Abstract
The possibility that bacteria may have evolved strategies to overcome host cell apoptosis was explored by using Rickettsia rickettsii, an obligate intracellular Gram-negative bacteria that is the etiologic agent of Rocky Mountain spotted fever. The vascular endothelial cell, the primary target cell during in vivo infection, exhibits no evidence of apoptosis during natural infection and is maintained for a sufficient time to allow replication and cell-to-cell spread prior to eventual death due to necrotic damage. Prior work in our laboratory demonstrated that R. rickettsii infection activates the transcription factor NF-κB and alters expression of several genes under its control. However, when R. rickettsii-induced activation of NF-κB was inhibited, apoptosis of infected but not uninfected endothelial cells rapidly ensued. In addition, human embryonic fibroblasts stably transfected with a superrepressor mutant inhibitory subunit IκB that rendered NF-κB inactivatable also underwent apoptosis when infected, whereas infected wild-type human embryonic fibroblasts survived. R. rickettsii, therefore, appeared to inhibit host cell apoptosis via a mechanism dependent on NF-κB activation. Apoptotic nuclear changes correlated with presence of intracellular organisms and thus this previously unrecognized proapoptotic signal, masked by concomitant NF-κB activation, likely required intracellular infection. Our studies demonstrate that a bacterial organism can exert an antiapoptotic effect, thus modulating the host cell’s apoptotic response to its own advantage by potentially allowing the host cell to remain as a site of infection.
Rickettsia rickettsii is an obligate intracellular bacterial parasite and the causative agent of Rocky Mountain spotted fever. During in vivo infection, the vascular endothelial cell (EC) is the primary target of infection and extensive studies using cultured human EC have documented that the host EC not only sustains damage but also actively responds to intracellular R. rickettsii infection. EC injury, manifested by widespread dilatation of membranes and loss of osmoregulatory control, likely occurs due to oxidant damage (1–6). EC surface adhesiveness to platelets increases (7), von Willebrand factor is released (8), and the pattern of EC gene expression changes early in the course of infection, with expression of both proinflammatory and procoagulant proteins (9–14). In vitro infection of ECs results in activation of members of the NF-κB family of transcription factors (15).
NF-κB is a ubiquitous transcription factor involved in expression of many early response genes (for review, see refs. 16 and 17). Because it exists in a preformed pool within the cytoplasm of eukaryotic cells, NF-κB is readily available for immediate activation in response to environmental stimuli. Nuclear localization signals and DNA-binding domains are masked via interaction with inhibitory proteins providing a mechanism for retention of NF-κB in an inactive pool in the cell cytoplasm. Upon appropriate stimulation, such as by cytokines or growth factors, the inhibitory proteins are phosphorylated, ubiquitinated, and degraded by a pathway involving the multisubunit protease complex known as the proteasome. Such activation is often transient and self-limiting, partly because NF-κB activation results in a rapid increase in expression of the inhibitory subunit IκB. R. rickettsii-induced activation likely requires intracellular infection and displays biphasic kinetics with an early transient phase peaking at 3 hr and a later more sustained phase evident at 18–24 hr (15). It has yet to be determined which cell signaling pathway(s) are involved in R. rickettsii-induced activation.
During the course of studies to determine the role of NF-κB activation in R. rickettsii-induced expression of certain EC genes, it was noted that blocking such activation resulted in extensive EC loss. Concomitant with this observation, NF-κB activation was reported to play a critical role in protection of cells from apoptosis induced by tumor necrosis factor α (TNF-α), radiation, and treatment with chemotherapeutic drugs (18–21). The studies described in this report were designed to explore the possibility that R. rickettsii-induced NF-κB activation protects the host cell from apoptosis during infection and thus is critical for host cell survival. This possibility was explored in the natural target cell, the EC, by using a specific pharmacologic inhibitor of NF-κB and in human embryonic fibroblasts (HEFs) stably transfected with a transdominant negative mutant IκB engineered to render NF-κB inactivatable (21). This study demonstrates that inhibition of apoptosis functions during the course of infection with a bacterial organism and that the mechanism involved requires Rickettsia-induced activation of host cell NF-κB.
MATERIALS AND METHODS
Cell Culture.
Human umbilical vein EC cultures were established as described (22, 23) and cultured in McCoy’s 5a medium (Flow Laboratories), to which 20% fetal bovine serum (FBS), EC growth supplement (50 μg/ml; Collaborative Research), and heparin (100 μg/ml; Sigma) were added. All experiments used ECs at passage 2 and ECs were plated to achieve 80–90% confluence after 4–5 days in culture. Wild-type HEFs were established as described (21, 24) and were cultured in DMEM (GIBCO/BRL/Life Technologies) supplemented with 10% FBS. T24 bladder carcinoma cells (21) were cultured in RPMI 1640 medium (GIBCO/BRL) with 10% FBS. HEFs and T24 cells were used that were stably transfected with a transdominant-negative mutant IκB as described (21). Mutant HEFs were maintained in DMEM in the presence of G418 (800 μg/ml; GIBCO/BRL). Mutant and sham-transfected T24 cells were maintained in RPMI 1640 medium in the presence of G418 (400 μg/ml).
