Abstract
In Bacillus subtilis, parE and parC were shown to be essential genes for the segregation of replicated chromosomes. Disruption of either one of these genes resulted in failure of the nucleoid to segregate. Purified ParE and ParC proteins reconstituted to form topoisomerase IV (topo IV), which was highly proficient for ATP-dependent superhelical DNA relaxation and decatenation of interlocked DNA networks. By immunofluorescence microscopy and by directly visualizing fluorescence by using green fluorescence protein fusions, we determined that ParC is localized at the poles of the bacteria in rapidly growing cultures. The bipolar localization of ParC required functional ParE, suggesting that topo IV activity is required for the localization. ParE was found to be distributed uniformly throughout the cell. On the other hand, fluorescence microscopy showed that the GyrA and GyrB subunits of gyrase were associated with the nucleoid. Our results provide a physiologic distinction between DNA gyrase and topo IV. The subcellular localization of topo IV provides physical evidence that it may be part of the bacterial segregation machinery.
Chromosome segregation is a fundamental process that ensures the equal distribution of newly replicated chromosomes into the two daughter cells. In eukaryotes, there are well defined structures and components that form a mitotic apparatus for the dynamic process of chromosome movement, which eventually positions the chromosomes in daughter cells (1). In bacteria, analogous structures such as centromeres (to provide sites for attachment), cytoskeletal or microtubular spindles, and motors (to provide the driving force for the chromosomal movement) have not been identified. How the apparently simpler bacteria segregate their chromosomes remains an unresolved problem. Early proposals of chromosome segregation relying on the growth of the membrane to which chromosomes were attached to provide the driving force are inadequate because of the randomness of membrane insertion. Recent experiments clearly point to the existence of a specific cellular machinery that is actively involved in bacterial chromosome segregation. The segregation machinery is likely to involve specific protein components, which recognize and bind to a special region(s) of the chromosome, and membrane components, which serve as an anchor (2). Identification and descriptions of some of these components are beginning to emerge. It has been known for some time that plasmid DNA segregation requires specific plasmid-encoded partition proteins and a specific DNA site (3). Chromosomal genes homologous to the plasmid partition genes have been identified and analyzed genetically in Bacillus and in Caulobacter (4, 5). More recently, a possible direct role for them in chromosome segregation was supported by the observation that fluorescence techniques localize them at the poles of these cells (5, 6). In Bacillus subtilis, it has been shown convincingly that the circular chromosome is oriented spatially in such a way that the replication–origin region is localized preferentially near the poles early in the cell cycle (7). Although no specific DNA recognition sequence has been identified for the B. subtilis partition protein SpoOJ, its preferred binding to the origin-containing one-third of the chromosome, combined with its polar localization, makes it attractive to postulate its direct involvement in the chromosome partition machinery.
Because of the extraordinary length of the chromosome, untangling the replicating chromosome is a prerequisite for chromosome segregation. Hence, DNA topoisomerase, the enzyme designed to resolve DNA topological problems, is expected to participate in this process. DNA gyrase, the prototypical type II DNA topoisomerase, because of its ability to effect duplex DNA strand passage through transient strand breaks, has been suggested to play a role in chromosome segregation almost from the time it was discovered (8, 9). Since the discovery of a second homologous type II topoisomerase, Escherichia coli topoisomerase IV (topo IV) (10), which is more proficient in unknotting and decatenating DNA than DNA gyrase, more emphasis has been placed on topo IV and its subunits ParE and ParC as the proteins that may be involved in plasmid and chromosome segregation. Morphologic defects seen in parE or parC mutants provide further support for its direct role in chromosome segregation because they fail to segregate the replicated nucleoids. Furthermore, E. coli topo IV and not gyrase was found to be necessary for the decatenation of linked, replicative plasmid DNA in vivo and in vitro (11, 12). Exactly how the enzyme, which presumably decatenates and untangles the replicating chromosome, might aid in the implementation of chromosome movement to distribute the replicated chromosomes into daughter cells is not known.
Because of the ease of chromosomal gene manipulation and the opportunity to investigate both symmetric and asymmetric cell division and sporulation (13, 14), we chose to examine the role of B. subtilis topo IV in this process. We first identified, cloned, and characterized the topo IV genes in B. subtilis. We then determined the subcellular location of both gyrase and topo IV in actively growing cells by using immunofluorescence microscopy with specific antibodies against these type II topoisomerases and by using fusion proteins with the Aequorea victoria green fluorescence protein (GFP) (15). We found that the subcellular distributions of gyrase and topo IV are different. GyrA and GyrB subunits of gyrase molecules are mainly nucleoid-associated. ParC is mainly bipolarly localized, and this distribution depends on functional ParE. The subcellular location of topo IV suggests that it is involved not only in the untangling of replicated chromosome but that it also may be part of the chromosome segregation machinery.
