Abstract
Enhancers can regulate designate promoters over long distances by forming chromatin loops. Whether chromatin loops are lost or reconfigured during gene repression is largely unexplored. We examined the chromosome conformation of the Kit gene that is expressed during early erythropoiesis, but is downregulated upon cell maturation. Kit expression is controlled by sequential occupancy of two GATA family transcription factors. In immature cells, a distal enhancer bound by GATA-2 is in physical proximity with the active Kit promoter. Upon cell maturation, GATA-1 displaces GATA-2 and triggers a loss of the enhancer/promoter interaction. Moreover, GATA-1 reciprocally increases the proximity in nuclear space among distinct downstream GATA elements. GATA-1-induced transitions in chromatin conformation are not simply the consequence of transcription inhibition and require the cofactor FOG-1. This work shows that a GATA factor exchange reconfigures higher order chromatin organization and suggests that de novo chromatin loop formation is employed by nuclear factors to specify repressive outcomes.
Keywords: chromatin, chromatin loops, GATA-1, GATA-2, gene expression, hematopoiesis, erythropoiesis, Kit
Introduction
Regulatory elements that are separated by large genomic distances can cluster in nuclear space to form chromatin loops. For enhancer elements, looping serves to establish contact with target promoters, which might augment local transcription factor concentrations (for review see (Chambeyron and Bickmore, 2004; Fraser, 2006)). Interactions can occur between regulatory sequences even when they are located on different chromosomes (Dekker et al., 2002; Spilianakis et al., 2005). Similar to enhancers, insulator elements can also cluster in nuclear space and tether chromatin loops to distinct sub-nuclear compartments ((Yusufzai et al., 2004) (for review see (Gaszner and Felsenfeld, 2006)). The resulting separation of enhancer and promoter elements into distinct looped domains is thought to block enhancer function. Thus, chromatin loops may be configured to either increase or inhibit gene expression.
Sequence-specific DNA-binding proteins have emerged as critical regulators of looped conformations in the nucleus. For example, the erythroid transcription factors GATA-1, its co-factor FOG-1 (friend of GATA-1), and EKLF (erythroid kruppel-like factor) directly occupy looped enhancers as well as target gene promoters at the β-globin locus (Drissen et al., 2004; Vakoc et al., 2005), indicating that several factors are necessary, although perhaps not sufficient (Kooren et al., 2007; Vernimmen et al., 2007), for looping to occur. Similarly, it has been suggested that GATA-3 and STAT6 cooperate in establishing long-range chromatin interactions at the TH2 cytokine locus (Spilianakis and Flavell, 2004).
An open question is whether transcription factors directly mediate loop formation, for example by forming dimers, or whether loops form because of secondary changes in transcriptionally active chromatin, such as histone hyper-acetylation, DNase 1 sensitivity, remodeled or repositioned nucleosomes, and the increased presence of general transcription factors or RNA polymerase II (pol2). Another potential mechanism for loop formation invokes the simultaneous association of a distal regulatory site and a promoter with a “transcription factory” (Osborne et al., 2004) (for review see (Marenduzzo et al., 2007). The strict requirement for each of these features in loop formation has not been resolved. We reasoned that evaluating chromosome conformation during gene repression might uncouple the roles of individual DNA-binding proteins from general features of active chromatin in the formation of looped chromatin domains. To better elucidate the molecular components that underlie chromatin loop formation, we examined the chromosome configuration at the Kit locus before and after repression by the transcription factor GATA-1 in erythroid cells. Possible scenarios are GATA-1-induced loss of enhancer-promoter interactions, the formation of new, perhaps repressive chromatin loops, or a combination of the two.
GATA-1 is essential for normal erythroid differentiation (Ferreira et al., 2005). Mutations in the GATA-1 gene are associated with congenital anemias and megakaryoblastic leukemias (for review see (Crispino, 2005)). While GATA-1 functions as a transcriptional activator at key erythroid-specific genes, it also can function as a transcriptional repressor (Grass et al., 2003; Hong et al., 2005; Rodriguez et al., 2005; Welch et al., 2004). GATA-1-directly represses several proto-oncogenes including Myc, and Kit, to coordinate decreased cell proliferation with terminal erythroid maturation (Munugalavadla et al., 2005; Rylski et al., 2003). Another GATA-family member, GATA-2, which is expressed in hematopoietic stem cells, multilineage progenitors and early committed erythroblasts, is likewise a direct repression target of GATA-1 (Grass et al., 2003; Letting et al., 2004). Upon cell maturation, GATA-1 levels increase leading to silencing of GATA-2 expression. Thus, erythroid differentiation is accompanied by an exchange of GATA factors.
We explored the impact of GATA-1 on higher order chromatin organization during gene repression at the proto-oncogene Kit. Kit encodes a receptor tyrosine kinase that is essential for normal hematopoiesis. Loss of Kit in mice leads to embryonic death from anemia (for review see (Broudy, 1997). In the hematopoietic system, Kit is expressed in stem cells and lineage progenitor cells, and is downregulated upon terminal erythroid differentiation. Moreover, forced expression of Kit in erythroid precursors impairs their maturation (Munugalavadla et al., 2005; Wessely et al., 1997).
We assessed the occupancy of GATA-1, GATA-2, RNA polymerase 2 (pol 2) and histone acetylation across ∼ 310 kilobases of the Kit locus in erythroid cells and identified several GATA factor-bound regulatory sites positioned at -114, +5, +58 and +73 kilobases with respect to the transcription start site. Using chromosome conformation capture (3C) (Dekker et al., 2002), we observed that prior to GATA-1 expression, the upstream -114 element, which functions as an enhancer and is bound by GATA-2, forms a loop with the promoter proximal region. Conditional activation of GATA-1 extinguished GATA-2 occupancy at the Kit locus coincident with disruption of the enhancer-promoter loop. Notably, de novo loop formation was observed among downstream GATA-1-bound elements. This indicates that different GATA proteins exert distinct effects on chromosome conformation and resultant transcriptional activity. In particular, we propose that GATA factor exchange at the Kit locus is critical for the developmental reconfiguration of the chromatin fiber where newly formed repressive chromatin loops lead to the exclusion of enhancer loops.
Results and Discussion
Profile of GATA factors at the Kit locus
Transcriptional activation and repression by GATA-1 have been studied extensively in the ES cell derived erythroid cell line G1E. G1E cells have been rendered GATA-1-deficient via gene targeting and are developmentally arrested at an early erythroblast stage (Weiss et al., 1997). Induction in these cells of an estradiol-dependent form of GATA-1 (GATA-1-ER) leads to synchronous erythroid maturation and cell cycle arrest, as reflected by activation of erythroid-specific genes, and concomitant repression of numerous genes, including the Kit and Gata2 (Grass et al., 2003; Munugalavadla et al., 2005; Welch et al., 2004). Thus, G1E cells represent a powerful system in which GATA-1 can be studied in its natural context. Prior to GATA-1 activation, GATA-2 occupies select GATA elements at the Gata2 locus, presumably enhancing transcription in a positive auto-regulatory circuit (Grass et al., 2003). Upon GATA-1 activation, GATA-2 expression is rapidly silenced, thus leaving GATA-1 as the sole GATA binding protein to activate and repress gene expression during terminal erythroid maturation. Importantly, transcriptional repression by GATA-1 requires the cofactor FOG-1 (Crispino et al., 1999) which recruits several co-repressor complexes including NuRD (Hong et al., 2005; Rodriguez et al., 2005) and CtBP-2 (Fox et al., 1999).