R. rickettsii Infection of Cultured Cell.
Near-confluent cell cultures were infected with R. rickettsii as described (15), by using a seed stock [1–5 × 107 plaque-forming units (pfu)/ml] prepared from infected Vero cells (7). Cells were infected with approximately 6 × 104 pfu/cm2 of cell culture area diluted in culture medium. After a 2-hr incubation at 37°C, the inoculum was removed and cell monolayers were washed three times with culture medium to remove Vero cell debris. Infection was monitored by using ECs plated on Thermanox coverslips (Ted Pella, Reading, CA) and stained by indirect immunofluorescence using a polyclonal anti-R. rickettsii antibody (kindly provided by Theodore Tzianobos, Centers for Disease Control, Atlanta, GA) as described (12, 23), which revealed that 60–80% of ECs were infected at 6 hr. The peptide-aldehyde proteasome inhibitor carbobenzoxylleucinylleucinyl-leucinal-H (MG 132, 50 μM; Peptide Institute, Osaka) and the antioxidant pyrrolidine dithiocarbamate (PDTC, 25 μM; Sigma) were added to EC cultures during infection. Recombinant human TNF-α (Becton Dickinson) was used at a final concentration of 0.5 ng/ml.
Nuclear Extraction and Gel Shift Analysis.
Approximately 5 × 106 cells were treated per experimental condition, nuclei were harvested, and nuclear proteins were extracted as described (15). Protein concentration in nuclear extracts ranged from 0.5 to 2 μg/μl as measured with the Bradford assay. HeLa cell nuclear extracts containing activated NF-κB (Promega) were used as controls for all gel shift studies. Gel shift assays were performed by using the gel shift assay system (Promega) by following the manufacturer’s instructions. Two micrograms of nuclear protein was added per gel shift reaction. The double-stranded consensus NF-κB oligonucleotide 5′-AGTTGAGGGGACTTTCCCAGGC-3′ was end-labeled using T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol; 10 mCi/ml; 1 Ci = 37 GBq; DuPont/NEN) by the manufacturer’s instructions (gel shift assay system, Promega). Competition studies were performed by addition of a 10-fold molar excess of unlabeled oligonucleotide probe to the gel shift reaction mixture before addition of labeled probe. Reactions were analyzed on 4% nondenaturing low-ionic-strength polyacrylamide gels prepared in 0.5× TBE (89 mM Tris⋅HCl, pH 8.0/89 mM boric acid/2 mM EDTA); 0.5× TBE was the running buffer. Gels were electrophoresed at 100 V for 2 hr and subjected to radiographic exposure for 24–65 hr.
In Situ Detection of DNA Fragmentation.
DNA fragmentation characteristic of apoptosis in cultured cells was monitored by terminal deoxynucleotidyltransferase (terminal deoxynucleotidyltransferase-mediated UTP end labeling, TUNEL) staining using the Apoptag kit (Oncor) by following manufacturer’s instructions. Cells cultured on plastic coverslips were fixed in 3.7% formaldehyde diluted in PBS for 20 min, postfixed in ethanol/acetic acid, 2:1 (vol/vol) at −20°C for 5 min, then rinsed twice in PBS. Coverslips were then incubated with terminal deoxynucleotidyltransferase and digoxigenin-dUTP and stained with fluorescein isothiocyanate-conjugated anti-digoxigenin antibody by the manufacturer’s instructions and then mounted (cells facing up) on glass microscope slides with Gelmount (Biomedia, Foster City, CA). Propidium iodide counterstain (3 μg/ml in PBS) was applied and cells were covered with a glass coverslip, the edges were sealed with rubber cement, and slides were stored at −20°C. Staining was viewed with a Nikon Eclipse E800 fluorescent microscope equipped with a dual wavelength filter cube. The percent of apoptotic cells in the populations was determined by counting the number of cells in randomly selected microscope fields with nuclei exhibiting green fluorescence (fluorescein isothiocyanate), indicative of DNA fragmentation, and dividing this by the total number of nuclei. Fifteen hundred to 3,000 cells were scored per experimental condition, and three to five experiments were conducted. Apoptotic change was associated with loss of adhesion only at the later time point (16–18 hr), and nonadherent cells were determined to be essentially 100% apoptotic as shown by TUNEL of nonadherent cell smears. The fraction of nonadherent cells in these samples was determined by comparing total number of cells treated under given conditions (e.g., 16–18 hr with R. rickettsii and MG 132) in a minimum of five randomly selected fields with the number of cells in the matched untreated cell population (e.g., 16–18 hr with R. rickettsii alone). Percent apoptosis was then calculated as follows: percent apoptosis = [(fraction adherent × percent adherent cells exhibiting fluorescein isothiocyanate fluorescence) + (fraction nonadherent cells)] × 100%. Immunofluorescence staining to detect R. rickettsii was performed as described (12) by using a rhodamine-conjugated secondary antibody. Cell nuclei were subsequently stained by incubating in Hoechst dye (1 μg/ml of bis-benzimide in PBS) for 5 min at room temperature and then rinsed twice in PBS. The plastic coverslips were mounted, cells up, between a glass slide and glass coverslip by using Fluoromount (Southern Biotechnology Associates). Photographs were taken with a Nikon H-III 35-mm camera.