MATERIALS AND METHODS
Plasmid and Strain Constructions.
B. subtilis topo IV subunit genes parE and parC (GenBank accession no. AF024713) and DNA gyrase subunit genes gyrB and gyrA (GenBank accession no. X02369) were cloned into pET21 or its derivatives (Novagen) for the over expression of these proteins in E. coli BL21(DE3) (Novagen). For gene disruption experiments, we cloned an internal fragment of parE (from residues 52–462) or parC (from residues 47–224) into the conditional integrational vector pKSV7 (16). A mutant of GFP with improved fluorescence signal without red-shifting the excitation maximum (17) was used to prepare fusion constructs with each of the four type II DNA topoisomerase subunit genes. In brief, the gfp sequence was fused in-frame to the C termini of these genes in the pET-based or pGEM4-based (Promega) plasmids for propagation in E. coli. A neomycin cassette (18) also was inserted into these plasmids for selectability in B. subtilis. pC–gfp containing parC (codons 243–806)–gfp, pE–gfp containing parE (codons 462–655)–gfp, and pA–gfp containing gyrA (codons 174–821)–gfp are pET21-based plasmid derivatives. pB–gfp containing gyrB (codons 283–638)–gfp was constructed similarly with the neomycin cassette but cloned into a pGEM4 vector. These plasmids do not replicate in B. subtilis. The four topoisomerase subunit gene–gfp fusions were introduced into B. subtilis by single crossover into the respective genes by using the above plasmids as integrative plasmids selecting for neomycin resistance. The integration of these plasmids at the correct sites was checked by PCR by using primers flanking the expected inserts for the presence of gfp in the chromosome. These constructs were designed such that the expressions of the fusion GFPs were driven by their endogenous promoter in these strains. The exclusive presence of the larger topoisomerase subunit fusion GFPs in place of the wild-type protein was confirmed by Western blot analyses probed with topoisomerase antibodies. The strains harboring the topoisomerase gene fusions were viable with growth rates in rich media similar to that of the wild-type cells.
CB10 (hisA1, trpC2) (19), a derivative of 168, was used as the wild-type B. subtilis strain. The parE mutant H528P, which carries a single substitution of Pro at residue His-528, was constructed by using PCR oligomutagenesis of a plasmid carrying the parE gene followed by transforming the mutated plasmids into CB10 and screening for temperature sensitivity for growth. The mutation was confirmed by sequencing a PCR product amplified from the genomic DNA with flanking sequences in the parE region. The method of construction and detailed characteristics of temperature-sensitive (ts) mutants will be described elsewhere.
Antibodies and Western Blot Analysis.
Anti-GyrA rabbit antibody was raised against a purified N-terminal fragment of gyrA, which harbors residues 1–607. An antibody against ParC was raised in a rabbit (HRP, Denver, PA) by using as an immunogen a purified his-tagged C-terminal fragment of ParC, which harbors residues 499–806-(his)6. Anti-ParE rabbit antibody (HRP) was raised against full length ParE. The anti-ParE antibody showed significant reaction to both ParE and GyrB, which were well resolved in SDS polyacrylamide gels. B. subtilis proteins from cell extracts were analyzed in SDS gels (7.5%) and were transferred to poly(vinylidene difluoride) membranes (Amersham) by using a Bio-Rad Trans-Blot apparatus. The immunoblots, after reacting with the primary antibodies against the individual topoisomerase subunits, were probed with cy5-conjugated goat anti-rabbit IgG secondary antibody (Amersham). The blots were scanned by using a Storm 860 Fluorimager (Molecular Dynamics), and the band intensities were quantified by comparison with known amounts of purified proteins analyzed on the same blot by using the imagequant software (Molecular Dynamics). We estimate that there are ≈750 copies of ParC, 4,000 copies of parE, 4,000 copies of GyrA, and 7,000 copies of GyrB per cell during exponential growth of B. subtilis cells in Luria–Bertani medium at 37°C.
Fluorescence Microscopy.
