1 (RIP140) is essential for female fertility. Nat Med. 2000;6:1368–74. doi: 10.1038/82183. [DOI] [PubMed] [Google Scholar]
- Xu J., Qiu Y., DeMayo F. J., Tsai S. Y., Tsai M. J., O'Malley B. W. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279:1922–5. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- Xu J., Liao L., Ning G., Yoshida-Komiya H., Deng C., O'Malley B. W. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc Natl Acad Sci U S A. 2000;97:6379–84. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T. P., Oh S. P., Fuchs M., Zhou N. D., Ch'ng L. E., Newsome D., Bronson R. T., Li E., Livingston D. M., Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–72. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- Yeom S. Y., Kim G. H., Kim C. H., Jung H. D., Kim S. Y., Park J. Y., Pak Y. K., Rhee D. K., Kuang S. Q., Xu J., Han D. J., Song D. K., Lee J. W., Lee K. U., Kim S. W. Regulation of insulin secretion and β-cell mass by activating signal cointegrator 2. Mol Cell Biol. 2006;26:4553–63. doi: 10.1128/MCB.01412-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kuang S. Q., Liao L., Zhou S., Xu J. Haploid inactivation of the amplified-in-breast cancer 3 coactivator reduces the inhibitory effect of peroxisome proliferator-activated receptor γ and retinoid X receptor on cell proliferation and accelerates polyoma middle-T antigen-induced mammary tumorigenesis in mice. Cancer Res. 2004;64:7169–77. doi: 10.1158/0008-5472.CAN-04-1176. [DOI] [PubMed] [Google Scholar]
- Zhang H., Liao L., Kuang S. Q., Xu J. Spatial distribution of the messenger ribonucleic acid and protein of the nuclear receptor coactivator, amplified in breast cancer-3, in mice. Endocrinology. 2003;144:1435–43. doi: 10.1210/en.2002-0018. [DOI] [PubMed] [Google Scholar]
- Zhao C., Toresson G., Xu L., Koehler K. F., Gustafsson J. A., Dahlman-Wright K. Mouse estrogen receptor β isoforms exhibit differences in ligand selectivity and coactivator recruitment. Biochemistry. 2005;44:7936–44. doi: 10.1021/bi047691m. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Qi C., Cao W. Q., Yeldandi A. V., Rao M. S., Reddy J. K. Cloning and characterization of PIMT, a protein with a methyltransferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proc Natl Acad Sci U S A. 2001;98:10380–5. doi: 10.1073/pnas.181347498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. J., Crawford S. E., Stellmach V., Dwivedi R. S., Rao M. S., Gonzalez F. J., Qi C., Reddy J. K. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. J Biol Chem. 2003;278:1986–90. doi: 10.1074/jbc.C200634200. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Qi C., Jain S., Rao M. S., Reddy J. K. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem. 1997;272:25500–6. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Kan L., Qi C., Kanwar Y. S., Yeldandi A. V., Rao M. S., Reddy J. K. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J Biol Chem. 2000;275:13510–6. doi: 10.1074/jbc.275.18.13510. [DOI] [PubMed] [Google Scholar]