Abstract
Recent genomic analyses of transcription factor binding, histone modification, and gene expression have provided a global view of transcriptional regulation by nuclear receptors (NRs) that complements an existing large body of literature on gene-specific studies. The picture emerging from these genomic studies indicates that NRs bind at promoter-proximal and promoter-distal enhancers in conjunction with other transcription factors (e.g., activator protein-1, Sp1 and FOXA1). This binding promotes the recruitment of coregulators that mediate the posttranslational modification of histones at promoters and enhancers. Ultimately, signaling through liganded NRs stimulates changes in the occupancy of RNA polymerase II (Pol II) or the activation of preloaded Pol II at target promoters. Chromosomal looping and/or Pol II tracking may underlie promoter-enhancer communication. Interestingly, the direct target genes of NR signaling represent a limited subset of all the genes regulated by NR ligands, with the rest being regulated through secondary effects. As suggested by previous gene-specific analyses, NR-mediated outcomes are highly cell type- and promoter-specific, highlighting the complexity of transcriptional regulation by NRs and the value of genomic analyses for identifying commonly shared patterns. Overall, NRs share common themes in their patterns of localization and transcriptional regulation across mammalian genomes. In this review, we provide an overview of recent advances in the understanding of NR-mediated transcription garnered from genomic analyses of gene expression, factor localization, and target DNA sequences.
Introduction
The spatial and temporal regulation of gene expression is an important means by which cells respond to physiological and environmental signals. DNA-binding transcription factors, non-DNA-binding coregulators, and the RNA polymerase II (Pol II) machinery are important for mediating proper (i.e., context-specific or developmentally appropriate) patterns of gene expression [Naar et al., 2001; Orphanides and Reinberg, 2002]. Nuclear receptors (NRs) comprise a superfamily of ligand-regulated, DNA-binding transcription factors, which can both activate and repress gene expression [Mangelsdorf et al., 1995]. Given the number of related factors in the superfamily (49 NR genes and more than 75 NR proteins in mammals; [Robinson-Rechavi et al., 2001]) and their physiological roles throughout the body, NRs make an interesting model to study the mechanisms of transcriptional regulation in response to cellular signals. Transcriptional regulation by NRs is a multistep process involving: (1) the association of NRs with regulatory sites in the genome (i.e., enhancers or silencers) in the context of chromatin, (2) the ligand-dependent recruitment and function of coregulators to modify chromatin and associated factors, (3) the regulation of Pol II binding and activity at target promoters, and (4) the termination or attenuation of NR-dependent signaling [Acevedo and Kraus, 2004; Glass and Rosenfeld, 2000; Kraus and Wong, 2002; McKenna et al., 1999; Metivier et al., 2006]. The complexity of transcriptional regulation by NRs provides many opportunities for exquisite regulatory control of signal-dependent transcriptional responses.
The mechanisms of transcriptional regulation by ligand-bound NRs have been studied extensively in numerous gene-specific studies over the past 30 years. Recently, the development of large-scale genomic methods to analyze gene expression and factor binding to DNA has generated an extensive amount of information about NR-regulated transcription. Some of these methods (i.e., gene expression microarray analyses, ChIP-chip, in silico binding site analyses) have been reviewed recently elsewhere [Kim and Ren, 2006; Tavera-Mendoza et al., 2006]. Here, we provide an overview of recent advances in our understanding of NR-mediated transcription focusing on genomic analyses of gene expression, factor localization, and target DNA sequences.
Genomic analyses of NR-regulated gene expression
Gene expression microarrays have been widely used to determine genes whose expression changes upon treatment with NR ligands, such as estrogens, androgens, glucocorticoids, vitamin D3, and lipid metabolites [Frasor et al., 2003; Lee et al., 2003; Quinn et al., 2005; Rogatsky et al., 2003; White, 2004]. Due to the large number of studies available, we will use the regulation of gene expression by ER ligands as an example to illustrate the use of expression microarrays to understand global features of NR-mediated transcription. The approaches and key concepts are similar for most global NR-mediated gene expression studies to date.
How many genes are regulated by estrogen signaling?
The first question addressed by expression microarray analyses of estradiol (E2)-dependent gene expression was "How many genes are regulated by E2 in human cells?" The answer turned out to be less straightforward than expected, and several studies currently available report different numbers of E2-regulated genes, ranging from ∼100 to ∼1,500 [Carroll and Brown, 2006; Coser et al., 2003; Frasor et al., 2003; Kian Tee et al., 2004; Kininis et al., 2007; Kwon et al., 2007; Levenson et al., 2002; Lin et al., 2004; Lin et al., 2007; Monroe et al., 2003; Rae et al., 2005; Stender et al., 2007] (Table 1). These discrepancies can be attributed to many factors including differences in: (1) the cell lines (tissue origin, ER expression, and growth conditions), (2) the length of the E2 treatment, (3) the microarray platforms and associated experimental variability, and (4) the data analysis protocols. Examples of such types of variation among recent global E2-dependent gene expression analyses are shown in Table 1.
Table 1. Microarray analyses of E2-regulated gene expression in human cell lines.
Published microarray analyses of E2-mediated gene expression report a variable number of E2-regulated genes. The discrepancies among these studies can be attributed to many factors including differences in (1) the cell lines used, (2) the length of the E2 treatment, (3) the microarray platforms used, and (4) the data analysis protocols used. Examples of these types of variation are indicated.
Global features of E2-regulated gene expression
Despite the differences noted above, genomic analyses of E2-regulated gene expression have provided a wealth of useful information for understanding the global features of NR-mediated gene expression. In summary, genomic expression analyses have indicated that: (1) only a limited subset (20-30%) of the E2-regulated genes represent direct E2 targets, as shown by co-treatment with the translation inhibitor cyclohexamide [Lin et al., 2004]; (2) the majority of E2-regulated genes after short (1-8 h) hormone treatments are upregulated, whereas most genes regulated after longer treatments (12-48 h) are downregulated [Frasor et al., 2003; Lin et al., 2004]; (3) a surprisingly limited overlap exists between the gene sets regulated by E2 in different cell lines (and even the same cell line grown in different laboratories) [Carroll and Brown, 2006; Frasor et al., 2003; Kininis et al., 2007; Lin et al., 2004; Rae et al., 2005; Stender et al., 2007] (also see Supplemental Table 1 in Kininis et al. [Kininis et al., 2007]); (4) the two subtypes of ER (ERα and ERβ) regulate diverse (>70% different) sets of genes [Kian Tee et al., 2004; Monroe et al., 2003]; (5) selective estrogen receptor modulators (SERMs) have both antagonistic and agonistic effects on global patterns of gene expression, in some cases overlapping with E2-regulated gene sets [Frasor et al., 2004; Levenson et al., 2002]; and (6) some E2-regulated genes are dependent on other DNA-bound transcription factors, such as AP-1, for their expression [DeNardo et al., 2005]. Collectively, the available gene expression microarrays have revealed new global features of NR-mediated transcription, complementing previous gene-specific studies.