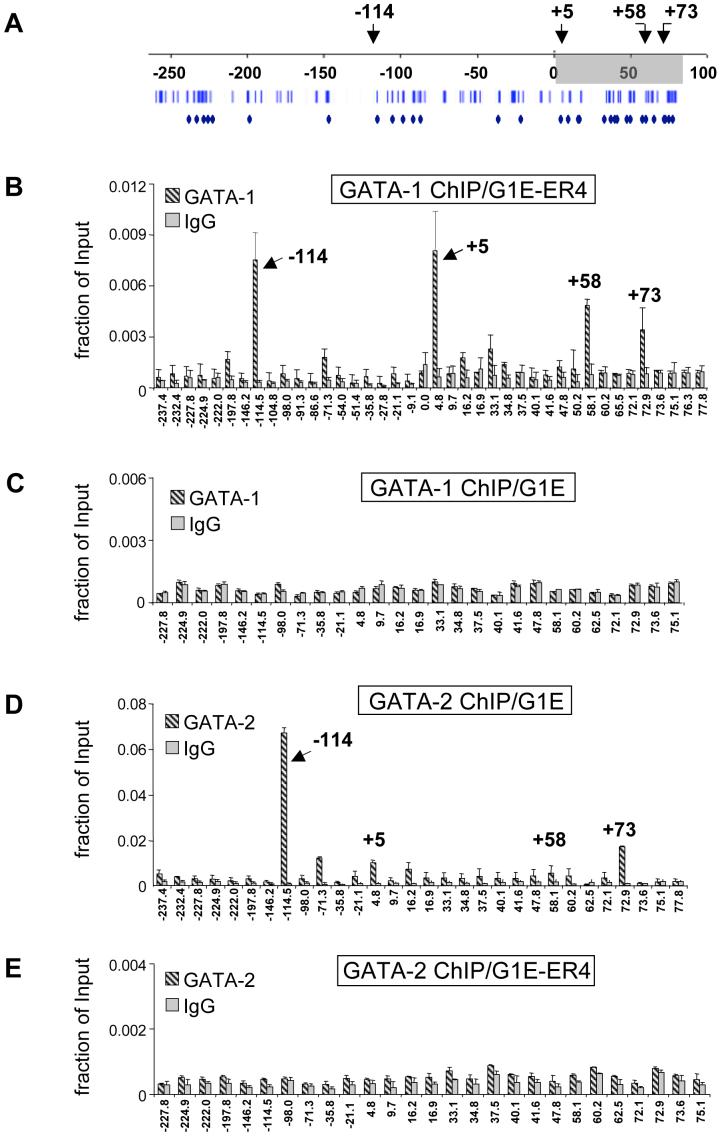
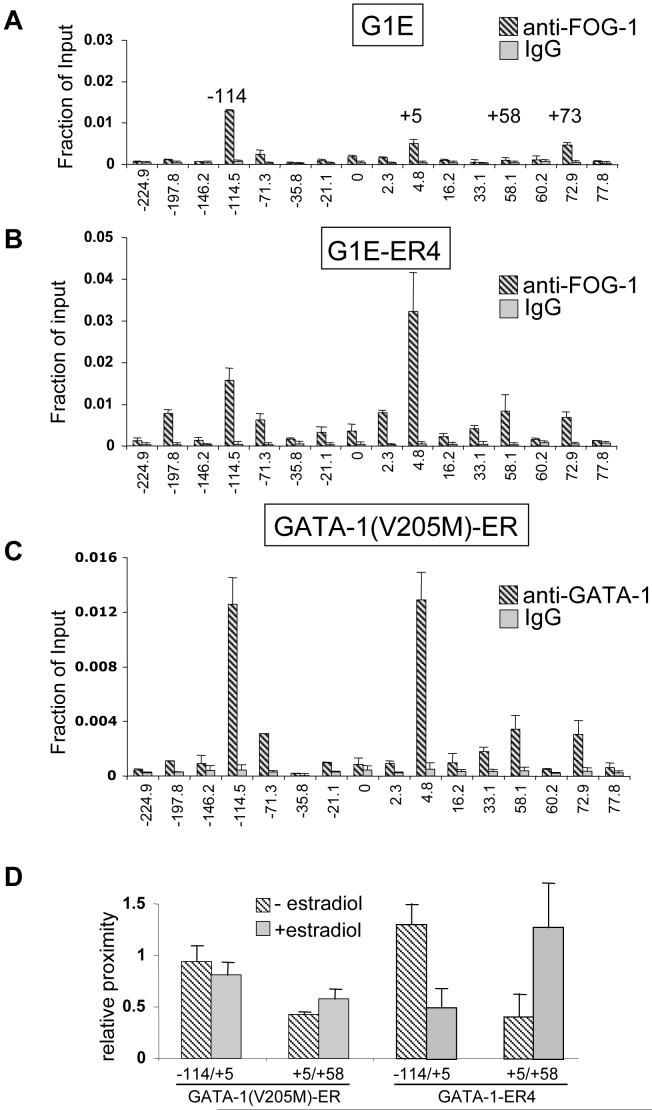
The murine Kit locus is controlled by proximal and distal regulatory sites that are spread over more than 300 kilobases to direct expression in diverse tissues (Berrozpe et al., 2006; Berrozpe et al., 1999; Cairns et al., 2003; Kluppel et al., 1997). In G1E cells stably expressing GATA-1-ER (G1E-ER4), estradiol treatment represses Kit expression as determined by quantitative real time RT-PCR [(suppl. Fig.1) and (Munugalavadla et al., 2005)]. Previous work employing chromatin immunoprecipitation (ChIP) in G1E-ER4 cells detected high level GATA-1 binding at a region in the Kit gene that resides approximately 5 kilobases (kb) downstream of the transcription start site (Munugalavadla et al., 2005). We extended these findings by profiling GATA-1 occupancy by ChIP in estradiol-treated G1E-ER4 cells across 310 kb of sequence from -230 to +80 kb with respect to the transcription start site. ChIP primer pairs were designed to cover conserved GATA motifs that were predicted to have high regulatory potential (RP score of >0.05, scale from -0.41 to +0.3, (http://gala.cse.psu.edu/ (Elnitski et al., 2003; Wang et al., 2006)) using bioinformatic approaches. 76 GATA-1 sites were found that are conserved between human, mouse and rat. 45 of these had an RP score of >0.05. Scores above zero are more likely to have patterns indicative of regulatory regions. All of these sites were covered by 31 PCR amplicons (Fig.1A). Additional primer pairs were used that spanned regions not containing obvious GATA elements, including the Kit promoter. In addition to the known site of GATA-1 occupancy at +5kb (Munugalavadla et al., 2005), we observed high enrichment of GATA-1-ER at -114kb, and +58kb with respect to the transcriptional start site (Fig.1B). Additional sites with lower GATA-1-ER occupancy were found at -198kb, -71kb, +33kb, and +73kb. ChIP experiments in G1E cells that lack GATA-1 served as control for specificity (Fig.1C). While little or no GATA-1-ER was detected throughout most of the Kit locus in untreated G1E-ER4 cells, considerable amounts of GATA-1-ER were present at the -114 site, reflecting “leakiness” of the conditional system (not shown). Therefore, in the following experiments we used for comparison G1E cells and induced G1E-ER4 cells. Finally, although our study covered approximately 310 kb of the Kit locus, it is possible that we missed additional elements that reside within or outside this region (Berrozpe et al., 2006).