For immunofluorescence microscopy, cells were grown at 37°C in Luria–Bertani broth to early exponential growth phase. After washing in M9 minimal medium (20), the cells were fixed in paraformaldehyde and glutaraldehyde and permeabilized by lysozyme as described by Harry et al. (21), except that the cells were not dehydrated in methanol and acetone before the immunoreactions. The cells were spread on poly-l-lysine (0.01%, Sigma)-coated slides. Rabbit antisera directed against topoisomerase subunit proteins were used at 1:100 to 1:1000 dilution as the primary antibodies. Fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit IgG (H+L) (Pierce) was used at 1:100 dilution as the secondary antibody. Finally, the slides were mounted by using a Prolong antifade kit (Molecular Probes) and were air-dried overnight. Samples were examined in two Nikon Diaphot fluorescence microscopes. One microscope was equipped with Nomarski: DIC attachments and dichroic filters that allowed for the examination of cell shape by Nomarski optics (Nikon) while simultaneously viewing with one of either FITC (or GFP) or 4′,6-diamidino-2-phenylindole (DAPI) emission conditions but did not allow for double fluorescence observation. The second microscope was equipped with a triple dichroic XF56 filter set (Omega Optical, Brattleboro, VT) with which FITC (or GFP) and DAPI double-fluorescence could be detected simultaneously. The intensities were lower in the latter arrangement. Samples were viewed with a Plan/Fluor ×100 objective lens (Omega Optical). Images were collected by using a Photometrics (Tucson, AZ) cooled charge-coupled device camera or a VE1000 video camera (MTI, Michigan City, IN) and digitized by using openlab software (version 1.7.2) (Improvision). Digitized images were false-colorized and were superimposed by using photoshop 4.0 (Adobe Systems, Mountain View, CA).
For observation of GFP fluorescence, cells were grown in Luria–Bertani medium with vigorous aeration to very early exponential phase. After washing in M9 minimal medium, the cells were spread on poly-l-lysine (0.01%)-coated slides for same-day observation or were resuspended in a buffer containing 40 mM NaN3, 50% sucrose, 0.5 M Tris (pH 7.5), and 0.77 M NaCl for storage on ice for next-day observation after the removal of the preservation buffer (22). The nucleoids were stained with DAPI (2.5 μg/ml, Sigma).
RESULTS
parE and parC Genes Are Required for Chromosome Segregation.
In B. subtilis, the parE and parC gene fragments were identified by PCR by using degenerate primers spanning the highly conserved regions of gyrB and gyrA followed by multiple rounds of forward and inverse PCR (23). The two par genes were found to be contiguous, transcribed in the clockwise direction, and located near the 1900-kbp position on the 143-kbp NotI fragment, opposite from where the homologous gyrB and gyrA are located near the origin of replication (data not shown). The predicted amino acid sequence of B. subtilis ParE (655 aa in size) is 56% identical to B. subtilis GyrB and 41% identical to E. coli ParE. The predicted amino acid sequence of B. subtilis ParC (806 aa) is 42% identical to B. subtilis GyrA and 32% identical to E. coli ParC. Purified B. subtilis ParE and ParC proteins, when mixed together in an equimolar ratio, had an ATP-dependent superhelical DNA relaxation (see Fig. 3) and kinetoplast DNA decatenation activities and were not capable of DNA supercoiling. These enzymatic activities are similar to those of E. coli topo IV; hence, the B. subtilis ParE and ParC are subunits of DNA topo IV (10).
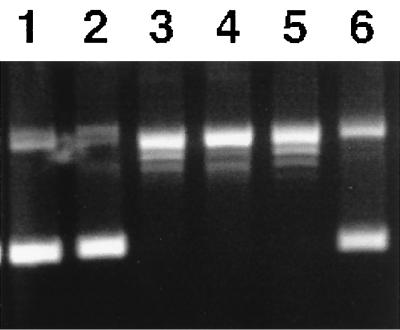
Figure 3.
Topo IV DNA relaxation activity of reconstituted wild-type ParE and tsParE(H528P) with ParC. The complete reaction mixture consisted of 40 mM Tris (pH 7.5), 6 mM MgCl2, 0.1 M K-glutamate, 1 mM DTT, 1 mM ATP, 50 μg of BSA, and 1 μg of supercoiled DNA. Incubation was for 15 min at 37°C unless specified. The product was analyzed on a 1% agarose gel. Lanes: (1) no enzyme; (2–4) with wild-type enzyme; (2) no ATP; (3) incubated at 37°C; (4) incubated at 45°C; (5–6) with mutant enzyme; (5) incubated at 37°C; (6) incubated at 45°C.