Facilitating comparisons between different studies of NR-regulated gene expression
Although it may be unrealistic to expect all researchers in the NR field to follow the same experimental protocols (e.g., RNA processing, microarray platform and handling, and data analysis), means are available to facilitate comparisons among different studies of NR-regulated gene expression. One approach is to evaluate expression microarray performance using complementary approaches, such as Northern blotting or quantitative reverse transcription-PCR, with clear reporting of the confirmation rates [Tan et al., 2003; Taniguchi et al., 2001]. Although this approach would help to control for false positives introduced by the microarray experimental variability, it would not account for false negatives (i.e., genes whose expression changes in the experimental samples, but is not detected by the microarrays). A second simple approach would be to use external controls, such as RNA molecules synthetically produced and spiked in defined amounts to biological samples before hybridization to microarrays [van Bakel and Holstege, 2004]. By using this protocol, differences in microarray sensitivity could be assessed, and proper normalization could be applied to the data. Furthermore, a direct comparison of each study’s results to previously published data would greatly facilitate our understanding of the extent and sources of variation. Finally, the deposition of raw (i.e., unprocessed) datasets in databases, such as GEO (http://www.ncbi.nlm.nih.gov/geo/) or NURSA (http://www.nursa.org/), would facilitate meta-analysis efforts aiming to compare and contrast data from different studies using the same analysis algorithms.
Global studies of NR localization
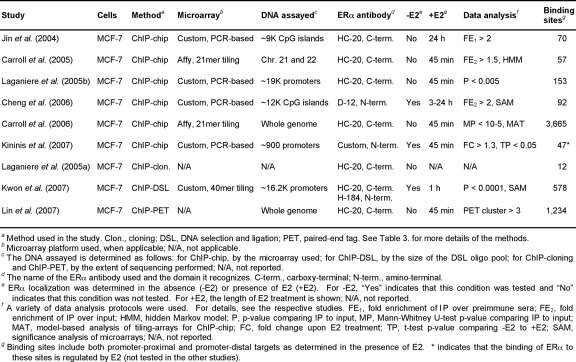
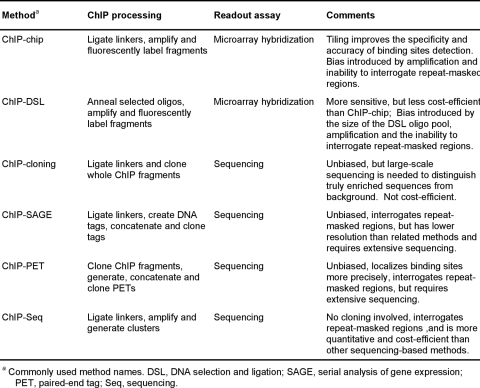
A number of recent studies determined the genomic binding sites of several NRs, including ERα, AR, GR, VDR, ERRα, and ERRβ. Given that most of these studies have focused on the localization of ERα, either to gene promoters, chromosomes, or the whole human genome [Carroll et al., 2005; Carroll et al., 2006; Cheng et al., 2006; Gao et al., 2008; Jin et al., 2004; Kininis et al., 2007; Kwon et al., 2007; Laganiere et al., 2005a; Laganiere et al., 2005b; Lin et al., 2007] (Table 2), we will mainly focus our review on ERα and compare the ERα-specific conclusions to findings about other NRs. The existing studies used a wide variety of chromatin immunoprecipitation (ChIP)-based methods, including combinations of ChIP with microarrays (e.g., ChIP-chip and ChIP-DSL) and with DNA sequencing (e.g., ChIP-cloning and ChIP-PET) (Table 3), and the results have not always been consistent.
Table 2. Genomic analyses of ERα localization in human cell lines.
Table summarizing the similarities and differences among the genomic analyses of ERα localization in human cell lines published to date.
Table 3. ChIP-based methods to study factor localization on a genomic scale.
A number of recent studies have determined the genomic binding sites of several NRs. These studies used a wide variety of chromatin immunoprecipitation (ChIP)-based methods, including combinations with microarrays (e.g., ChIP-chip and ChIP-DSL) and DNA sequencing (e.g., ChIP-cloning and ChIP-PET). Important technical features, as well as advantages and disadvantages of each method, are summarized.
Similarities and differences among the available genomic ERα location analyses
Remarkably, all genomic analyses of ERα localization performed to date in human cells used the same cell line (MCF-7) and most of them used a similar length of E2 treatment (∼45 min.) (Table 2). However, the overlap of ERα binding sites among these studies is strikingly limited. For example, only 624 of the 1,234 (50.6%) ChIP-PET ERα binding sites identified by Lin et al., 2007 [Lin et al., 2007] were common to the 3,665 ChIP-chip ERα binding sites identified by Carroll et al., 2006 [Carroll et al., 2006], representing an overlap of less than 20% for the union of ERα binding sites from the two studies. In addition, only one common ERα-bound promoter (TFF1) from the overlapping chromosome 21 and 22 promoter set was identified in two recent ChIP-chip studies, although the same antibody was used [Carroll et al., 2005; Laganiere et al., 2005b; Tavera-Mendoza et al., 2006]. Some of these differences may be attributed to the specific microarray platform used and other experimental variations, but cross-validation of the conflicting results in the available experimental systems is necessary to determine whether the overlap or the union of the identified binding sites represents more accurately the true landscape of ERα targets in MCF-7 cells.
Despite their differences, the ERα localization studies mentioned collectively provide an increasingly clearer view of the general patterns of ERα binding across the genome. For many E2-regulated genes, the ERα binding sites are located at great distances (>100 Kb) from target promoters, while for other E2-regulated genes, the receptor binding sites are located within or near proximal promoter regions [Carroll et al., 2006; Kininis et al., 2007; Kwon et al., 2007; Laganiere et al., 2005b; Lin et al., 2007]. A recent study in mice confirmed the binding of ERα both proximally and distally from promoters in vivo [Gao et al., 2008]. Interestingly, genes upregulated by E2 contain more promoter-proximal ERα binding sites than genes downregulated by E2 [Kininis et al., 2007; Kwon et al., 2007; Lin et al., 2007]. Furthermore, genes upregulated by shorter E2 treatments are enriched in ERα binding sites adjacent to their promoters in contrast to genes upregulated by longer E2 treatments, suggesting secondary regulatory effects for the latter gene set [Kininis et al., 2007; Kwon et al., 2007; Lin et al., 2007]. In some cases, ERα-bound distal enhancers were shown to physically interact with the nearest promoters, presumably regulating their E2-dependent expression [Carroll et al., 2005]. The available genomic studies suggest that this long-range regulation may be particularly important for some subsets of genes. Further studies are needed, however, to understand the role of ERα binding to distal enhancers and the generality of the proposed enhancer-promoter looping mechanisms.