Figure 1. Profile of GATA-1-ER and GATA-2 occupancy at the Kit locus.
(A) Organization of the Kit locus. Numbers indicate distance in kb from the transcriptional start site. Shading delineates the transcribed region. Bars denote conserved GATA sites. Diamonds denote GATA elements within regions of high regulatory potential. Arrows denote sites of highest GATA factor occupancy. (B, C) ChIP analysis of GATA-1-ER in estradiol-treated G1E-ER4 cells (B) and G1E cells (C). (D, E) ChIP for GATA-2 in G1E cells (D), and in estradiol-treated G1E-ER4 cells (E). Results are averages of 3 independent experiments. Error bars represent standard deviation. IgG, isotype matched control.
Since GATA-2 is expressed in immature erythroid cells prior to its repression by GATA-1 (Weiss et al., 1994), we examined whether GATA-2 occupies the Kit locus in G1E cells. The highest levels of GATA-2 were present at -114 while lower levels of GATA-2 were found at -71, +5, and +73 (Fig.1D). Thus, despite similar in vitro DNA binding specificities, GATA-1 and GATA-2 exhibit distinct binding profiles in vivo, which likely contributes to their disparate functions at the Kit locus. Since GATA-2 occupancy is associated with active Kit expression, the -114 element might represent a GATA-2-dependent enhancer (see below). Following activation of GATA-1-ER, GATA-2 was depleted from the Kit locus (Fig.1E). Thus, repression of Kit is associated with a complete exchange of GATA-1 for GATA-2.
We failed to detect substantial amounts of GATA-1 or GATA-2 at the proximal promoter where it has been proposed that the transcription factor Sp-1 recruits a GATA-1/GATA-2-containing transcription factor complex (Lecuyer et al., 2002). It is possible that indirect recruitment of GATA-1 lowers the signal obtained by ChIP.
Histone acetylation and RNA polymerase II at the Kit locus
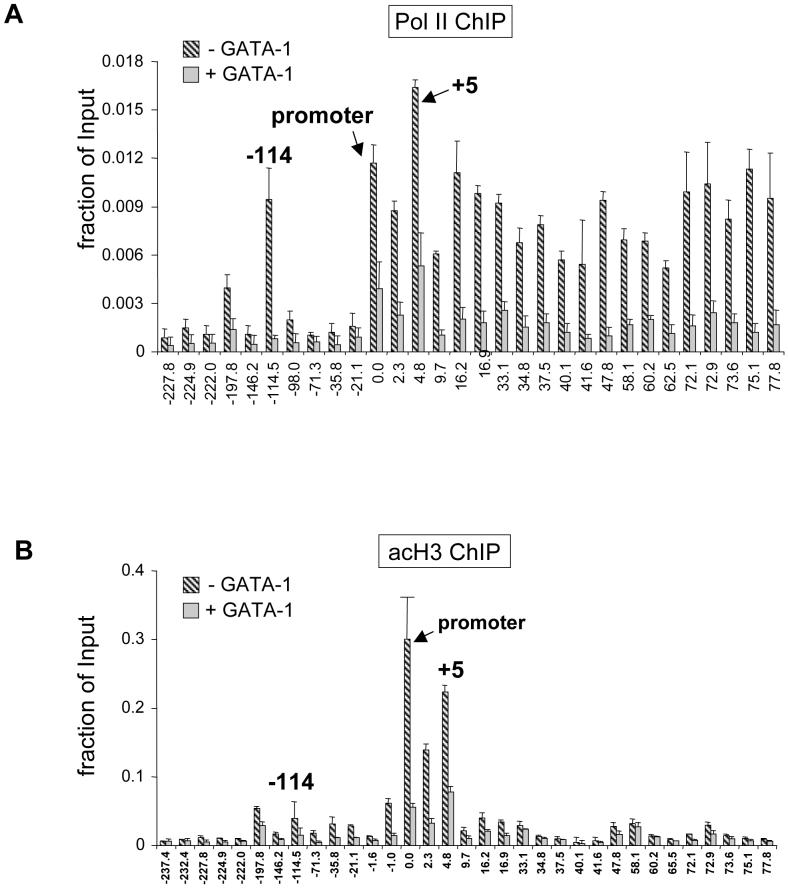
Histone hyperacetylation and pol 2 are often found at enhancers and locus control regions. To examine whether the sites of high GATA-2 occupancy represent enhancer sequences, we measured pol2 and histone acetylation across the Kit locus by ChIP. As expected, pol 2 was found at high levels throughout the transcribed portion of the Kit gene and was markedly reduced when Kit expression was silenced (Fig.2A). Notably, high levels of pol 2 were found at the -114 region and to a lesser extent at -198 (Fig.2A). Using anti-acetyl histone H3 antibodies we found H3 acetylation to be highest around the promoter region that was lost upon Kit repression (Fig.2B). Modest enrichments of H3 acetylation at the active Kit gene were also observed at -114 and -198. The combined presence of GATA-2, pol2 and acetylated histones at -114 suggests that this region is a long-range GATA-2-dependent enhancer.
Figure 2. ChIP assays of pol2.
(A) and acetylated histone H3 (acH3, B) at the Kit locus in G1E cells (-GATA-1, striped bars) or estradiol-treated G1E-ER4 cells (+GATA-1, grey bars). Results are averages of 2 independent experiments.
The -114 region functions as GATA-2-selective enhancer
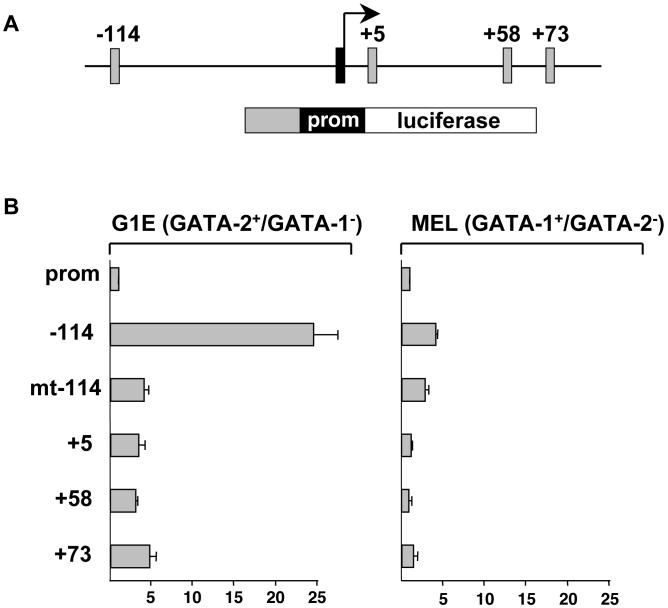
Although both GATA-1 and GATA-2 bind to the -114 region, only the presence of GATA-2 is associated with active Kit transcription (Fig.1). This suggests that the -114 region might function as a GATA-2 selective enhancer. To examine this directly, we constructed a firefly luciferase reporter gene containing the Kit core promoter and the -114 region placed upstream (Fig.3A). This construct was introduced into erythroid cells expressing only GATA-2 (parental G1E cells) or only GATA-1 (MEL cells). Luciferase activity was normalized to that of a cotransfected Renilla luciferase plasmid. We found that the -114 region substantially increased transcription in G1E cells with much lower activity in MEL cells (Fig.3B). To determine whether enhancer activity is mediated by GATA-2 and not other factors bound to this sequence, we mutated both GATA elements within the -114 fragment (mt-114). These mutations dramatically reduced reporter gene expression in G1E cells, indicating that GATA-2 is required for enhancer activity (Fig.3B). Notably, the activities of the other regions bound by GATA factors, i.e. +5, +58 and +73, were substantially lower than that of -114 (Fig.3B). Together, these results suggest that the -114 region is a GATA-2 dependent enhancer. In its natural context the GATA-2-bound enhancer might function by establishing physical contacts with the promoter proximal region. This was examined below.