Two types of experiments were used to assess the in vivo function of these proteins. We tested whether these genes are essential for B. subtilis viability and determined the morphological phenotype associated with defects in parE or parC by gene disruption experiments and by the use of temperature-sensitive mutants. We performed gene disruption experiments using a conditional integrational plasmid, pKSV7, (16) carrying an internal fragment of either the parE (from residues 52–462) or parC (from residues 47–224). The chloramphenicol-resistant integrative plasmid is temperature-sensitive for plasmid replication. Under nonpermissive conditions (45°C) and in the presence of chloramphenicol, the plasmid carrying the B. subtilis DNA insert cannot replicate in B. substilis. Hence, it must integrate into the chromosome via homologous recombination between the plasmid and chromosomal copies of the parE or parC sequences, thereby disrupting the reading frame of the chromosomal gene. In B. subtilis cells harboring these integrative plasmid derivatives, on temperature shift from 30 to 45°C, cell viability dropped by about four orders of magnitude with both parE and parC disruptions. After 2 hr at 45°C, ≈20% of the cells were elongated to about double length with a single enlarged and asymmetrically located nucleoid, as revealed by microscopic examination under DAPI staining [data not shown; the morphology was similar to that described for ts parE mutants described below (Fig. 2C) and those of ts parE or parC mutants in E. coli and in Salmonella typhimurium (24)]. This morphology clearly indicates a failure to segregate replicated chromosomes. In temperature-sensitive mutants of B. subtilis parE, after a temperature shift from 30 to 45°C for 2 hr, ≈15–30% of the cells also became elongated, and the nucleoid, revealed by DAPI-staining, was asymmetrically located. Also seen were ≈5% of anucleate cells. An example of the ts parE defective phenotype (parE H528P) under nonpermissive conditions is depicted in Fig. 2C, where the location of the elongated nucleoid relative to the cell shape is visualized clearly by using a combination of DAPI fluorescent staining and Nomarski cell imaging for the same field. Anucleate minicells also are seen in the Nomarski image. This morphology is in contrast to a wild-type nucleoid-staining pattern exemplified by the parE–gfp fusion cells shown in Fig. 2G. (A temperature-sensitive mutant of parC is not available.) Based on the biochemical activity of the purified proteins and the morphology of cells defective in these genes, we conclude that, analogous to E. coli, B. subtilis parE, and parC, the subunit genes for topo IV are required for the proper segregation of replicated chromosomes into daughter cells.
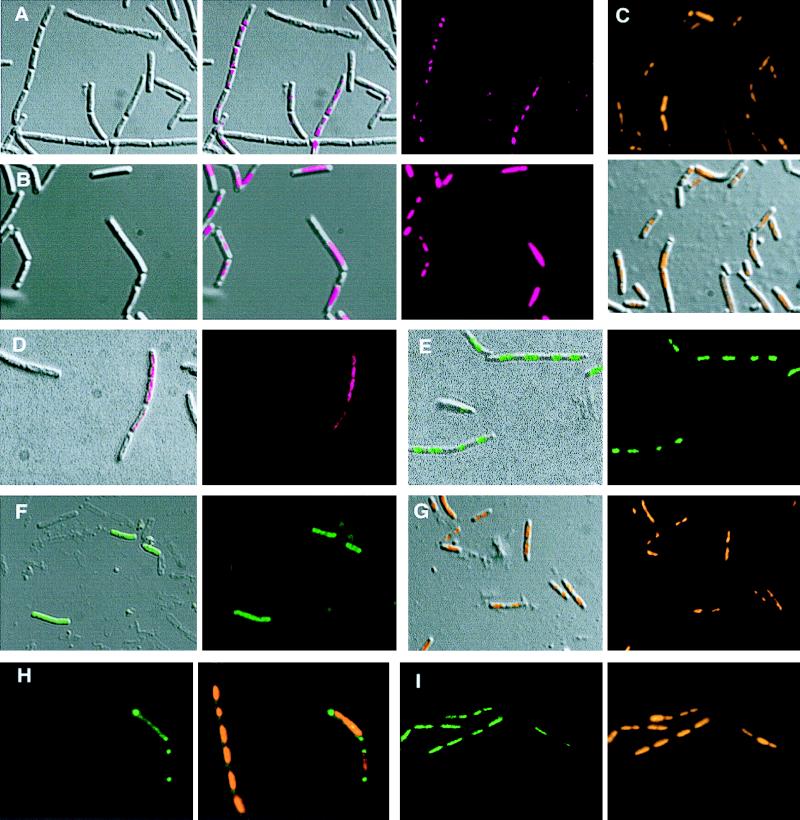
Figure 2.