Genomic localization themes shared by NRs
In addition to ERα, recent ChIP-chip analyses have examined the localization of AR, GR, VDR, ERRα, and ERRγ at selected regions of the human genome [Bolton et al., 2007; Dufour et al., 2007; Pike et al., 2007; So et al., 2007; Wang et al., 2007]. The global features of AR, GR and VDR binding to DNA are remarkably similar to those identified for ERα in that the receptors localize both distally and proximally to gene promoters, and enhancer-promoter looping may be an important mode of transcriptional regulation [Pike et al., 2007; So et al., 2007; Wang et al., 2007]. The ERR location analysis focused on gene promoters and identified a significant overlap in the ERRα- and ERRγ-bound targets, suggesting that the two ERR subtypes may function as heterodimers in human cells [Dufour et al., 2007]. Overall, NRs share some common themes in their patterns of localization across the human genome (i.e., promoter-proximal and promoter-distal binding, long-range enhancer-promoter interactions), although more studies covering a wider range of NRs are needed to fully establish the generality of these patterns.
NR binding sites: lessons from the underlying DNA sequences
Gene-specific studies have shown that NRs can associate with their target DNA sequences through at least two different mechanisms: (1) direct binding to specific response elements as monomers, homodimers, or heterodimers [Klein-Hitpass et al., 1988; Mangelsdorf et al., 1995], and (2) indirect binding (or "tethering") through other classes of DNA-bound transcription factors (e.g., activator protein-1, AP-1) [Kushner et al., 2000]. In addition to directing NRs to specific regions of the genome, the target DNA sequences can also regulate the specificity of NR-mediated responses [Klinge et al., 2004; Kurokawa et al., 1995; Lefstin and Yamamoto, 1998; Loven et al., 2001; O'Lone et al., 2004; So et al., 2007]. Interestingly, although the NR binding sequences may vary extensively from a consensus, the precise sequences at individual NR binding sites are often well conserved among mammalian species [So et al., 2007]. Furthermore, the direct or indirect association of NRs with their target sites may allosterically regulate receptor activity and coregulator recruitment [Shang and Brown, 2002].
Many of the recent genome-wide localization studies have used bioinformatic approaches to search for common motifs in the DNA sequences underlying the identified binding sites. As expected, estrogen-, androgen-, glucocorticoid-, vitamin D3- and estrogen related-response elements (EREs, AREs, GREs, VDREs and ERREs, respectively) were enriched in the binding sites of their cognate NRs [Carroll et al., 2005; Carroll et al., 2006; Cheng et al., 2006; Dufour et al., 2007; Gao et al., 2008; Kininis et al., 2007; Kwon et al., 2007; Laganiere et al., 2005b; Lin et al., 2007; Pike et al., 2007; So et al., 2007; Wang et al., 2007]. Notably, for ERRα and ERRγ, the same consensus ERRE sequence was found for both receptor subtypes, consistent with the binding of these receptors as heterodimers to common sites [Dufour et al., 2007]. In addition to NR response elements, binding elements for various other transcription factors (e.g., AP-1, Sp1, FOXA1, Oct1, CREB, C/EBPα, Myc) were also enriched in NR binding sites [Carroll et al., 2005; Carroll et al., 2006; Cheng et al., 2006; Dufour et al., 2007; Gao et al., 2008; Kininis et al., 2007; Laganiere et al., 2005b; Lin et al., 2007; Wang et al., 2007]. In some cases, the binding elements for the transcription factors were found adjacent to NR response elements (e.g., FOXA1, Myc, SF1, and PAX3 with ERα) [Carroll et al., 2005; Cheng et al., 2006; Laganiere et al., 2005b; Lin et al., 2007], while in other cases, they were found in lieu of NR response elements (e.g., AP-1 with ERα) [Carroll et al., 2006; Kininis et al., 2007]. This latter result provides genomic support for the existence of a tethering mechanism in vivo. Gene-specific ChIP assays have confirmed the binding of the transcription factors to their cognate elements, while RNAi-mediated knockdown and mutagenesis of the binding element has demonstrated a role for these transcription factors in ligand-mediated signaling by NRs [Carroll et al., 2005; Carroll et al., 2006; Cheng et al., 2006; Dufour et al., 2007; Kininis et al., 2007; Laganiere et al., 2005b; Lin et al., 2007; Wang et al., 2007]. Collectively, the combination of bioinformatic sequence analyses and experimental validation has greatly extended our understanding of NR binding to DNA and associated crosstalk with other signaling pathways.
Connecting NR binding to gene expression through RNA Pol II
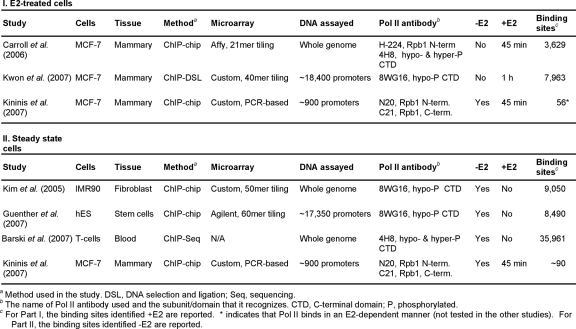
Although the mechanisms of NR binding to cognate binding sites have been well characterized, the underlying mechanisms connecting NR functions to Pol II activity at target promoters are less well understood. Gene-specific studies examining ligand-dependent changes in Pol II occupancy at a limited set of NR-regulated promoters have provided a view of Pol II regulation by NRs that is usually assumed to be true for all NR-regulated genes. In this “classical” view, the binding of NRs and associated coregulators to enhancers ultimately regulates the recruitment of Pol II and general transcription factors to target promoters, thus modulating the expression of the associated genes [Dilworth and Chambon, 2001; McKenna et al., 1999; Metivier et al., 2006; Orphanides et al., 1996] (Figure 1). To examine the generality of this view, several studies have used ligand-treated cells to examine the localization of Pol II to NR-regulated genes using ChIP-chip [Carroll et al., 2006; Kininis et al., 2007; Kwon et al., 2007]. These analyses, summarized in Table 4, have addressed two important questions: (1) where does Pol II bind and (2) how are Pol II binding and activity regulated by NR signaling.
Figure 1. Transcriptional regulation by nuclear receptors.
Transcriptional regulation by NRs is a multistep process involving: (1) the binding of liganded NRs to promoter-proximal and promoter-distal enhancers in conjunction with other transcription factors (e.g., AP1, Sp1 and FOXA1), (2) the ligand-dependent recruitment and actions of coregulators (coactivators and corepressors) to modify chromatin and associated factors, and (3) the regulation of Pol II binding or the activation of preloaded Pol II at target promoters.