Electron Microscopy.
T-25 flasks of ECs were infected for 38 hr with R. rickettsii alone or R. rickettsii with MG 132. At 38 hr, cells were collected and pelleted by centrifugation at 500 × g for 10 min. Cells were pipetted into a 10× vol of 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, swirled, and allowed to fix for 1 hr at room temperature. Cells were washed for four 10-min periods in 0.1 M sodium cacodylate buffer, after which they were pipetted into hematocrit centrifuge tubes, centrifuged, and postfixed for 1 hr with 1% osmium tetroxide in 0.1 M sodium cacodylate buffer. Cell pellets were removed from the tubes, dehydrated in a graded ethanol series, followed by propylene oxide, and embedded in epoxy resin. Thin sections were cut with a diamond knife, stained with uranyl acetate and lead citrate, and photographed on a Zeiss 10C transmission electron microscope.
Statistics.
Standard error of the mean for percent apoptosis in the cell populations was calculated with unpaired two-tailed Student’s t test. χ2 test for independence was used to determine the relationship between intracellular infection and apoptosis.
RESULTS
To determine the role of NF-κB activation in cell survival during infection, ECs were infected in the presence and absence of MG 132, a peptide-aldehyde protease inhibitor that blocks NF-κB activation via its effect on the proteasome (25, 26). ECs infected for 7–18 hr in the presence of MG 132 (50 μM) showed increased numbers of cells that exhibited evidence of nuclear DNA fragmentation (Fig. 1Ab), an early marker of apoptotic change, when compared with ECs infected in the absence of inhibitor (Fig. 1Aa). Presence of the inhibitor alone did not result in apoptosis above background levels (data not shown). The TUNEL staining technique was used, which detects free 3′ hydroxyl DNA ends indicative of DNA fragmentation. ECs were considered TUNEL-positive if they displayed green fluorescence when visualized through a dual wavelength filter, whereas normal nuclei exhibited orange fluorescence emitted by the propidium iodide counterstain. Nuclear condensation and disruption of cell adhesion was typical of TUNEL-positive cells.
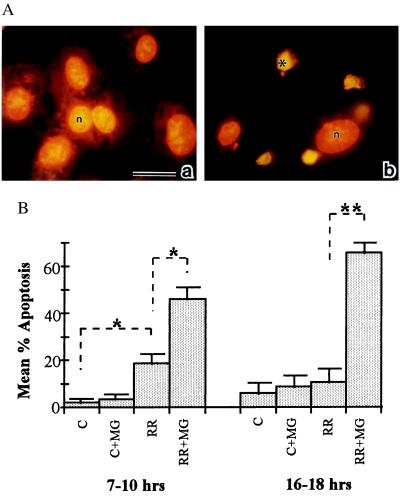
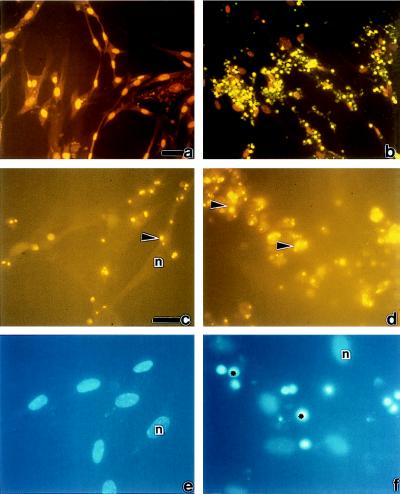
Figure 1.
(A) In situ detection of EC apoptosis during R. rickettsii infection. Infected human umbilical vein ECs were incubated in the presence (b) or absence (a) of the NF-κB inhibitor MG 132. At 18 hr, ECs were fixed and processed for in situ detection of DNA fragmentation by TUNEL and then counterstained with propidium iodide. When visualized under dual wavelength fluorescence, normal nuclei exhibited orange fluorescence (n), whereas apoptotic nuclei displayed green fluorescence and appeared condensed (∗). MG 132 treatment did not induce apoptosis in uninfected ECs (data not shown). (Bar = 20 μm.) (B) The percentages of apoptotic ECs were determined by scoring cells as apoptotic (exhibiting green fluorescence) or normal (exhibiting orange fluorescence) in randomly chosen microscopic fields. Fifteen hundred to 3,000 cells were scored per experimental condition. P values comparing mean percent apoptotic cells between experimental conditions indicated by brackets were determined by Student’s t test and are indicated by asterisks (n = 3 to 5). ∗, P < 0.05; ∗∗, P < 0.00005.