Figure 3. The -114 kb region is a GATA-2 dependent enhancer.
(A) Structure of the reporter constructs. Grey boxes represent regions that were placed upstream of the Kit core promoter (prom). (B) G1E and MEL cells were transfected with reporter constructs along with a plasmid expressing Renilla luciferase. The activity of the enhancer-less Kit promoter was set as 1. mt-114 represents a construct in which both GATA sites were destroyed. Results are averages of 3 independent experiments.
Dynamic changes in chromosome configuration upon GATA-1 induced gene repression at the Kit locus
GATA-1 binds to the β-globin locus control region and the β-globin promoter in vivo and triggers increased proximity between these sites, indicative of chromatin looping (Vakoc et al., 2005). Here we studied the effects of GATA-1 on higher order chromatin organization during repression of the Kit gene. Specifically, we considered two scenarios that might occur alone or in combination. First, activation of GATA-1 might disrupt pre-existing enhancer/promoter loops. Second, GATA-1 might trigger the formation of new distinct loops that are repressive in nature. To test these possibilities, we employed 3C technology (Dekker et al., 2002), also known in varied form as nuclear ligation assay (Cullen et al., 1993), measuring the relative distances among the regions bound by GATA-1 and GATA-2.
Briefly, cells were fixed with formaldehyde, lysed, and nuclei treated with the restriction enzyme Bgl2. Following the inactivation of the enzyme, sample dilution and ligation, the resulting DNA was purified, and selected ligation products quantitated by PCR with primer pairs spanning the ligation junctions. The amount of the ligation product is proportional to the proximity of the examined DNA fragments. The experimental details of this method are described elsewhere (Dekker et al., 2002; Vakoc et al., 2005). The extent of restriction digestion influences the number of free DNA ends available for ligation. Therefore, we determined the digestion efficiency by measuring the residual amounts of undigested DNA using real time PCR (suppl. Fig. 2, see also Experimental Procedures). Only samples in which the Blg2 digests had occurred to a similar extent were examined in comparative studies. To accurately quantify the PCR products we employed Taqman real time PCR with probes near the restriction sites (Vernimmen et al., 2007). To account for possible variation in template quality, all data were normalized to 3C products of two restriction fragments from the α-tubulin gene whose expression does not change upon GATA-1 induction. To account for potential differences in amplification efficiency among primer pairs and Taqman probes, defined amounts of each 3C ligation product were used to generate standard curves against which all PCR data were normalized.
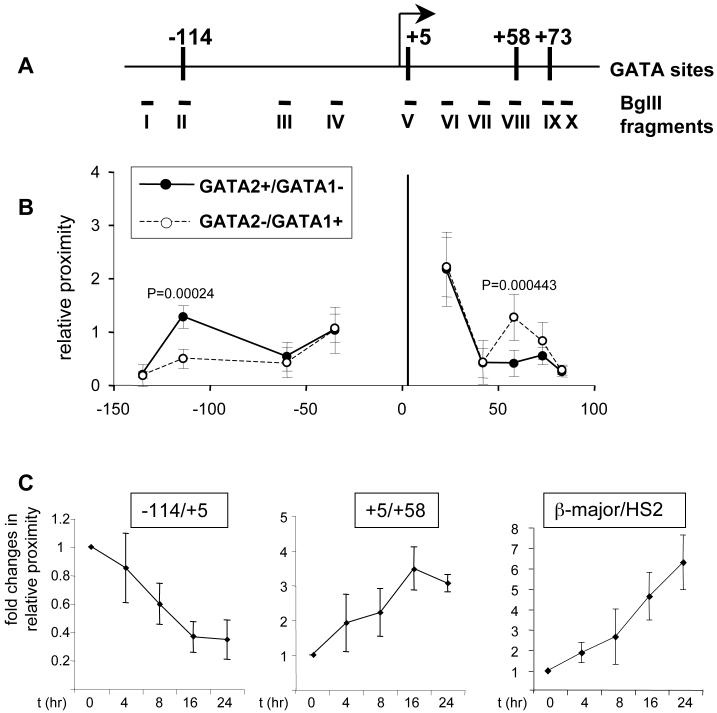
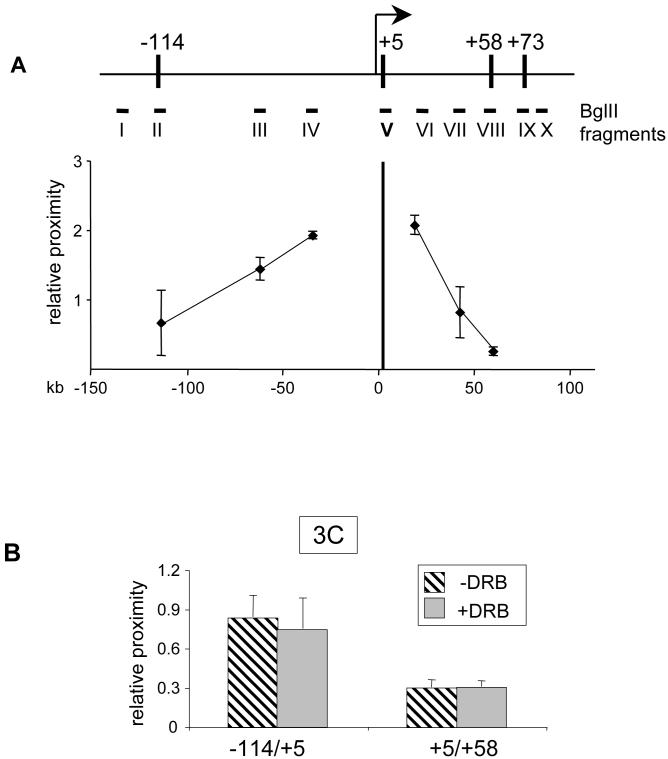
We began by comparing the relative distances among the GATA-1-ER- or GATA-2-bound elements in estradiol-treated G1E-ER4 cells and G1E cells, using as reference point a promoter-proximal Bgl2 fragment spanning +1.5 to +5 that contains the +5 GATA elements. In G1E cells where GATA-2 is present and Kit expression is high, we found the proximities of Bgl2 fragments encompassing the +58, and +73 sites to be low and similar to a control fragment positioned at +42 (Fig.4B, closed circles). This is consistent with the transcribed region being in a linear configuration with few significant interactions among the sites bound by GATA-2. We did not display on this graph numbers obtained with fragments that are adjacent or very close to the promoter-proximal restriction fragment since their 3C ligation products are constitutively very abundant (not shown). When upstream fragments were examined, we found no interaction between the +5 and the -36 and -60 regions (Fig.4B, closed circles). However, we observed a substantial increase in proximity between the +5 region and the -114 region. The resolution limits of 3C preclude us from distinguishing whether the -114 region is in direct proximity with the Kit promoter or a region slightly downstream thereof. Nevertheless, these data suggest that the -114 region is closer to the Kit promoter than the intervening -36 and -60 sequences, consistent with the possibility of an activating chromatin loop that juxtaposes a long-range enhancer element with the promoter.