Table 4. Genomic analyses of RNA polymerase II localization in human cell lines.
Table summarizing the similarities and differences among genomic analyses of RNA polymerase II localization in E2-treated (I) and steady-state (II) human cells.
Pol II binding at promoters and distal enhancers
As expected, genome-wide studies showed that in E2-treated cells, Pol II was localized to the proximal promoter regions of most E2-stimulated genes [Carroll et al., 2006; Kininis et al., 2007; Kwon et al., 2007]. In some cases, however, Pol II was also bound to distal ERα enhancers [Carroll et al., 2006; Kininis et al., 2007; Kwon et al., 2007]. Based on gene-specific studies with ER- and AR-regulated genes, two models have been proposed for the actions of Pol II at distal enhancers. In the first, Pol II is thought to track from a promoter-distal enhancer to the transcriptional start site (TSS) upon treatment with ligand [Louie et al., 2003; Wang et al., 2005]. In the second, enhancer-bound Pol II associates with the TSS through chromosomal looping [Carroll et al., 2005]. Although these suggestions are intriguing, it is difficult to eliminate the possibility that the Pol II detected at distal enhancers results from (1) crosslinking of the enhancers with Pol II-bound promoters (as opposed to the specific association of Pol II with the enhancer) or (2) the presence of unannotated TSSs throughout the genome [Birney et al., 2007; Hatzis and Talianidis, 2002]. Furthermore, the analysis of Pol II localization only after, but not before, treatment with NR ligands in many of the available studies increases the difficulty in assessing the ligand-dependent effects of NR signaling on Pol II function (see Table 4, Part I).
Regulation of Pol II binding and activity by NR signaling
As noted above, in the “classical” view of NR-regulated transcription, the binding of NRs and coregulators to enhancers regulates the ligand-dependent recruitment of Pol II to target promoters and modulation of gene expression [Dilworth and Chambon, 2001; McKenna et al., 1999; Metivier et al., 2006; Orphanides et al., 1996]. However, recent genomic analyses have indicated that stable Pol II-containing complexes exist at the promoters of many unexpressed genes prior to activation (Table 4, Part II) [Barski et al., 2007; Guenther et al., 2007; Kim et al., 2005; Radonjic et al., 2005]. Accordingly, a recent genomic analysis of Pol II occupancy in both the presence and absence of E2 identified E2-stimulated genes with "preloaded" Pol II at their promoters prior to gene activation [Kininis et al., 2007]. For these genes, E2 signaling stimulates the phosphorylation, but not the recruitment, of Pol II, promoting transcription elongation through the gene rather than transcription initiation. In this regard, NRs, including ERα, have been shown to interact with proteins that regulate Pol II phosphorylation and activity [Aiyar et al., 2004; Kinyamu and Archer, 2007; Wittmann et al., 2005]. Although the generality of this mechanism remains to be determined, it is intriguing to suggest that, in some cases, NRs may control gene expression by regulating the activity, and not the recruitment, of Pol II at target promoters, as shown for other DNA-bound transcription factors [Saunders et al., 2006] (Figure 1).
Defining the direct target genes of NR signaling
As the first genomic analyses of NR-regulated gene expression showed, ligand-dependent regulation of gene expression itself is not sufficient to distinguish between direct and secondary targets of NR signaling. Over the past few years, genomic studies have used various criteria to define the direct target genes of NR signaling, including: (1) regulation of gene expression by short treatments with ligand [Frasor et al., 2003; Lin et al., 2004]; (2) dependence of gene regulation on the receptors, as determined by receptor antagonists and siRNA-mediated knockdown of NRs [Lin et al., 2004; Wang et al., 2004]; (3) identification of NR binding sites at candidate promoters [Kwon et al., 2007; Lin et al., 2007]; (4) loss of ligand-regulated gene expression after co-treatment with protein translation inhibitors [Lin et al., 2004; Rogatsky et al., 2003]; and (5) evidence for changes in Pol II promoter occupancy (an early step in transcriptional regulation) prior to changes in mRNA accumulation [Kininis et al., 2007]. Although each of the above criteria alone is not sufficient, their use in combination can provide a powerful means for determining the direct genomic effects of NRs and their ligands on gene expression.
Genomic analyses of coregulator localization and chromatin modifications
After binding to their genomic targets, NRs mediate their ligand-dependent actions by recruiting positive or negative coregulators. These coregulators, which are shared by various other DNA-binding transcription factors, include: (1) histone- and factor-modifying enzymes, such as the histone acetyltransferase p300 and the histone demethylase LSD1, and (2) bridging factors, such as the steroid receptor coactivators (SRCs), which function to recruit the histone- and factor-modifying enzymes to ligand-bound receptors [Jepsen and Rosenfeld, 2002; Lonard and O'Malley, 2006; McKenna et al., 1999; Perissi and Rosenfeld, 2005]. Although the ligand-dependent recruitment and release of NR coregulators has been examined in detail for a number of genes, the localization of NR coregulators on a global scale has not been studied extensively.
The available genomic localization studies for NR coregulators, including SRCs, p300/CBP, and LSD1, have provided an initial view of the global function of these factors with respect to NR-dependent signaling. All of the aforementioned NR coregulators were found to bind to both promoter-proximal and promoter-distal regions, mirroring the binding patterns observed for NRs [Gamble and Kraus, 2007; Garcia-Bassets et al., 2007; Heintzman et al., 2007; Kininis et al., 2007; Kwon et al., 2007; Labhart et al., 2005]. In recent studies, nearly all of the SRC-bound sites were also occupied by ERα, suggesting a strong link between the recruitment of coregulators and the binding of NRs to sites across the genome [Kininis et al., 2007; Labhart et al., 2005]. A given NR coregulator, however, is not located at all NR binding sites, suggesting that NRs recruit a variety of coregulators in a target gene-specific manner [Garcia-Bassets et al., 2007; Kininis et al., 2007]. Accordingly, the correlation between ERα and SRC recruitment held only for E2-stimulated genes, and no SRC was detected at the promoters of E2-repressed genes [Kininis et al., 2007]. Further studies are necessary to understand the mechanisms regulating the selective recruitment of coregulators to some, but not other, NR binding sites in the same cell.