To quantitate apoptotic change in the EC populations, ECs were cultured on coverslips and infected in the presence or absence of MG 132, and the percentage of TUNEL-positive cells was determined by scoring randomly selected microscope fields (Fig. 1B). ECs incubated under control conditions or in the presence of MG 132 alone exhibited a minimal amount of apoptosis. ECs infected in the absence of MG 132 exhibited 19 ± 4% apoptosis, which represented a statistically significant increase when compared with uninfected EC at 7–10 hr. The percent apoptotic cells in infection alone was not significantly different from control cultures at 16–18 hr. In contrast, the percent of apoptotic ECs in the populations infected in the presence of MG 132 averaged 46 ± 5% at 7–10 hr, which represented a statistically significant increase over infection alone. The percent of apoptosis in these populations increased to a mean of 66 ± 4% at 16–18 hr. Inhibition of NF-κB activation with PDTC (25 μM) produced similar results, with an average 75 ± 10% apoptosis (n = 2) observed at 16 hr.
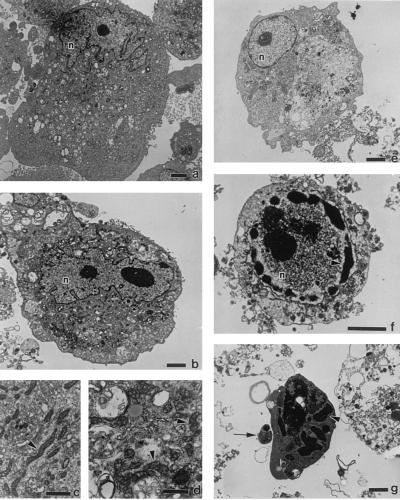
A morphologic study was performed to confirm that cellular changes consistent with apoptosis occurred in NF-κB-inhibited infected cells. ECs were infected for 38 hr in the presence and absence of MG 132 (50 μM), collected, and processed for transmission electron microscopic evaluation. ECs infected in the presence of MG 132 had a spectrum of morphologic changes consistent with both early and advanced stages of apoptosis (Fig. 2). Nuclei in uninfected ECs typically displayed an irregular and convoluted shape (Fig. 2a). ECs with round nuclei and condensed cytoplasm were commonly observed when infection was carried out in the presence of MG 132 and, although not usually seen in apoptosis, may be typical of early apoptotic change in this cell type (Fig. 2e). ECs in more advanced stages of apoptosis had compaction and segregation of chromatin into dense masses, convolution and segmentation of nuclei, and formation of apoptotic bodies (Fig. 2 f and g). In contrast, ECs infected in the absence of MG 132 (Fig. 2b) displayed the intact irregularly shaped nuclei and chromatin distribution typical of cultured EC (Fig. 2a) but exhibited signs of necrotic cell damage including mitochondrial swelling (Fig. 2d) compared with mitochondria in uninfected cells (Fig. 2c).
Figure 2.
Transmission electron micrographs of R. rickettsii-infected ECs. Shown are uninfected ECs (a and c) or ECs infected for 38 hr in the absence (b and d) or presence (e–g) of MG 132. EC nuclei are denoted by n. Arrowheads in c and d point to mitochondria. Arrowheads in f and g point to areas of condensed chromatin; the arrow in g points to an apoptotic body. [Bars = 2.5 μm (a, b, and e–g) and 1 μm (c and d).]
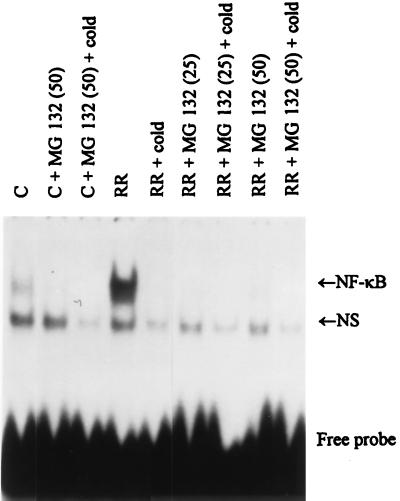
Gel shift analysis was performed to verify that R. rickettsii-induced activation of NF-κB was inhibited by treatment with MG 132 (Fig. 3). Nuclear extracts were prepared from uninfected ECs and from ECs infected in the presence and absence of 25 and 50 μM MG 132, and gel shift analysis was conducted with a 32P-labeled NF-κB oligonucleotide probe. The presence of activated NF-κB in the nuclear extracts results in formation of noncovalent complexes with the labeled DNA probe and causes a retardation in migration of the labeled probe in nondenaturing gels. As reported (15), R. rickettsii infection resulted in activation of EC NF-κB. This activation was completely inhibited with 25 and 50 μM MG 132 (Fig. 3) or 25 μM PDTC (data not shown). MG 132 and PDTC had no effect on levels of activation of the unrelated transcription factor AP-1 (data not shown).