Figure 4. 3C analysis of the Kit locus.
(A) Schematic of the Kit locus. Vertical bars demarcate sites bound by GATA-1 or GATA-2. Bgl II fragments used for 3C assay are indicated by Roman numerals.
(B) 3C assay in G1E cells or estradiol-treated (21 hours) G1E-ER4 cells. Fragment V (vertical bar at position +5) served as “fixed” fragment. GATA-2+/GATA-1- represents parental G1E cells. GATA-2-/GATA-1+ represents estradiol-treated G1E-ER4 cells. Results are averages of 5 independent experiments. Error bars represent standard deviation.
(C). Time course 3C analysis of the interaction between -114/+5 (left panel), +5/+58 (middle panel) and HS2/β-major (right panel) in G1E-ER4 cells treated with estradiol for 0, 4, 8, 16 and 24hrs. Results are averages of 4 (left panel), 3 (middle panel), and 3 (right panel) independent experiments.
When we examined estradiol-treated G1E-ER4 cells where GATA-1 activity is present and Kit is transcriptionally repressed, we observed a striking difference in chromosome conformation. First, the DNA fragment at +58 moved significantly closer to the promoter proximal +5 region (Fig.4B, open circles), suggesting that GATA-1-ER triggers clustering of chromosomal regions not only during gene activation, as we described previously, but also during gene repression. The extent of GATA-1-induced loop formation was comparable to that found between the LCR and the β-globin promoter (data not shown, (Vakoc et al., 2005)). Second, the proximity between the -114 region and the promoter proximal fragment was lost (Fig.4B, open circles). Thus, the pattern of interactions among upstream sequences and the promoter proximal region is more similar to that expected for a linear chromosome fragment. Collectively, these results suggest that induction of GATA-1 and loss of GATA-2 lead to a reciprocal change in proximities of regulatory elements.
To confirm the data obtained by Taqman real time PCR, we analyzed several 3C products by conventional PCR. Specifically, we examined the interaction of both ends of the +58 Bgl2 fragment with the +5 region and with a fragment positioned at the Kit promoter. We found that GATA-1-ER specifically triggered an increase in proximity among the +5 and +58 fragments to an extent similar to that observed with Taqman PCR (suppl. Fig.3). No substantial change in proximity was observed between the +5 and +73 region. The interaction frequency between +58 and +5 and that between +58 and the Kit promoter are similar, and forward and reverse primers of the +58 fragment produced essentially the same results (suppl. Fig.3). Thus, both methods to quantify the ligation products produced consistent results.
To examine more closely the reciprocal relationship between upstream and downstream chromatin loops, we performed a time course 3C analysis. For these experiments we used G1E-ER4 cells exposed to estradiol for varying amounts of time. Within 4 hours, the interaction between the +5 and the +58 downstream fragments was increased and showed a further upward trend at later time points (Fig.4C middle panel). Conversely, we observed a progressive decline in proximity among the -114 and +5 fragments (Fig.4C left panel). Because of the above-mentioned leakiness of this system, the absolute changes in GATA-1-induced loop formation were somewhat lower than those in the previous experiments that compared induced G1E-ER4 cells to parental G1E cells. However, with regard to extent and kinetics these changes were comparable to those observed at the β-globin locus under the same conditions (Fig.4C right panel).
In concert, these data show that the sequential occupancy of distinct GATA factors is associated with defined transitions in chromatin configuration leading to removal of an upstream loop at the expense of a loop among the downstream GATA elements. Notably, together with our previous work (Vakoc et al., 2005), the current data show that GATA-1 appears capable of clustering regulatory sites both during gene activation and repression. The ability of stage-specific GATA factors to induce unique chromatin configurations at the same gene locus may represent a general mechanism to modulate gene expression during tissue development.
GATA factor-dependent chromatin loops are expected to be absent in cells that do not express Kit and lack GATA-1 and GATA-2. To test this, we performed 3C in NIH3T3 fibroblasts. Neither the +5/-114 nor the +5/+58 interaction was detected (Fig.5A). Instead, we observed a linear decline in the proximities between the +5 region and fragments that reside further upstream (II, III, and IV) and downstream (VI, VII and VIII), consistent with the absence of loop formation. The analysis of fragments that reside further away from +5 (I and IX) produced even lower PCR signals that approached the threshold of reliable detection and were not included in the graph. It is of interest that the decline in proximities is steeper among the downstream fragments when compared to the upstream ones. We speculate that this might reflect non-random folding of the repressed chromatin fiber. Nevertheless, these data show that the critical interactions among GATA factor-bound regulatory sites are absent at the silent Kit locus in heterologous cells.
Figure 5.
A) 3C analysis of indicated fragments in NIH 3T3 cells. Results are averages of 3 independent experiments. B) 3C analysis of indicated BglII fragments in G1E cells in the presence or absence of 75μM DRB (6 hours). Results are averages of 2 independent experiments.
GATA-1-induced changes in chromosome configuration are not a consequence of transcription inhibition
We considered the possibility that the observed changes in chromatin organization result from loss of Kit transcription and might therefore reflect an indirect consequence of GATA-1 action. To address this point we performed 3C on G1E cells treated with 75 μM 5,6 dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) that inhibits p-TEFb-mediated phosphorylation of RNA pol2 at serine 2 of the C-terminal domain. As expected, DRB treatment for six hours led to loss of pol2 from the transcribed region, but not from the promoter and the upstream -114 region, verifying the effectiveness of the compound (Suppl. Fig.4). Using 3C, we found no changes in the relative distances between Bgl2 fragments at +5 and -114, and between +5 and +58 in DRB treated cells when compared to untreated cells (Fig.5B). Thus, loss of Kit transcription per se is not sufficient to induce changes in chromatin conformation, further supporting a direct role for GATA-1 in this process.
The GATA-1 cofactor FOG-1 is required for chromatin loop formation
FOG-1 interacts physically and functionally with GATA-1 and GATA-2 (Chang et al., 2002; Tevosian et al., 1999; Tsang et al., 1997). Mutations in GATA-1 that disrupt the FOG-1 interaction compromise both transcriptional activation and repression by GATA-1 (Crispino et al., 1999; Nichols et al., 2000). Notably, such mutations cause congenital anemias and thrombocytopenias in patients and genetically modified mice (Nichols et al., 2000), for review see (Crispino, 2005)). Through the use of a specific GATA-1 mutant (GATA-1(V205M)-ER) that is defective for FOG-1 binding, we found previously that FOG-1 is required for GATA-1-induced loop formation at the β-globin locus (Vakoc et al., 2005). However, loss of FOG-1 binding also compromised the ability of GATA-1 to stably associate with the β-globin promoter in vivo (Letting et al., 2004). Therefore, a direct role for FOG-1 during loop formation could not be evaluated.
To examine whether FOG-1 is required for GATA-1-induced loop formation at the Kit locus we first measured FOG-1 occupancy in G1E cells, in induced G1E-ER4 cells, and in G1E cells expressing comparable amounts of GATA-1(V205M)-ER (Letting et al., 2004). In the absence of GATA-1, FOG-1 distribution is similar to that of GATA-2 with the highest levels found at -114 and little or no FOG-1 occupying the +5 and +58 regions (Fig.6A). In contrast, in induced G1E-ER4 cells, FOG-1 distribution more closely resembles that of GATA-1 with peak levels found at -114 and +5, and intermediate levels at +58 (Fig.6B). Hence, the presence of FOG-1 correlates well with GATA factor occupancy.