In addition to studying the localization of histone-modifying coregulators, several studies have examined the state of chromatin, as indicated by histone modifications, at promoters of NR-regulated genes. Histone modifications, such as acetylation and methylation, can regulate NR-mediated transcription by creating (1) a chromatin landscape more or less favorable to gene expression and (2) binding sites on histones for regulatory proteins, with specific modifications specifying transcriptional activation or transcriptional repression [Fischle et al., 2003; Jenuwein and Allis, 2001; Seet et al., 2006; Shogren-Knaak et al., 2006]. Acetylation of histones, primarily H3 acetylated at lysines 9 and 14 (AcH3K9/14), was observed at both promoter-proximal and promoter-distal ERα binding enhancers [Kininis et al., 2007; Kwon et al., 2007]. The ligand-dependent changes in histone acetylation at the promoters of E2-stimulated or -repressed genes correlated with Pol II recruitment or release, respectively [Kininis et al., 2007]. In contrast to histone acetylation, histone methylation showed a more complicated pattern. While some histone methylation marks for active genes (e.g., H3 mono- and di-methylated at lysine 4 ; H3K4me1 and H3K4me2) were detected at both promoter-proximal and promoter-distal enhancers, other marks for active genes (e.g., H3 tri-methylated at lysine 4; H3K4me3) were found exclusively at promoter-proximal NR binding sites [Kwon et al., 2007], as reported for other DNA-bound transcription factors [Barski et al., 2007; Guenther et al., 2007; Heintzman et al., 2007]. Furthermore, histone methylation marks previously associated with inactive genes (e.g., H3 tri-methylated at lysine 9 and H3 di-methylated at lysine 79; H3K9me3 and H3K79me2, respectively) were enriched in some NR-regulated genes after gene activation, suggesting that the role of these modifications is more complicated than previously thought [Kwon et al., 2007]. Interestingly, the ratio of AcH3K9 to H3K9me2 at promoters was shown to be a good marker for ERα recruitment and E2-dependent regulation of gene expression [Cheng et al., 2006]. Collectively, these results suggest that NR signaling regulates the chromatin state of its genomic targets through histone modification. Certain modifications, however, have distinct gene-specific roles and their effects on gene expression cannot be generalized [Berger, 2007].
Summary and future perspectives
Recent genomic analyses of transcription factor binding, histone modification, and gene expression have provided a global view of transcriptional regulation by NRs, which complements the existing literature of gene-specific studies. These genomic analyses have revealed some common themes for the molecular regulation of gene expression by NRs. These themes include: (1) direct or indirect binding of NRs at promoter-proximal and promoter-distal enhancers in conjunction with other transcription factors (e.g., AP-1, Sp1 and FOXA1), (2) NR-dependent recruitment of coregulators and subsequent modification of histones at both enhancers and promoters, and (3) ultimately, NR-dependent regulation of the recruitment of Pol II or the activity of preloaded Pol II at the promoters of target genes. This regulation may be facilitated by chromosomal looping and Pol II tracking, which can promote the enhancer-promoter communication. Overall, the direct targets of NR signaling seem to be limited to a subset of all the genes regulated by NR ligands, with the rest likely to be regulated through secondary effects. In addition, NR-dependent gene regulation occurs in a highly cell type- and promoter-specific manner, indicating the complexity of transcriptional regulation by NRs and the value of genomic analyses in identifying commonly shared patterns. Despite their differences, NRs share common themes in their patterns of localization and transcriptional regulation in the human genome. Further studies are needed to confirm the generality of these patterns and examine the associated mechanisms in more detail.
Although the global picture of NR-mediated transcription provided by the studies described herein has advanced our understanding of NR-dependent transcription, many questions remain unanswered and new ones have emerged from the wealth of available genomic data. One of the key challenges in the NR field today is to determine the in vivo functionality of NR binding sites that have been identified in genomic analyses, especially with respect to enhancer-promoter interactions and cell type-specificity. A common theme from the numerous available chromatin- and transcription-related genome-wide factor localization analyses is that the binding of a factor to a specific site in the genome does not always correlate in an obvious way with a functional outcome for a specific target gene (as determined, for example, by RNAi-mediated knockdown of the relevant factor, coupled with gene-specific expression analyses). In some cases, a particular binding event may only have a detectable functional outcome in a particular cell type. Thus, validation of the functionality of individual NR binding sites in vivo is essential (e.g., by mutating the associated DNA elements in their native genomic and chromatin environment and examining the outcome in cells and animals).
A number of other areas of NR biology will benefit from additional genomic analyses. For example, ongoing efforts to map long-range interactions between distal NR binding sites and target promoters on a genomic scale should greatly facilitate our understanding of target genes regulated by NR binding. Moreover, additional genomic studies of NR coactivator and corepressor localization will help to provide a broader picture of the ligand-dependent effects of NR binding to genomic sites. Furthermore, genomic analyses of factors involved in the tethering of NRs to target enhancers (e.g., such as AP-1 and NF-κB) will provide new insights into this poorly understood, but physiologically important, mechanism. Finally, additional global analyses of Pol II localization and activity are needed to determine the generality of Pol II recruitment and Pol II activation at NR-regulated genes. Collectively, genomic analyses of gene expression, factor localization, and target DNA sequences provide a new set of tools for examining the generality of previously studied mechanisms, as well as identifying new mechanisms by which NRs mediate their physiologically important actions.
Acknowledgments
The authors would like to thank the following members of the Kraus lab for comments and suggestions on this review: Kris Frizzell, Matt Gamble, Nina Heldring, Gary Isaacs, and Raga Krishnakumar. The authors’ research is supported in part by grants from the NIH/NIDDK (to W.L.K.) and the Department of Defense Breast Cancer Research Program (to M.K).