Figure 3.
Effect of MG 132 on R. rickettsii induced NF-κB activation. Gel shift analysis using a 32P-labeled oligonucleotide probe was performed on nuclear extracts prepared from control ECs (C) and ECs infected for 3 hr with R. rickettsii (RR) in the absence and presence of 25 μM (25) or 50 μM (50) MG 132. Analysis of complex formation was performed on 4% nonreducing polyacrylamide gels followed by autoradiography. An R. rickettsii-inducible gel-shifted complex representing interaction of dimeric NF-κB molecules with the labeled probe is indicated in the gel (NF-κB) as is the presence of a nonspecific noninducible gel-shifted complex (NS). Specificity of the NF-κB gel-shifted complexes was demonstrated by inclusion of excess unlabeled oligonucleotide probe (+cold).
During in vitro infection, a heterogenous population of ECs exists with regard to presence of intracellular organisms and the number of organisms per EC (8). Double-label fluorescence staining was used to determine whether an association existed between the presence of intracellular infection and induction of apoptosis (data not shown). After infection in the presence and absence of MG 132, ECs were fixed and stained by indirect immunofluorescence with anti-R. rickettsii antibody and a rhodamine-labeled secondary antibody, and the nuclei were stained with Hoechst dye. One hundred to 300 ECs per condition were then scored for presence or absence of intracellular R. rickettsii (ECs were counted as infected if they contained three or more rickettsia organisms) and the same cells were scored for evidence of apoptotic nuclear change, i.e., decreased nuclear diameter and increased intensity of Hoechst staining. A χ2 test for independence was performed. For ECs infected in the presence of MG 132, apoptosis was dependent on intracellular infection (χ2 = 19.05 at α = 0.05). In contrast, for ECs infected in the absence of MG 132, the low rate of apoptosis observed occurred independently of the presence or absence of intracellular organisms (χ2 = 0.499 at α = 0.05). MG 132 did not influence the initial rate or extent of EC infection as determined at three hr (data not shown).
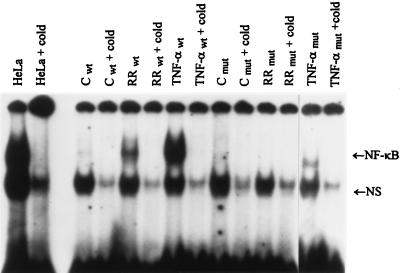
To confirm the role of NF-κB activation in host cell survival during infection, mutant HEF cells were used that had been transfected with a transdominant-negative superrepressor IκB rendering NF-κB inactivatable in response to TNF-α stimulation (21). As reported (21), stimulation of wild-type HEFs with TNF-α resulted in NF-κB activation, whereas minimal activation was seen in the mutant HEF cells. Likewise, R. rickettsii infection resulted in NF-κB activation in wild-type but not mutant HEF cells (Fig. 4). TUNEL staining and double-label immunofluorescence staining of wild-type and mutant HEFs with and without infection revealed that wild-type cells were highly resistant to apoptosis even when heavily infected (Fig. 5 a, c, and e). In striking contrast, extensive apoptosis occurred in infected mutant cells (Fig. 5 b, d, and f). Likewise, using T24 bladder carcinoma cells transfected with this dominant-negative IκB mutant (21), extensive apoptosis was observed in mutant but not in sham-transfected T24 cells (data not shown).
Figure 4.
Gel shift analysis of NF-κB induction in HEF cells. Gel shift analysis was performed on nuclear extracts prepared from control (C), R. rickettsii-infected (RR), and TNF-α-treated (10 ng/ml) wild-type (wt) and mutant (mut) HEFs incubated for 3 hr. Complexes were analyzed on 4% nondenaturing polyacrylamide gels followed by autoradiography. HeLa cells were included as positive markers. Other symbols are as in Fig. 3.
Figure 5.
Induction of apoptosis in wild-type and mutant HEF cells during R. rickettsii infection. Wild-type HEFs (a, c, and e) and HEFs stably transfected with a transdominant-negative IκBα mutant (b, d, and f) were infected with R. rickettsii for 18 hr and then subjected to TUNEL (a and b), immunofluorescence staining for R. rickettsii (c and d), and nuclear staining using Hoechst dye (e and f). c and e are identical fields as are d and f. Nuclei with normal morphology are denoted by n. Apoptotic nuclei in b and f are indicated by ∗. Arrowheads in c and d point to R. rickettsii organisms. (a and b Insets) Uninfected wild-type HEFs and uninfected mutant HEFs, respectively. [Bars = 5 μm (a for a and b) and 20 μm (c for c–f). Scoring of cells in these populations after TUNEL revealed that 74% of infected mutant HEFs were apoptotic and that 21% of infected wild-type HEFs were apoptotic.