Figure 6. ChIP analysis of FOG-1 in G1E cells.
(A) and in estradiol-treated G1E-ER4 cells (B). (C) anti-GATA-1 ChIP in estradiol-treated cells expressing GATA-1(V205M)-ER. (D) 3C analysis of -114/+5, +5/+58 in GATA-1(V205M)-ER expressing cells and G1E-ER4. Bars denote standard deviation and results are averages of 2 (GATA-1(V205M)-ER) and 5 (G1E-ER4) independent experiments.
We next examined by ChIP the chromatin occupancy of GATA-1(V205M)-ER across the Kit locus. We found that the levels of GATA-1(V205M)-ER and its spatial distribution at the Kit locus are highly similar to that observed with wild-type GATA-1 (Fig.6C, compare with Fig.1B). This suggests that, in contrast to the β-globin promoter, FOG-1 binding is not essential for the association of GATA-1 with the Kit locus. Nevertheless, GATA-1(V205)-ER failed to extinguish Kit expression, implying a requirement of FOG-1 during GATA-1-induced Kit gene repression (suppl. Fig.1).
We also determined by ChIP the profile of FOG-1 occupancy across the Kit locus in estradiol-treated and untreated GATA-1(V205M)-ER expressing cells. As expected, FOG-1 occupancy was low in untreated cells similar to that observed in parental G1E cells (suppl. Fig.5A, compare with Fig.6A). Upon exposure to estradiol, FOG-1 recruitment increased slightly, possibly because the V205M mutation disrupts FOG1 binding incompletely (Nichols et al., 2000). Importantly, however, FOG-1 recruitment was substantially below that observed in induced GATA-1ER4 cells, especially at positions +5 and +58 (suppl. Fig.5B compare with Fig.6B). This allowed us to examine by 3C whether FOG-1 is required for the dynamic chromatin reconfiguration by GATA-1. Indeed, GATA-1(V205M)-ER failed to reduce the interaction between -114 and +5 (Fig.6D). Moreover, it also failed to induce clustering of the downstream GATA elements +5 and +58 (Fig.6D). These experiments suggest a direct role for FOG-1 in GATA-1-induced chromatin loop formation that cannot be explained by regulating GATA-1 occupancy.
FOG-1 is required for the exchange of GATA-1 for GATA-2 at the GATA-2 gene (Pal et al., 2004). In agreement with this finding, GATA-2 is not completely displaced from the Kit locus in GATA-1(V205M)-ER expressing cells (data not shown). Thus, it is possible that the FOG-1 requirement for chromatin reconfiguration is simply a reflection of an incomplete GATA factor exchange. However, we favor a more direct role for FOG-1 in chromatin reorganization since GATA-1(V205M)-ER occupancy is virtually the same as GATA-1-ER. If residual GATA-2-mediated loops were to compete with those formed by GATA-1-ER, one might expect a looping phenotype intermediate between GATA-1 and GATA-2 expressing cells. However, loss of FOG-1 binding essentially abrogates any changes in chromosome conformation, which argues in favor of a direct role for FOG-1 in GATA-1-ER-induced loop formation. Of course, additional proteins bound to FOG-1 or GATA-1-ER might be required for establishing physical interactions at the base of the loops.
FOG-1 contains nine zinc fingers, four of which can bind GATA factors (Fox et al., 1999). Therefore, it is possible that FOG-1 might physically link GATA-1 proteins bound at distinct regions of the Kit locus. However, mutant forms of FOG-1 that retain just one GATA-1-binding zinc finger are biologically active in erythroid cells (Cantor et al., 2002), suggesting that intermediary factors might be involved in linking GATA-1/FOG-1 complexes between distant sites.
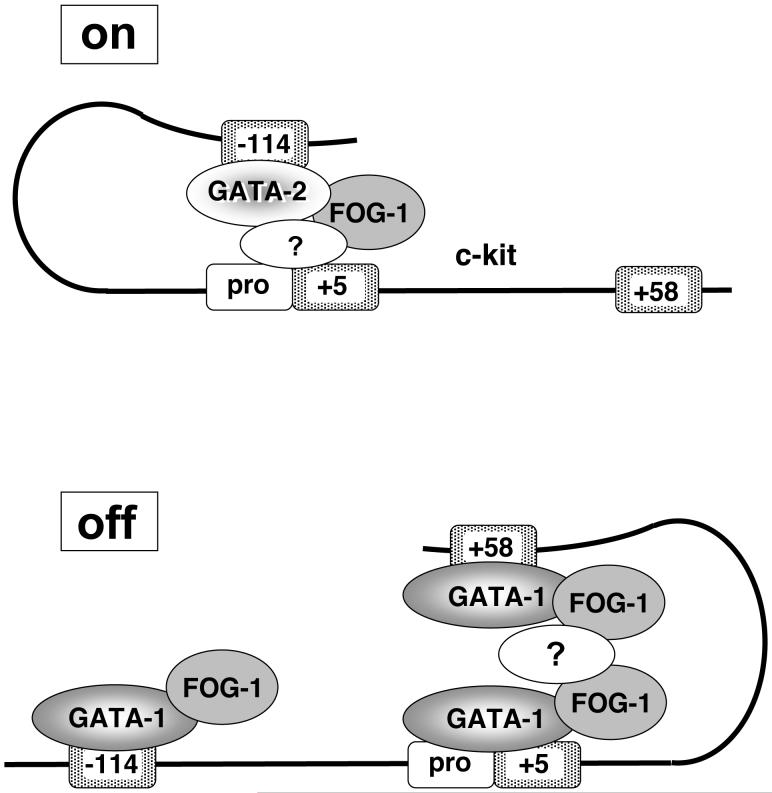
For erythropoiesis to occur normally, Kit must be expressed in early progenitors and then downregulated during late maturation. Our studies demonstrate that this dynamic, biologically essential pattern of regulation is accompanied by substantial structural reorganization induced by a GATA factor switch. During terminal erythroid differentiation, the exchange of GATA-1 for GATA-2 is associated with loss of a chromatin loop between a distal upstream enhancer and the promoter proximal region. Conversely, downstream elements bound by GATA-1 are physically close in nuclear space (Fig.7). Thus, although GATA-1 and GATA-2 target an overlapping set of GATA sites, they confer opposite functions on the activity of the Kit locus. Accordingly, GATA-1 and GATA-2 are associated with distinct chromosome configurations. One likely interpretation of these findings is that the reorganization of gene topology induced by GATA-1 causes transcriptional repression by disrupting the physical interaction between the promoter proximal region and the upstream enhancer.
Figure 7.
Model of the chromosomal configuration of the Kit gene. In immature erythroid cells GATA-2 binds the -114 enhancer to form an activating chromatin loop. Upon maturation, GATA-1 replaces GATA-2 and increases the interaction among downstream elements at the expense of the enhancer/promoter interaction. Loss of the enhancer loop might contribute to repression of Kit. The effects of GATA-1 require FOG-1. The resolution limits of 3C preclude us from distinguishing whether the enhancer loops to the transcription start site or the +5 region. We favor the former possibility as enhancers commonly target promoter elements. The question mark represents possible intermediary protein complexes involved in loop formation.