Abbreviations
- AcH
acetylated histones H3/H4
- AcH3K9/14
H3 acetylated at lysines 9 and 14
- AP-1
activator protein-1
- AR
androgen receptor
- CBP
CREB binding protein
- ChIP
chromatin immunoprecipitation
- CREB
cAMP response element binding protein
- DSL
DNA selection and ligation
- E2
estradiol
- ERα
estrogen receptor α
- ERE
estrogen response elements
- ERR
estrogen-related receptor
- GR
glucocorticoid receptor
- H3K4me1
H3 mono-methylated at lysine 4
- H3K4me2
H3 di-methylated at lysine 4
- H3K4me3
H3 tri-methylated at lysine 4
- H3K9me3
H3 tri-methylated at lysine 9
- H3K79me2
H3 di-methylated at lysine 79
- NR
nuclear receptor
- PET
paired-end tag
- Pol II
RNA polymerase II
- qPCR
quantitative real-time polymerase chain reaction
- SAGE
serial analysis of gene expression
- SRC
steroid receptor coactivator
- TSS
transcription start site
- VDR
vitamin D receptor
References
- Acevedo M. L., Kraus W. L. Transcriptional activation by nuclear receptors. Essays Biochem. 2004;40:73–88. doi: 10.1042/bse0400073. [DOI] [PubMed] [Google Scholar]
- Aiyar S. E., Sun J. L., Blair A. L., Moskaluk C. A., Lu Y. Z., Ye Q. N., Yamaguchi Y., Mukherjee A., Ren D. M., Handa H., Li R. Attenuation of estrogen receptor α-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 2004;18:2134–46. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Berger S. L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Birney E., Stamatoyannopoulos J. A., Dutta A., Guigo R., Gingeras T. R., Margulies E. H., Weng Z., Snyder M., Dermitzakis E. T., Thurman R. E., Kuehn M. S., Taylor C. M., Neph S., Koch C. M., Asthana S., Malhotra A., Adzhubei I., Greenbaum J. A., Andrews R. M., Flicek P., Boyle P. J., Cao H., Carter N. P., Clelland G. K., Davis S., Day N., Dhami P., Dillon S. C., Dorschner M. O., Fiegler H., Giresi P. G., Goldy J., Hawrylycz M., Haydock A., Humbert R., James K. D., Johnson B. E., Johnson E. M., Frum T. T., Rosenzweig E. R., Karnani N., Lee K., Lefebvre G. C., Navas P. A., Neri F., Parker S. C., Sabo P. J., Sandstrom R., Shafer A., Vetrie D., Weaver M., Wilcox S., Yu M., Collins F. S., Dekker J., Lieb J. D., Tullius T. D., Crawford G. E., Sunyaev S., Noble W. S., Dunham I., Denoeud F., Reymond A., Kapranov P., Rozowsky J., Zheng D., Castelo R., Frankish A., Harrow J., Ghosh S., Sandelin A., Hofacker I. L., Baertsch R., Keefe D., Dike S., Cheng J., Hirsch H. A., Sekinger E. A., Lagarde J., Abril J. F., Shahab A., Flamm C., Fried C., Hackermuller J., Hertel J., Lindemeyer M., Missal K., Tanzer A., Washietl S., Korbel J., Emanuelsson O., Pedersen J. S., Holroyd N., Taylor R., Swarbreck D., Matthews N., Dickson M. C., Thomas D. J., Weirauch M. T., Gilbert J., Drenkow J., Bell I., Zhao X., Srinivasan K. G., Sung W. K., Ooi H. S., Chiu K. P., Foissac S., Alioto T., Brent M., Pachter L., Tress M. L., Valencia A., Choo S. W., Choo C. Y., Ucla C., Manzano C., Wyss C., Cheung E., Clark T. G., Brown J. B., Ganesh M., Patel S., Tammana H., Chrast J., Henrichsen C. N., Kai C., Kawai J., Nagalakshmi U., Wu J., Lian Z., Lian J., Newburger P., Zhang X., Bickel P., Mattick J. S., Carninci P., Hayashizaki Y., Weissman S., Hubbard T., Myers R. M., Rogers J., Stadler P. F., Lowe T. M., Wei C. L., Ruan Y., Struhl K., Gerstein M., Antonarakis S. E., Fu Y., Green E. D., Karaoz U., Siepel A., Taylor J., Liefer L. A., Wetterstrand K. A., Good P. J., Feingold E. A., Guyer M. S., Cooper G. M., Asimenos G., Dewey C. N., Hou M., Nikolaev S., Montoya-Burgos J. I., Loytynoja A., Whelan S., Pardi F., Massingham T., Huang H., Zhang N. R., Holmes I., Mullikin J. C., Ureta-Vidal A., Paten B., Seringhaus M., Church D., Rosenbloom K., Kent W. J., Stone E. A., Batzoglou S., Goldman N., Hardison R. C., Haussler D., Miller W., Sidow A., Trinklein N. D., Zhang Z. D., Barrera L., Stuart R., King D. C., Ameur A., Enroth S., Bieda M. C., Kim J., Bhinge A. A., Jiang N., Liu J., Yao F., Vega V. B., Lee C. W., Ng P., Shahab A., Yang A., Moqtaderi Z., Zhu Z., Xu X., Squazzo S., Oberley M. J., Inman D., Singer M. A., Richmond T. A., Munn K. J., Rada-Iglesias A., Wallerman O., Komorowski J., Fowler J. C., Couttet P., Bruce A. W., Dovey O. M., Ellis P. D., Langford C. F., Nix D. A., Euskirchen G., Hartman S., Urban A. E., Kraus P., Van Calcar S., Heintzman N., Kim T. H., Wang K., Qu C., Hon G., Luna R., Glass C. K., Rosenfeld M. G., Aldred S. F., Cooper S. J., Halees A., Lin J. M., Shulha H. P., Zhang X., Xu M., Haidar J. N., Yu Y., Ruan Y., Iyer V. R., Green R. D., Wadelius C., Farnham P. J., Ren B., Harte R. A., Hinrichs A. S., Trumbower H., Clawson H., Hillman-Jackson J., Zweig A. S., Smith K., Thakkapallayil A., Barber G., Kuhn R. M., Karolchik D., Armengol L., Bird C. P., de Bakker P. I., Kern A. D., Lopez-Bigas N., Martin J. D., Stranger B. E., Woodroffe A., Davydov E., Dimas A., Eyras E., Hallgrimsdottir I. B., Huppert J., Zody M. C., Abecasis G. R., Estivill X., Bouffard G. G., Guan X., Hansen N. F., Idol J. R., Maduro V. V., Maskeri B., McDowell J. C., Park M., Thomas P. J., Young A. C., Blakesley R. W., Muzny D. M., Sodergren E., Wheeler D. A., Worley K. C., Jiang H., Weinstock G. M., Gibbs R. A., Graves T., Fulton R., Mardis E. R., Wilson R. K., Clamp M., Cuff J., Gnerre S., Jaffe D. B., Chang J. L., Lindblad-Toh K., Lander E. S., Koriabine M., Nefedov M., Osoegawa K., Yoshinaga Y., Zhu B., de Jong P. J. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton E. C., So A. Y., Chaivorapol C., Haqq C. M., Li H., Yamamoto K. R. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–17. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J. S., Liu X. S., Brodsky A. S., Li W., Meyer C. A., Szary A. J., Eeckhoute J., Shao W., Hestermann E. V., Geistlinger T. R., Fox E. A., Silver P. A., Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carroll J. S., Brown M. Estrogen receptor target gene: an evolving concept. Mol Endocrinol. 2006a;20:1707–14. doi: 10.1210/me.2005-0334. [DOI] [PubMed] [Google Scholar]
- Carroll J. S., Meyer C. A., Song J., Li W., Geistlinger T. R., Eeckhoute J., Brodsky A. S., Keeton E. K., Fertuck K. C., Hall G. F., Wang Q., Bekiranov S., Sementchenko V., Fox E. A., Silver P. A., Gingeras T. R., Liu X. S., Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006b;38:1289–97. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Cheng A. S., Jin V. X., Fan M., Smith L. T., Liyanarachchi S., Yan P. S., Leu Y. W., Chan M. W., Plass C., Nephew K. P., Davuluri R. V., Huang T. H. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-α responsive promoters. Mol Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Coser K. R., Chesnes J., Hur J., Ray S., Isselbacher K. J., Shioda T. Global analysis of ligand sensitivity of estrogen inducible and suppressible genes in MCF7/BUS breast cancer cells by DNA microarray. Proc Natl Acad Sci U S A. 2003;100:13994–9. doi: 10.1073/pnas.2235866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo D. G., Kim H. T., Hilsenbeck S., Cuba V., Tsimelzon A., Brown P. H. Global gene expression analysis of estrogen receptor transcription factor cross talk in breast cancer: identification of estrogen-induced/activator protein-1-dependent genes. Mol Endocrinol. 2005;19:362–78. doi: 10.1210/me.2004-0267. [DOI] [PubMed] [Google Scholar]