Localization of topo IV and gyrase subunits in vegetatively growing B. subtilis cells. (A and B) Three images each: Nomarski (Left), fluorescence (Right), and an overlay of the left and right images (Center). (A) Immunostaining of ParC in wild-type cells. (B) Immunostaining of ParC in ts parE(H528P) grown under nonpermissive conditions (45°C) for 2 hr. (C–G) Two images each: a fluorescent image and an overlay of fluorescence onto Nomarski cell shape image. (C) DAPI staining of cells grown in B. (D) Immunostaining of GyrA in wild-type cells. (E) Visualization of GyrA–GFP. (F) ParE–GFP. (G) DAPI staining of cells containing parE–gfp fusion. (H and I) Images from simultaneous viewing with two different fluorescences. (H) ParC–GFP fluorescence (Left) and an overlay of DAPI fluorescence for nucleoid and GFP (Right). (I) GyrA–GFP fusion fluorescence (Left) and DAPI staining of the same field (Right). Color code for different fluorescence: magenta for FITC-conjugated secondary antibody used in immunofluorescence, green for GFP fluorescence, and orange for DAPI.)
ParC Has Bipolar Localization.
To identify the subcellular localization of these partition proteins, we used immunofluorescence microscopy techniques. Rabbit antibody raised against the C-terminal third of the ParC (from residues 499–806) was specific to ParC, and, in particular, it did not cross react with the paralogous GyrA. Similarly, rabbit antibody raised against the N-terminal three-quarters of GyrA (from residues 1–607) was specific for GyrA and did not cross react with ParC. Fig. 1 shows immunoblots probed with anti-ParC, anti-GyrA, and anti-ParE antibodies. In rapidly growing wild-type vegetative cells (doubling time ≈25 min), ParC antibody complexes were visualized by the fluorescence of a secondary antibody conjugated to FITC, and the cell shapes were depicted clearly by using Nomarski optics (Nikon) (Fig. 2A). Although many bacteria grew as chains, the septations that define the individual bacteria were clearly visible. Superposition of the fluorescent and Nomarski images showed that the highest concentration of ParC was located at both poles of these rod-shaped bacteria. There was a gradient of ParC with a lower concentration of proteins toward midcell. Indirect ParC fluorescence was seen in a majority (≈80%) of the cells, although with varying intensities, and the highest concentration was always at the poles.
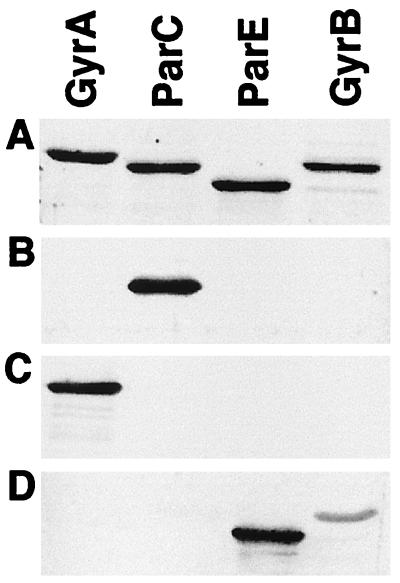
Figure 1.
Specificity of antisera against subunits of type II topoisomerases. Purified B. subtilis GyrA, ParC, ParE, and GyrB (0.75 μg each) were analyzed on an SDS gel. The gel either was stained for total protein or was processed for immunoblotting. (A) Stained with SYPRO Red (Molecular Dynamics). (B) Probed with antiserum against ParC (residues 499–806). (C) Probed with antiserum against GyrA (residues 1–607). (D) Probed with antiserum against full length ParE.
When the GyrA-specific antibody was used to visualize its subcellular localization in the same population of wild-type B. subtilis cells, the distribution pattern was different. GyrA appeared distributed within the interior of the cell and was not concentrated in the polar region (Fig. 2D). The distribution was indistinguishable from the wild-type nucleoid staining pattern (represented by Fig. 2G).
Visualization of Topoisomerase-GFP.