DISCUSSION
During the complex interaction between an infectious agent and the host organism, induction or prevention of apoptosis could be a critical determinant in the outcome of infection. Indeed, many viral gene products have been identified that exert either a pro- or antiapoptotic effect on the host cell (27). Intracellular parasites such as Leishmanias have also acquired mechanisms to evade host cell apoptosis (27) and this likely serves to prevent elimination of the essential living host cell. Much interest has developed recently concerning the role of apoptosis in the pathogenesis of bacterial disease, although to date there have been no reports to demonstrate evasion of host cell apoptosis by bacterial organisms (28, 29). Several bacterial agents have been shown to exert a proapoptotic influence on host cells (28, 30, 31), and it can only be speculated whether the capacity to induce host cell apoptosis evolved as a cellular host defense strategy or whether it provided the bacterial organism with a selective advantage. Members of the genus Rickettsia, are unique among bacterial organisms in that they are strict obligate intracellular parasites requiring a viable host cell for survival and replication. It is plausible, therefore, that Rickettsia have evolved mechanisms to ensure maintenance of the host cell by thwarting potential host defense mechanisms involving programmed cell death. Studies described in this report provide evidence that during intracellular infection, R. rickettsii can inhibit induction of host cell apoptosis and does so via a mechanism involving activation of the transcription factor NF-κB.
EC apoptosis has not previously been observed as a consequence of rickettsial infection in vivo or in vitro, and in fact necrotic cell injury has been considered to be a hallmark of infection (1, 32, 33). Descriptions of EC changes during infection include dilatation of intracellular membranes, swelling of mitochondria, and loss of osmoregulatory capacity, although nuclear integrity is typically maintained until the time of total cellular disruption (1, 32). The present study demonstrates that an entirely different picture emerges when EC NF-κB activation is blocked. When in vitro infection was carried out in the presence of MG 132, a peptide-aldehyde proteasome pathway inhibitor of EC NF-κB activation, EC apoptosis was observed as early as 7 hr, as was revealed by in situ detection of nuclear DNA fragmentation and correlated with the intracellular presence of Rickettsia organisms. The percent apoptotic cells increased with time of infection and correlated with substantial loss of cell adhesion. Morphologic analysis of cellular architecture in NF-κB-inhibited infected ECs revealed typical apoptotic changes (34) including nuclear condensation and compaction, segregation of nuclear chromatin, and formation of apoptotic bodies (Fig. 2). Treatment of ECs with MG 132 alone exerted no proapoptotic influence, and the background level of apoptosis seen in infected ECs in which NF-κB activation was allowed was likely nonspecific because it occurred independently of the presence of intracellular infection. The mechanism governing this apoptotic response is unknown but perhaps derives from unknown factors present at the initial time of infection. The fact that there was no significant apoptosis detected in cultures infected in the absence of inhibitor at the later time point (16–18 hr) suggests that the ECs that had been induced to undergo apoptosis at the onset of infection had lost adhesion and/or had been reduced to undetectable cellular debris.
Although the vascular EC is the primary target of R. rickettsii infection in vivo, the organism does not exhibit strict host cell specificity and is capable of infecting many cell types in culture. To circumvent the need for pharmacologic manipulation, the relationship between NF-κB activation and host cell survival was explored in HEFs (Fig. 4) and in T24 bladder carcinoma cells (data not shown) that had been stably transfected with a transdominant-negative mutant IκB protein (21). This mutation, which involves both N- and C-terminal signals for phosphorylation (17), renders NF-κB inactivatable by TNF-α. These cell types were readily infected by R. rickettsii (Fig. 5), and although infection resulted in NF-κB activation in the wild-type cells, such activation failed to occur in the mutant cells (Fig. 4). Extensive apoptosis due to infection occurred in the mutant cells and correlated with intracellular presence of organisms (Fig. 5), whereas wild-type cells avidly survived even when heavily infected. Thus, the presence of a proapoptotic stimulus and a protective mechanism involving NF-κB activation is not restricted to the ECs and, therefore, can be generalized to other cell types.
NF-κB activation clearly plays a critical role in protecting against apoptosis during R. rickettsii infection, but the nature of host cell signaling resulting in induction of apoptosis remains elusive. Several bacteria possess the ability to stimulate host cell apoptosis by specific activation of molecules involved in the apoptotic process. For example, Shigella and Salmonella initiate host cell apoptosis by directly activating interleukin 1β converting enzyme (28, 35). The R. rickettsii-induced “death signal” likely requires intracellular presence of the organism and could involve oxidant production or interaction with any number of regulatory host cell molecules involved in induction of apoptosis. Apoptosis in NF-κB-inhibited ECs correlated strongly with intracellular infection, suggesting that the proapoptotic stimulus does not involve a soluble intermediate. Preliminary studies indicate that inclusion of a neutralizing antibody against TNF-α, a known inducer of apoptosis in many cell types, did not block R. rickettsii-induced apoptosis in NF-κB-inhibited ECs. Also, culture medium “conditioned” by infected ECs did not contain a proapoptotic stimulus when used to treat NF-κB-inhibited uninfected ECs.