- Dilworth F. J., Chambon P. Nuclear receptors coordinate the activities of chromatin remodeling complexes and coactivators to facilitate initiation of transcription. Oncogene. 2001;20:3047–54. doi: 10.1038/sj.onc.1204329. [DOI] [PubMed] [Google Scholar]
- Dufour C. R., Wilson B. J., Huss J. M., Kelly D. P., Alaynick W. A., Downes M., Evans R. M., Blanchette M., Giguere V. Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRalpha and γ. Cell Metab. 2007;5:345–56. doi: 10.1016/j.cmet.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Fischle W., Wang Y., Allis C. D. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–83. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Frasor J., Danes J. M., Komm B., Chang K. C., Lyttle C. R., Katzenellenbogen B. S. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–74. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- Frasor J., Stossi F., Danes J. M., Komm B., Lyttle C. R., Katzenellenbogen B. S. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004;64:1522–33. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- Gamble M. J., Kraus W. L. Visualizing the histone code on LSD1. Cell. 2007;128:433–4. doi: 10.1016/j.cell.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Gao H., Falt S., Sandelin A., Gustafsson J. A., Dahlman-Wright K. Genome-Wide Identification of Estrogen Receptor {α}-Binding Sites in Mouse Liver. Mol Endocrinol. 2008;22:10–22. doi: 10.1210/me.2007-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bassets I., Kwon Y. S., Telese F., Prefontaine G. G., Hutt K. R., Cheng C. S., Ju B. G., Ohgi K. A., Wang J., Escoubet-Lozach L., Rose D. W., Glass C. K., Fu X. D., Rosenfeld M. G. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–18. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C. K., Rosenfeld M. G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzis P., Talianidis I. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol Cell. 2002;10:1467–77. doi: 10.1016/s1097-2765(02)00786-4. [DOI] [PubMed] [Google Scholar]
- Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C. D. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jepsen K., Rosenfeld M. G. Biological roles and mechanistic actions of co-repressor complexes. J Cell Sci. 2002;115:689–98. doi: 10.1242/jcs.115.4.689. [DOI] [PubMed] [Google Scholar]
- Jin V. X., Leu Y. W., Liyanarachchi S., Sun H., Fan M., Nephew K. P., Huang T. H., Davuluri R. V. Identifying estrogen receptor α target genes using integrated computational genomics and chromatin immunoprecipitation microarray. Nucleic Acids Res. 2004;32:6627–35. doi: 10.1093/nar/gkh1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kian Tee M., Rogatsky I., Tzagarakis-Foster C., Cvoro A., An J., Christy R. J., Yamamoto K. R., Leitman D. C. Estradiol and selective estrogen receptor modulators differentially regulate target genes with estrogen receptors α and β. Mol Biol Cell. 2004;15:1262–72. doi: 10.1091/mbc.E03-06-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H., Barrera L. O., Zheng M., Qu C., Singer M. A., Richmond T. A., Wu Y., Green R. D., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–80. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H., Ren B. Genome-wide analysis of protein-DNA interactions. Annu Rev Genomics Hum Genet. 2006;7:81–102. doi: 10.1146/annurev.genom.7.080505.115634. [DOI] [PubMed] [Google Scholar]
- Kininis M., Chen B. S., Diehl A. G., Isaacs G. D., Zhang T., Siepel A. C., Clark A. G., Kraus W. L. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol. 2007;27:5090–104. doi: 10.1128/MCB.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinyamu H. K., Archer T. K. Proteasome activity modulates chromatin modifications and RNA polymerase II phosphorylation to enhance glucocorticoid receptor-mediated transcription. Mol Cell Biol. 2007;27:4891–904. doi: 10.1128/MCB.02162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Hitpass L., Ryffel G. U., Heitlinger E., Cato A. C. A 13 bp palindrome is a functional estrogen responsive element and interacts specifically with estrogen receptor. Nucleic Acids Res. 1988;16:647–63. doi: 10.1093/nar/16.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge C. M., Jernigan S. C., Mattingly K. A., Risinger K. E., Zhang J. Estrogen response element-dependent regulation of transcriptional activation of estrogen receptors α and β by coactivators and corepressors. J Mol Endocrinol. 2004;33:387–410. doi: 10.1677/jme.1.01541. [DOI] [PubMed] [Google Scholar]
- Kraus W. L., Wong J. Nuclear receptor-dependent transcription with chromatin. Is it all about enzymes? Eur J Biochem. 2002;269:2275–83. doi: 10.1046/j.1432-1033.2002.02889.x. [DOI] [PubMed] [Google Scholar]
- Kurokawa R., Soderstrom M., Horlein A., Halachmi S., Brown M., Rosenfeld M. G., Glass C. K. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–4. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- Kushner P. J., Agard D. A., Greene G. L., Scanlan T. S., Shiau A. K., Uht R. M., Webb P. Estrogen receptor pathways to AP-1. J Steroid Biochem Mol Biol. 2000;74:311–7. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- Kwon Y. S., Garcia-Bassets I., Hutt K. R., Cheng C. S., Jin M., Liu D., Benner C., Wang D., Ye Z., Bibikova M., Fan J. B., Duan L., Glass C. K., Rosenfeld M. G., Fu X. D. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor α-binding program on human gene promoters. Proc Natl Acad Sci U S A. 2007;104:4852–7. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Karmakar S., Salicru E. M., Egan B. S., Alexiadis V., O'Malley B. W., Smith C. L. Identification of target genes in breast cancer cells directly regulated by the SRC-3/AIB1 coactivator. Proc Natl Acad Sci U S A. 2005;102:1339–44. doi: 10.1073/pnas.0409578102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganiere J., Deblois G., Giguere V. Functional genomics identifies a mechanism for estrogen activation of the retinoic acid receptor alpha1 gene in breast cancer cells. Mol Endocrinol. 2005a;19:1584–92. doi: 10.1210/me.2005-0040. [DOI] [PubMed] [Google Scholar]
- Laganiere J., Deblois G., Lefebvre C., Bataille A. R., Robert F., Giguere V. From the Cover: Location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci U S A. 2005b;102:11651–6. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. H., Olson P., Evans R. M. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 2003;144:2201–7. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]
- Lefstin J. A., Yamamoto K. R. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–8. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- Levenson A. S., Svoboda K. M., Pease K. M., Kaiser S. A., Chen B., Simons L. A., Jovanovic B. D., Dyck P. A., Jordan V. C. Gene expression profiles with activation of the estrogen receptor α-selective estrogen receptor modulator complex in breast cancer cells expressing wild-type estrogen receptor. Cancer Res. 2002;62:4419–26. [PubMed] [Google Scholar]