An open question remains as to why GATA-2, but not GATA-1, is capable of establishing a chromatin loop between -114 and the promoter proximal region. GATA-2 binding throughout the Kit locus is highest at the -114 region while GATA-1 binding occurs at high levels also at +5, +58 and at lower levels at +73. Interactions among the downstream elements might be of higher affinity thereby competing for any possible upstream interaction. However, even in the absence of such competing interactions, the -114 region functions as enhancer only in GATA-2 expressing cells. Thus, competing chromatin loops by themselves are insufficient to explain GATA-2 selectivity of the enhancer. It will be interesting to examine the sequence context that confers GATA-2 selectivity on the -114 enhancer.
If the -114 region bound by GATA-2 is close to the Kit promoter, why is GATA-2 not detected at the promoter by ChIP? The likely answer is that transcription factors are crosslinked more efficiently to sites where they contact DNA directly. For example, at the β-globin locus, deletion of the LCR has little or no effect on the amount of detectable GATA-1 at the β-globin promoter despite the fact that the LCR harbors multiple GATA sites and loops to the promoter (Vakoc et al., 2005).
Generally, interactions among distal genomic regions could reflect the formation of chromatin loops that engage only sequences that regulate the expression of a single gene. Alternatively, interactions among regulatory elements might result from localization to shared subnuclear structures such as transcription factories (Marenduzzo et al., 2007; Osborne et al., 2004). In the latter case, proximity among regulatory regions would not be limited to one gene. Indeed, the actively transcribing Ahsp (Eraf) and Hbb (β-globin) genes which reside ∼24 megabases apart on mouse chromosome 7 colocalize to sites enriched for pol2 (Osborne et al., 2004). However, in the case of the Kit locus, colocalization to shared pol2 factories is insufficient to explain the observed chromosome configuration. Localization transcription factories might account for the proximity between the -114 region and the promoter at the active Kit gene, but it does not explain the clustering of the downstream elements bound by GATA-1 when Kit expression is extinguished and pol2 is lost. While it remains possible that repressive GATA-1 complexes move the Kit locus to subnuclear sites of shared co-repressor complexes, we favor a model in which specific transcription factors such as GATA-1 and GATA-2 convey unique intragenic chromosome structures. In the presence of GATA-2, upstream enhancer/promoter loops are favored; in the presence of GATA-1, enhancer and promoter elements are partitioned on different loops. The latter scenario might facilitate transcriptional repression. Indeed, previous work using episomal constructs showed that separating enhancer and promoter elements by forced loop formation can abrogate enhancer function (Ameres et al., 2005). More generally, in contrast to co-localization to transcription factories or other nuclear compartments, it is easy to imagine why intragenic long-range interactions are formed to maintain a gene-specific expression pattern and avoid unwanted cross-regulation among sequences belonging to different gene loci. It is noteworthy that while tissue-specific genes can be in close physical proximity to each other, clustering in nuclear space also occurs between active tissue-specific and widely expressed genes, suggesting that clustering per se does not convey gene-specific control of transcription (Simonis et al., 2006). Thus, regulation of gene expression likely involves multiple levels of higher order chromatin organization. Juxtaposition among cis-acting elements in a gene-specific manner is accompanied by additional interactions that result from localization to shared sites of transcription. 3C does not reliably distinguish among these layers of interactions.
Another corollary of our work is that general features of transcriptionally active chromatin such as histone modifications, DNase hypersensitivity, and the presence of pol2 and general transcription factors are insufficient to explain the chromosome conformation at the Kit locus. First, specific interactions among downstream GATA elements increase upon gene repression, while pol2 and histone acetylation are reduced. Importantly, loss of transcription per se does not explain the observed changes in chromatin organization as revealed by the use of the elongation blocker DRB. Second, while it might be argued that features of silent chromatin (i.e. loss of acetylation) might account for changes in chromosome folding, the spatial distribution of histone marks and pol2 in the transcribed region of the Kit locus does not demarcate in any way the base of the observed chromatin loops. In contrast, the distribution of GATA-1 coincides with the base of the loops. Therefore, the simplest interpretation of these results is that DNA binding by GATA-1 serves as anchor for a specific chromatin loop in the transcribed region of Kit. FOG-1 and perhaps other protein intermediates might serve to bridge these interactions in the nucleus.
Distinct chromatin configurations between active and inactive alleles have been observed at the imprinted Igf2 and H19 genes (Kurukuti et al., 2006; Murrell et al., 2004; Yoon et al., 2007). Moreover, methyl-CpG binding protein 2 (MeCP2) is required for select long-range chromosomal interactions at the repressed imprinted Dlx5 and Dlx 6 genes (Horike et al., 2005). Configurations of imprinted loci are established early in development according to epigenetic specifications and are subsequently maintained throughout ontogeny. In contrast, the Kit locus is rapidly reconfigured upon repression, indicating that chromatin loops are highly dynamic and might be employed as a means to dynamically regulate gene expression.
Forced loop formation can result in loss of enhancer function under certain conditions (Ameres et al., 2005). Clustering of insulator elements in nuclear space has been invoked as a wide-spread mechanism to inhibit enhancer function (Gaszner and Felsenfeld, 2006). However, it is likely that GATA-1 represses transcription by additional means. For example, GATA-1 via FOG-1 binds to the nucleosome remodeling and histone deacetylase (NuRD) complex (Hong et al., 2005; Rodriguez et al., 2005). A challenging but rewarding effort will be to assess the individual contribution of loop formation and co-repressor recruitment to gene repression. While the disruption of enhancer/promoter loops might eliminate enhancer function, it might not suffice to fully repress transcription. Most promoters retain a basal level of activity even in the absence of distal enhancers. Moreover, sustained transcription in the absence of a transcriptional enhancer might be facilitated by the presence of epigenetic marks within chromatin. The recruitment of co-repressor complexes such as NuRD might serve to erase epigenetic marks to fully repress transcription and to stably maintain the repressed state. Thus, recruitment of co-repressors complexes and the formation of chromatin loops might represent distinct but cooperative mechanisms to silence gene expression during development.
Experimental Procedures
Cell Culture
G1E cells were maintained as described (Letting et al., 2004; Weiss et al., 1997). Where indicated, cells were treated with 1 μM estradiol (21 hours) or with 75 μM 5,6 dichloro-1-β-d-ribofuranosylbenzimidazole (DRB, Sigma).
ChIP Assay
ChIP was performed as described (Letting et al., 2003). Antibodies against GATA-1 (N6), FOG-1 (M20), GATA-2 (H116), and RNA pol II (N20) were from Santa Cruz Biotechnology (Santa Cruz, CA) and anti-ER (Ab10) from Lab Vision Neomarkers (Fremont, CA). DNA was quantified by using real-time PCR with SYBR Green dye on an ABI Prism 7900 system (PE Applied Biosystems). ChIP primer sequences are listed in Supplemental Materials. Results were normalized to unprecipitated chromatin (input).
3C assay
The 3C assay was performed as described (Vakoc et al., 2005). Amounts of 3C ligation products were measured in triplicates by quantitative Taqman real-time PCR described recently (Vernimmen et al, 2007). For a detailed description and primer sequences see Supplemental Materials.
Transient transfections
Reporter constructs were generated by PCR and cloned into the pGL4.10 luciferase reporter vector (Promega, Madison WI). For details see Supplemental Materials.