To observe the subcellular localization of the topoisomerase proteins directly, without fixation, and to determine the subcellular localization of GyrB and ParE against which specific antibodies were not available, we constructed gfp fusions in each of the four type II topoisomerase genes, parE and parC for topo IV and gyrB and gyrA for gyrase. All four fusion proteins appear to be functional because cells harboring each of these fusions replacing the wild-type copy of the topoisomerase subunit genes are viable, with growth rates comparable to those of the wild-type cells propagated at 37°C. (ParC–gfp cells grew poorly at temperatures below 30°C.) By using the fluorescence of ParC–GFP as a reporter, ParC was again seen accumulated at the poles not overlapping with the DAPI-stained nucleoid. Some weak fluorescence signals along the length of the cell were seen sometimes (Fig. 2H). These signals were in contrast to the GyrA–GFP signal, which coincided with the nucleoid staining (Fig. 2I). ParC–GFP fluorescence was seen in ≈6% of the cells, sometimes not with equal intensity at both poles. The reason for the low frequency of fluorescence signal in parC–gfp cells is not clear. The number of copies of ParC per cell was the lowest among the four type II topoisomerase subunits. It is present at ≈750 copies per cell, corresponding to ≈20% of ParE, GyrA, or GyrB. Perhaps ParC–GFP is often below a threshold of detectable GFP fluorescence under these experimental conditions. Because ParC was found to be concentrated at the membranous poles of the cell based on immunofluorescence (Fig. 2A), this property may have further decreased the quantum efficiency of GFP fluorescence emission because it is believed that the autofluorescence of GFP requires molecular oxygen and that highly soluble proteins are more fluorescent.
ParE–GFP exhibited fluorescence throughout the entire cell as shown in the superimposed image overlapping the cell shape and the fluorescence signal (Fig. 2F). In some images of ParE–GFP, the fluorescence was enriched at both poles similarly to but less intensely than those seen with ParC–GFP (data not shown). For comparison, parE–gfp cells also were stained with DAPI to display the nucleoid location (Fig. 2G). The dispersion of ParE–GFP throughout the cell is distinct from the concentration of DAPI stain in the nucleoid bodies. We interpret these images to mean that the ParE proteins are distributed throughout the cytoplasm, with perhaps a slight concentration at the poles of the cell.
The gyrase–GFP fusions again showed a different distribution. GyrA–GFP was associated with a nucleoid (Fig. 2E). The fluorescence is highest at distinct packets spaced along the chains of cells. This pattern appears to be the same as that of the DAPI-stained nucleoid. (represented by Fig. 2G). The GyrB–GFP fluorescence pattern was similar to that displayed by GyrA–GFP and also was found to be associated with the nucleoid (data not shown).
Bipolar Localization of ParC Requires Functional Topo IV.
To test whether complete topo IV is required for the observed bipolar localization of ParC, we performed immunofluorescence microscopy of ParC in B. subtilis cells that are temperature-sensitive for parE. This mutant harbors a Pro substitution at His-528 at the C-terminal region of ParE. Purified ParE-H528P, when mixed with the wild-type ParC, reconstituted a topo IV enzyme that provided full ATP-dependent supercoiled DNA relaxation activity at 37°C but was inactive at 45°C (Fig. 3). This result is consistent with the ts growth phenotype. The reconstituted ts topo IV was also inactive at decatenating interlocked kinetoplast DNA at 45°C (data not shown). In this mutant, when the cells were shifted from 37°C to 45°C for 2 hr, ParC immunofluorescence was no longer localized to the poles. It was associated mainly with the nucleoid (Fig. 2 B and C). Western blotting experiments further showed that, after temperature shifts, ParE accumulation in these cells decreased to <25% of the permissive level, whereas ParC and the GyrB and GyrA levels remained unchanged (data not shown). We conclude that the bipolar localization seen in wild-type cells requires a functional topo IV that consists of both ParE and ParC subunits.