- Lin C. Y., Strom A., Vega V. B., Kong S. L., Yeo A. L., Thomsen J. S., Chan W. C., Doray B., Bangarusamy D. K., Ramasamy A., Vergara L. A., Tang S., Chong A., Bajic V. B., Miller L. D., Gustafsson J. A., Liu E. T. Discovery of estrogen receptor α target genes and response elements in breast tumor cells. Genome Biol. 2004;5:R66. doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. Y., Vega V. B., Thomsen J. S., Zhang T., Kong S. L., Xie M., Chiu K. P., Lipovich L., Barnett D. H., Stossi F., Yeo A., George J., Kuznetsov V. A., Lee Y. K., Charn T. H., Palanisamy N., Miller L. D., Cheung E., Katzenellenbogen B. S., Ruan Y., Bourque G., Wei C. L., Liu E. T. Whole-genome cartography of estrogen receptor α binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard D. M., O'Malley B. W. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–4. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Louie M. C., Yang H. Q., Ma A. H., Xu W., Zou J. X., Kung H. J., Chen H. W. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor-p160 coactivator complex. Proc Natl Acad Sci U S A. 2003;100:2226–30. doi: 10.1073/pnas.0437824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven M. A., Likhite V. S., Choi I., Nardulli A. M. Estrogen response elements alter coactivator recruitment through allosteric modulation of estrogen receptor β conformation. J Biol Chem. 2001;276:45282–8. doi: 10.1074/jbc.M106211200. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna N. J., Lanz R. B., O'Malley B. W. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–44. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- Metivier R., Reid G., Gannon F. Transcription in four dimensions: nuclear receptor-directed initiation of gene expression. EMBO Rep. 2006;7:161–7. doi: 10.1038/sj.embor.7400626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe D. G., Getz B. J., Johnsen S. A., Riggs B. L., Khosla S., Spelsberg T. C. Estrogen receptor isoform-specific regulation of endogenous gene expression in human osteoblastic cell lines expressing either ERalpha or ERbeta. J Cell Biochem. 2003;90:315–26. doi: 10.1002/jcb.10633. [DOI] [PubMed] [Google Scholar]
- Naar A. M., Lemon B. D., Tjian R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- O'Lone R., Frith M. C., Karlsson E. K., Hansen U. Genomic targets of nuclear estrogen receptors. Mol Endocrinol. 2004;18:1859–75. doi: 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–51. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Lagrange T., Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–83. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Perissi V., Rosenfeld M. G. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;6:542–54. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- Pike J. W., Meyer M. B., Watanuki M., Kim S., Zella L. A., Fretz J. A., Yamazaki M., Shevde N. K. Perspectives on mechanisms of gene regulation by 1,25-dihydroxyvitamin D3 and its receptor. J Steroid Biochem Mol Biol. 2007;103:389–95. doi: 10.1016/j.jsbmb.2006.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn D. I., Henshall S. M., Sutherland R. L. Molecular markers of prostate cancer outcome. Eur J Cancer. 2005;41:858–87. doi: 10.1016/j.ejca.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Radonjic M., Andrau J. C., Lijnzaad P., Kemmeren P., Kockelkorn T. T., van Leenen D., van Berkum N. L., Holstege F. C. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol Cell. 2005;18:171–83. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Rae J. M., Johnson M. D., Scheys J. O., Cordero K. E., Larios J. M., Lippman M. E. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat. 2005;92:141–9. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M., Carpentier A. S., Duffraisse M., Laudet V. How many nuclear hormone receptors are there in the human genome? Trends Genet. 2001;17:554–6. doi: 10.1016/s0168-9525(01)02417-9. [DOI] [PubMed] [Google Scholar]
- Rogatsky I., Wang J. C., Derynck M. K., Nonaka D. F., Khodabakhsh D. B., Haqq C. M., Darimont B. D., Garabedian M. J., Yamamoto K. R. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci U S A. 2003;100:13845–50. doi: 10.1073/pnas.2336092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A., Core L. J., Lis J. T. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–67. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Seet B. T., Dikic I., Zhou M. M., Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–83. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- Shang Y., Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–8. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., Peterson C. L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- So A. Y., Chaivorapol C., Bolton E. C., Li H., Yamamoto K. R. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender J. D., Frasor J., Komm B., Chang K. C., Kraus W. L., Katzenellenbogen B. S. Estrogen-regulated gene networks in human breast cancer cells: involvement of E2F1 in the regulation of cell proliferation. Mol Endocrinol. 2007;21:2112–23. doi: 10.1210/me.2006-0474. [DOI] [PubMed] [Google Scholar]
- Tan P. K., Downey T. J., Spitznagel E. L., Jr., Xu P., Fu D., Dimitrov D. S., Lempicki R. A., Raaka B. M., Cam M. C. Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res. 2003;31:5676–84. doi: 10.1093/nar/gkg763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M., Miura K., Iwao H., Yamanaka S. Quantitative assessment of DNA microarrays--comparison with Northern blot analyses. Genomics. 2001;71:34–9. doi: 10.1006/geno.2000.6427. [DOI] [PubMed] [Google Scholar]
- Tavera-Mendoza L. E., Mader S., White J. H. Genome-wide approaches for identification of nuclear receptor target genes. Nucl Recept Signal. 2006;4:e018. doi: 10.1621/nrs.04018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H., Holstege F. C. In control: systematic assessment of microarray performance. EMBO Rep. 2004;5:964–9. doi: 10.1038/sj.embor.7400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Li W., Liu X. S., Carroll J. S., Janne O. A., Keeton E. K., Chinnaiyan A. M., Pienta K. J., Brown M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380–92. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C., Derynck M. K., Nonaka D. F., Khodabakhsh D. B., Haqq C., Yamamoto K. R. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci U S A. 2004;101:15603–8. doi: 10.1073/pnas.0407008101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Carroll J. S., Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–42. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- White J. H. Profiling 1,25-dihydroxyvitamin D3-regulated gene expression by microarray analysis. J Steroid Biochem Mol Biol. 2004;89-90:239–44. doi: 10.1016/j.jsbmb.2004.03.074. [DOI] [PubMed] [Google Scholar]
- Wittmann B. M., Fujinaga K., Deng H., Ogba N., Montano M. M. The breast cell growth inhibitor, estrogen down regulated gene 1, modulates a novel functional interaction between estrogen receptor α and transcriptional elongation factor cyclin T1. Oncogene. 2005;24:5576–88. doi: 10.1038/sj.onc.1208728. [DOI] [PubMed] [Google Scholar]