Supplementary Material
Acknowledgements
We thank Drs. Douglas Vernimmen and Douglas Higgs for advice on Taqman 3C analysis, and Drs. Steve Liebhaber and Mitch Weiss for critically reading this manuscript. This work was supported by NIH grants DK58044 and DK54937. H.J. was supported by a fellowship from the American Heart Foundation #0625482U, and C.R.V by T32 HL007150-31.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ameres SL, Drueppel L, Pfleiderer K, Schmidt A, Hillen W, Berens C. Inducible DNA-loop formation blocks transcriptional activation by an SV40 enhancer. Embo J. 2005;24:358–367. doi: 10.1038/sj.emboj.7600531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrozpe G, Agosti V, Tucker C, Blanpain C, Manova K, Besmer P. A distant upstream locus control region is critical for expression of the Kit receptor gene in mast cells. Mol Cell Biol. 2006;26:5850–5860. doi: 10.1128/MCB.01854-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrozpe G, Timokhina I, Yukl S, Tajima Y, Ono M, Zelenetz AD, Besmer P. The W(sh), W(57), and Ph Kit expression mutations define tissue-specific control elements located between -23 and -154 kb upstream of Kit. Blood. 1999;94:2658–2666. [PubMed] [Google Scholar]
- Broudy VC. Stem cell factor and hematopoiesis. Blood. 1997;90:1345–1364. [PubMed] [Google Scholar]
- Cairns LA, Moroni E, Levantini E, Giorgetti A, Klinger FG, Ronzoni S, Tatangelo L, Tiveron C, De Felici M, Dolci S, et al. Kit regulatory elements required for expression in developing hematopoietic and germ cell lineages. Blood. 2003;102:3954–3962. doi: 10.1182/blood-2003-04-1296. [DOI] [PubMed] [Google Scholar]
- Cantor AB, Katz SG, Orkin SH. Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakaryocytic maturation. Mol Cell Biol. 2002;22:4268–4279. doi: 10.1128/MCB.22.12.4268-4279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Does looping and clustering in the nucleus regulate gene expression? Curr Opin Cell Biol. 2004;16:256–262. doi: 10.1016/j.ceb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Chang AN, Cantor AB, Fujiwara Y, Lodish MB, Droho S, Crispino JD, Orkin SH. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc Natl Acad Sci U S A. 2002;99:9237–9242. doi: 10.1073/pnas.142302099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispino JD. GATA1 in normal and malignant hematopoiesis. Semin Cell Dev Biol. 2005;16:137–147. doi: 10.1016/j.semcdb.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Crispino JD, Lodish MB, MacKay JP, Orkin SH. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol Cell. 1999;3:219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- Cullen KE, Kladde MP, Seyfred MA. Interaction between transcription regulatory regions of prolactin chromatin. Science. 1993;261:203–206. doi: 10.1126/science.8327891. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnitski L, Hardison RC, Li J, Yang S, Kolbe D, Eswara P, O’Connor MJ, Schwartz S, Miller W, Chiaromonte F. Distinguishing regulatory DNA from neutral sites. Genome Res. 2003;13:64–72. doi: 10.1101/gr.817703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 Function, a Paradigm for Transcription Factors in Hematopoiesis. Mol Cell Biol. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Liew C, Holmes M, Kowalski K, Mackay J, Crossley M. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO Journal. 1999;18:2812–2822. doi: 10.1093/emboj/18.10.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P. Transcriptional control thrown for a loop. Curr Opin Genet Dev. 2006;16:490–495. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR, Rakowski C, Blobel GA. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. Embo J. 2005;24:2367–2378. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- Kluppel M, Nagle DL, Bucan M, Bernstein A. Long-range genomic rearrangements upstream of Kit dysregulate the developmental pattern of Kit expression in W57 and Wbanded mice and interfere with distinct steps in melanocyte development. Development. 1997;124:65–77. doi: 10.1242/dev.124.1.65. [DOI] [PubMed] [Google Scholar]
- Kooren J, Palstra RJ, Klous P, Splinter E, von Lindern M, Grosveld F, de Laat W. Beta-globin active chromatin Hub formation in differentiating erythroid cells and in p45 NF-E2 knock-out mice. J Biol Chem. 2007;282:16544–16552. doi: 10.1074/jbc.M701159200. [DOI] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Herblot S, Saint-Denis M, Martin R, Begley CG, Porcher C, Orkin SH, Hoang T. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 2002;100:2430–2440. doi: 10.1182/blood-2002-02-0568. [DOI] [PubMed] [Google Scholar]
- Letting DL, Chen YY, Rakowski C, Reedy S, Blobel GA. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc Natl Acad Sci U S A. 2004;101:476–481. doi: 10.1073/pnas.0306315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letting DL, Rakowski C, Weiss MJ, Blobel GA. Formation of a Tissue-Specific Histone Acetylation Pattern by the Hematopoietic Transcription Factor GATA-1. Mol Cell Biol. 2003;23:1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenduzzo D, Faro-Trindade I, Cook PR. What are the molecular ties that maintain genomic loops? Trends Genet. 2007;23:126–133. doi: 10.1016/j.tig.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Munugalavadla V, Dore LC, Tan BL, Hong L, Vishnu M, Weiss MJ, Kapur R. Repression of c-kit and its downstream substrates by GATA-1 inhibits cell proliferation during erythroid maturation. Mol Cell Biol. 2005;25:6747–6759. doi: 10.1128/MCB.25.15.6747-6759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, Weiss MJ. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000;24:266–270. doi: 10.1038/73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Pal S, Cantor AB, Johnson KD, Moran TB, Boyer ME, Orkin SH, Bresnick EH. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc Natl Acad Sci U S A. 2004;101:980–985. doi: 10.1073/pnas.0307612100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJ, Vyas P, de Boer E, Grosveld F, Strouboulis J. GATA-1 forms distinct activating and repressive complexes in erythroid cells. Embo J. 2005;24:2354–2366. doi: 10.1038/sj.emboj.7600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylski M, Welch JJ, Chen YY, Letting DL, Diehl JA, Chodosh LA, Blobel GA, Weiss MJ. GATA-1-mediated proliferation arrest during erythroid maturation. Mol Cell Biol. 2003;23:5031–5042. doi: 10.1128/MCB.23.14.5031-5042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006 doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, Deconinck AE, Cantor AB, Rieff HI, Fujiwara Y, Corfas G, Orkin SH. FOG-2: A novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc Natl Acad Sci U S A. 1999;96:950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang AP, E VJ, A TC, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among Distant Regulatory Elements at the beta-Globin Locus Requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. Embo J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang Y, Cheng Y, Zhou Y, King DC, Taylor J, Chiaromonte F, Kasturi J, Petrykowska H, Gibb B, et al. Experimental validation of predicted mammalian erythroid cis-regulatory modules. Genome Res. 2006;16:1480–1492. doi: 10.1101/gr.5353806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MJ, Keller G, Orkin SH. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1- embryonic stem cells. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properities of transcription factor GATA-1 revealed by phenotypic rescue of a gene targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Wessely O, Mellitzer G, von Lindern M, Levitzki A, Gazit A, Ischenko I, Hayman MJ, Beug H. Distinct roles of the receptor tyrosine kinases c-ErbB and c-Kit in regulating the balance between erythroid cell proliferation and differentiation. Cell Growth Differ. 1997;8:481–493. [PubMed] [Google Scholar]
- Yoon YS, Jeong S, Rong Q, Park KY, Chung JH, Pfeifer K. Analysis of the H19ICR insulator. Mol Cell Biol. 2007;27:3499–3510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.