It has yet to be shown whether or not the NF-κB protective mechanism described in this study functions during EC infection in vivo and how it may influence the process of disease. The NF-κB protective mechanism induced by R. rickettsii infection may not only protect the host cell from apoptosis induced by the intracellular infection itself but perhaps also from apoptotic signals generated by host defense mechanisms such as interaction with cells of the immune system or inflammatory cytokines. The capacity of certain cell types to evade host cell apoptosis via NF-κB activation may contribute to tissue tropism of the organism. The ability to induce host cell NF-κB activation is common to several infectious agents, including viruses, bacteria, and intracellular parasites (36). For example, human cytomegalovirus, HIV, human T cell leukemia virus, and adenovirus 5 have been shown to cause activation of host cell NF-κB during infection, and proteolytic activity of purified HIV protease has been shown to result in exposure of NF-κB DNA-binding domains in a cell-free assay system (37). HeLa cell NF-κB activation occurs after invasion of Shigella flexneri (38), and activation of NF-κB occurs in a macrophage cell line within minutes of exposure to Listeria monocytogenes (39). Infection of T lymphocytes with the intracellular protozoan parasite Theileria parva, results in sustained NF-κB activation (40). The ability of an infectious agent to block host cell apoptosis via NF-κB activation, however, may not be a general phenomenon. For example, the NF-κB activation that occurs during Shigella infection apparently does not protect the host cell from apoptosis (35). This could be due to differences in the kinetics of NF-κB activation, the mode of induction of the apoptotic response, or simply to variability among different cell types.
This report demonstrates that during infection, R. rickettsii overrides host EC apoptosis via a mechanism dependent on NF-κB activation. As a result, R. rickettsii can modulate the host cell’s apoptotic response to its own advantage and allow the cell to continue as the site of infection. Thus a bacterial organism protects the host cell from apoptosis by a NF-κB-dependent mechanism. This finding may provide a key element dependent in understanding the interplay between host cell defense strategies and parasite survival.
Acknowledgments
We thank Loel Turpin, Li Hua Rong, and Lisa Domotor for excellent technical assistance; Robert Freeman, Ph.D. for critical reading of the manuscript; Michael O’Reilly, Ph.D., Brian Rybarczyk, and Raymond Rancourt for helpful discussions; and Carol Weed for help in preparation of the manuscript. This work was supported in part by Grants HL-30616, AI-40689, AI-17416, ES-01247, and ES-07026 from the National Institutes of Health.
Note Added in Proof:
Fan et al. (41) recently reported that cells infected with the obligate intracellular bacterium chlamydia are profoundly resistant to apoptosis induced by exogenous stimuli, suggesting the existence of a chlamydia-induced, protective antiapoptotic mechanism.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: EC, endothelial cell; HEF, human embryonic fibroblast; TNF-α, tumor necrosis factor α; PDTC, pyrrolidine dithiocarbamate; TUNEL, terminal deoxynucleotidyltransferase-mediated UTP end labeling.