DISCUSSION
The subcellular localization of topo IV suggests that this decatenating enzyme is located strategically in a position compatible with its being part of a machinery responsible for equal partitioning of the chromosome into the progeny cells. The location is reminiscent of the observed subcellular localization of Caulobacter crescentus ParB, a cell cycle-dependent partition protein (5), and is reminiscent of the observed subcellular localization of the B. subtilis SpoOJ, which is required for chromosome segregation in vegatatively growing and in sporulating cells (4). [The bipolar localization of the B. subtilis SpoOJ was depicted relative to the septation protein FtsZ (25) and not to the whole cell image (6). Based on its high sequence identity to the Caulobacter ParB, we consider SpoOJ to be similarly polar-localized for this discussion]. We therefore favor the proposal that both the topo IV subunits and the SpoOJ-like origin region binding proteins are part of the membranous partition apparatus that may be a counterpart to the mitotic apparatus of eukaryotic cells. The identification of an enzyme that is responsible for the decatenation of replicating chromosomes as part of the membranous segregation machinery provides a continuity in chromosome decatenation and subsequent segregation and suggests that chromosomal untangling and movement could be linked processes. The bipolar localization of ParC also hints at an intriguing notion that the cell poles of this bacterium may be in communication with or integrated into the segregation apparatus. Cell poles can be thought of as remnants of former sites of cell division. In chains of cells, as rapidly growing B. subtilis cells tend to form (Fig. 2), the neighboring poles of adjacent cells are generated from the site of the most recently completed cell division. The location of the ParC at the poles of the chain of cells may be interpreted as the leftover ParC from the recently completed chromosomes partition event.
The subcellular localizations of gyrase are markedly different from those of topo IV. GyrA and GyrB are associated mainly with the nucleoid. These localizations can be seen with fixed cells by using immunofluorescence examination or in cells without fixation by using fusion GFPs. The subcellular localizations are consistent with the expected function of gyrase in maintaining the superhelicity of DNA and acting during transcription and the chain-elongation phase of replication. The different subcellular localizations of the homologous gyrase and topo IV also point to a rationale for why bacteria require both type II enzymes for growth: Gyrase, by its association with the nucleoid, functions at the level of DNA maintenance, whereas topo IV has a role as part of the chromosome segregation machinery. This report provides physical evidence for the distinct physiologic roles of these topoisomerases. Mutations in some alleles of E. coli gyrB and gyrA cause elongated cell morphology with unsegregated nucleoids (26, 27). Based on the subcellular localization described here and on other biochemical evidence (12), it is possible that failure to propagate replication forks because of inactivation of DNA gyrase is also sufficient to cause the replicated portions of a chromosome to remain congregated and thereby to provide the appearance of a defective chromosome partition.
The subcellular localizations of B. subtilis GyrA and GyrB observed in this report appear to be different from those of E. coli. Using immunogold preparations of fixed and resin-imbedded E. coli, electron microscopic images showed that GyrA and GyrB were distributed randomly throughout the cytoplasm (28). Whether the antibodies used in the analysis could cross react with the homologous ParE or ParC subunits, respectively, was not addressed. GFP fusion and immunofluorescence in light microscopy offered a more direct visualization of subcellular localization, albeit at lower resolution. However, the minimal handling of cells may be less likely to cause distortion. It is also possible that, depending on the growth conditions, the methods of sample preparations, and the relative abundance of the topoisomerases in different species, different populations of the proteins may be visualized preferentially.
ParE–GFP (Fig. 2G) is found distributed throughout the cell, making it distinct from the bipolar localization of the another topo IV subunit protein, ParC. Perhaps the lower abundance of the steady-state level of ParC (one-fifth the level of ParE as determined by quantitative Western analysis) makes it difficult to detect any uniformly distributed ParC. We hypothesize that, at some stages of the cell cycle, by yet unidentified mechanisms, ParC may become aggregated and migrate to the poles. Only in this concentrated state was ParC detected by either immunofluorescence staining or as signals generated with GFP fusion. The observation that, in the absence of ParE, ParC bipolar localization was abolished and that instead it colocalized with the nucleoid lends support to the notion that ParC localization may follow a dynamic pattern. It may migrate to the poles from a more general distribution. It is also possible that only a subpopulation of the ParC and ParE complex is involved in the polar localization. Little is known about the intracellular regulation of DNA gyrase levels, and even less in known about ParE or ParC regulation in any bacteria. Studies are in progress to determine whether there is cell cycle-dependent regulation of the topo IV expression. In eukaryotic cells, recent reexaminations of the localization of topo II on mitotic chromosomes suggest that it has a dynamic pattern of distribution following the cell cycle and that it is concentrated at the centromeres at metaphase (29).