References
- 1.Silverman D J. Infect Immun. 1984;44:545–553. doi: 10.1128/iai.44.3.545-553.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silverman D J, Santucci L A. Infect Immun. 1988;56:3110–3115. doi: 10.1128/iai.56.12.3110-3115.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silverman D J, Santucci L A. Ann NY Acad Sci. 1990;590:111–117. doi: 10.1111/j.1749-6632.1990.tb42213.x. [DOI] [PubMed] [Google Scholar]
- 4.Silverman D J, Santucci L A, Sekeyova Z. Infect Immun. 1991;59:4505–4510. doi: 10.1128/iai.59.12.4505-4510.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santucci L A, Gutierrez P L, Silverman D J. Infect Immun. 1992;60:5113–5118. doi: 10.1128/iai.60.12.5113-5118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devamanoharan P S, Santucci L A, Hong J E, Tian X, Silverman D J. Infect Immun. 1994;62:2619–2621. doi: 10.1128/iai.62.6.2619-2621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silverman D J. J Infect Dis. 1986;153:694–700. doi: 10.1093/infdis/153.4.694. [DOI] [PubMed] [Google Scholar]
- 8.Sporn L A, Shi R-J, Lawrence S O, Silverman D J, Marder V J. Blood. 1991;78:2595–2601. [PubMed] [Google Scholar]
- 9.Drancourt M, Alessi M-C, Levy P-Y, Juhan-Vague I, Raoult D. Infect Immun. 1990;58:2459–2463. doi: 10.1128/iai.58.8.2459-2463.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teysseire N, Arnoux D, George F, Sampol J, Raoult D. Infect Immun. 1992;60:4388–4393. doi: 10.1128/iai.60.10.4388-4393.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sporn L A, Lawrence S O, Silverman D J, Marder V J. Blood. 1993;81:2406–2412. [PubMed] [Google Scholar]
- 12.Sporn L A, Haidaris P J, Shi R-J, Nemerson Y, Silverman D J, Marder V J. Blood. 1994;83:1527–1534. [PubMed] [Google Scholar]
- 13.Kaplanski G, Teysseire N, Farnarier C, Kaplanski S, Lissitsky J-C, Durand J-M, Soubeyrand J, Dinarello C A, Bongrand P. J Clin Invest. 1995;96:2839–2844. doi: 10.1172/JCI118354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sporn L A, Marder V J. Infect Immun. 1996;64:1609–1613. doi: 10.1128/iai.64.5.1609-1613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sporn L A, Sahni S K, Lerner N B, Marder V J, Silverman D J, Turpin L C, Schwab A L. Infect Immun. 1997;65:2786–2791. doi: 10.1128/iai.65.7.2786-2791.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyamoto S, Verma I M. Adv Cancer Res. 1995;66:255–292. [PubMed] [Google Scholar]
- 17.Baldwin A S., Jr Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 18.Barinaga M. Science. 1996;274:724. doi: 10.1126/science.274.5288.724. [DOI] [PubMed] [Google Scholar]
- 19.Beg A A, Baltimore D. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 20.Wang C-Y, Mayo M W, Baldwin A S., Jr Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 21.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 22.Gimbrone M A, Jr, Cotran R S, Folkman J. J Cell Biol. 1974;60:673–680. doi: 10.1083/jcb.60.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner D D, Olmsted J B, Marder V J. J Cell Biol. 1983;95:355–360. doi: 10.1083/jcb.95.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palmer T, Rosman G J, Osborne W R A, Miller A D. Proc Natl Acad Sci USA. 1991;88:1330–1334. doi: 10.1073/pnas.88.4.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen T J, Loo M A, Pind S, Williams D B, Goldberg A L, Riordan J R. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 26.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 27.Shen Y, Shenk T E. Curr Opin Gene Dev. 1995;5:105–111. doi: 10.1016/s0959-437x(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 28.Zychlinsky A, Sansonetti P. J Clin Invest. 1997;100:493–496. doi: 10.1172/JCI119557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox J. ASM News. 1997;63:412–413. [Google Scholar]
- 30.Kasuya S, Nagano I, Ikeda T, Goto C, Shimokawa K, Takahashi Y. Infect Immun. 1996;64:3937–3941. doi: 10.1128/iai.64.9.3937-3941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merien F, Baranton G, Perolat P. Infect Immun. 1997;65:729–738. doi: 10.1128/iai.65.2.729-738.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverman D J. In: Annals de l’Institut Pasteur/Microbiologie 137A. Popoff M Y, Sansonetti P J, editors. Paris: Elsevier; 1986. pp. 307–356. [Google Scholar]
- 33.de Brito, T., Hoshino-Shimizu, S., Pereira, M. O. & Rigolon, N. Virchows Arch. Abt. A Pathol. Anat. 358, 205–214. [DOI] [PubMed]
- 34.Kerr J F R, Gobe G C, Winterford C M, Harmon B V. In: Cell Death. Schwartz L M, Osborne B A, editors. New York: Academic; 1995. pp. 1–27. [Google Scholar]
- 35.Chen Y, Smith M R, Thirumalai K, Zychlinsky A. EMBO J. 1996;15:3853–3860. [PMC free article] [PubMed] [Google Scholar]
- 36.Siebenlist U, Franzoso G, Brown K. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 37.Riviere Y, Blank V, Kourilsky P, Israel A. Nature (London) 1991;350:625–626. doi: 10.1038/350625a0. [DOI] [PubMed] [Google Scholar]
- 38.Dyer R B, Collaco C R, Niesel D W, Herzog N K. Infect Immun. 1993;61:4427–4433. doi: 10.1128/iai.61.10.4427-4433.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hauf N, Goebel W, Serfling E, Kuhn M. Infect Immun. 1994;62:2740–2747. doi: 10.1128/iai.62.7.2740-2747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanov V, Stein B, Baumann I, Dobbelaere D A E, Herrlich P, Williams R O. Mol Cell Biol. 1989;9:4677–4686. doi: 10.1128/mcb.9.11.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan T, Lu H, Hu H, Shi L, McClarty G A, Nance D M, Greenberg A H, Zhong G. J Exp Med. 1998;187:487–496. doi: 10.1084/jem.187.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]