The observed apical bipolar localization of B. subtilis ParC implicitly suggests that it is connected in some manner to the cell membrane. However, purified topoisomerase subunit proteins from E. coli and B. subtilis are soluble in solution without added detergent (refs. 12 and 30; this report). On the other hand, intracellular E. coli ParE and ParC or B. subtilis ParC appeared to be membrane-bound, based on their association with fast sedimenting material or in sucrose gradient analysis (ref. 30; J.L.L. and W.M.H., unpublished observation). In the absence of a convincing cytoskeletal-like structure, bacterial membrane has become accepted as the likely support and anchor for DNA and DNA complex in many proposed segregation mechanisms (2). DNA, and specifically oriC DNA, is thought to be membrane-associated based on its sedimentation properties (31). However, attachment to the membrane is not limited to the origin of replication. In E. coli, there are genetic arguments suggesting that oriC may not be the bacterial centromere-like sites for chromosome segregation because oriC containing plasmids segregated randomly and did not interfere with chromosome segregation. We favor the idea that the apparent membrane association of ParC may be indirect, involving specific unidentified DNA sites, cellular components, and proteins in promoting or facilitating this association. The segregation machinery may consist of other components such as SpoOJ-like DNA binding proteins and many others. E. coli MukB, an ATP-binding filamentous protein with globular domains at both ends, has been proposed to provide motor force for chromosome segregation (32). However, the universal presence of the MukB-like protein in bacteria has not been substantiated. Other universally conserved proteins, such as some of the Min-proteins (33) required for proper positioning of replicated chromosomes and the HU proteins (34) required for chromosome organization, may be possible players in the assembly and regulation of this machinery. The identification of new genes associated with chromosome segregation will be aided greatly by the completion of the genomic sequence of B. subtilis (35), and topo IV may provide access for the discovery of new elements in this process. One of the key issues in understanding bacterial segregation mechanisms is the identification of components that provide the driving force for chromosome movement. Topo IV is an attractive addition to the segregation apparatus because it is a DNA-dependent ATPase capable of DNA translocation.
Acknowledgments
We thank Steve Wiley and the Cell Imaging Core Facility for advice and use of the fluorescence microscopes; S. Casjens for help with image processing and for comments on the manuscript; P. Youngman for Bacillus integrational vector; and Wen-qing Yu for DNA sequencing. Part of this work was supported by a grant from the National Institutes of Health (GM21960).
ABBREVIATIONS
- topo IV
topoisomerase IV
- GFP
green fluorescence protein
- FITC
fluorescein isothiocyanate
- DAPI
4′,6-diamidino-2-phenylindole
- ts
temperature-sensitive
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF024713).
References
- 1.Barton N R, Goldstein L. Proc Natl Acad Sci USA. 1996;93:1735–1742. doi: 10.1073/pnas.93.5.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wheeler R, Shapiro L. Cell. 1997;88:577–579. doi: 10.1016/s0092-8674(00)81898-x. [DOI] [PubMed] [Google Scholar]
- 3.Hiraga S. Annu Rev Biochem. 1992;61:283–306. doi: 10.1146/annurev.bi.61.070192.001435. [DOI] [PubMed] [Google Scholar]
- 4.Ireton K, Gunther N W, IV, Grossman A D. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohl D A, Gober J W. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 6.Lin D C, Levin P A, Grossman A D. Proc Natl Acad Sci USA. 1997;94:4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webb C D, Teleman A, Gordon S, Straight A, Belmont A, Lin D C, Grossman A D, Wright A, Losick R. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 8.Steck T R, Drlica K. Cell. 1984;36:1081–1088. doi: 10.1016/0092-8674(84)90058-8. [DOI] [PubMed] [Google Scholar]
- 9.Wang J C. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 10.Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H. Cell. 1990;63:393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 11.Adams D, Shekhtman E, Zechiedrich E, Schmid M, Cozzarelli N R. Cell. 1992;71:277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- 12.Peng H, Marians K. Proc Natl Acad Sci USA. 1993;90:8571–8575. doi: 10.1073/pnas.90.18.8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wake R G, Errington J. Annu Rev Genet. 1995;29:41–67. doi: 10.1146/annurev.ge.29.120195.000353. [DOI] [PubMed] [Google Scholar]
- 14.Sharpe M E, Errington J. Proc Natl Acad Sci USA. 1995;92:8630–8634. doi: 10.1073/pnas.92.19.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chalfie M, Tu Y, Euskirchen G, Ward W, Prasher D. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 16.Smith K, Youngman P. Biochimie (Paris) 1992;74:705–711. doi: 10.1016/0300-9084(92)90143-3. [DOI] [PubMed] [Google Scholar]
- 17.Crameri A, Whitehorn E A, Tate E, Stemmer W. Nature Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 18.Itaya M, Kondo K, Tanaka T. Nucleic Acids Res. 1989;11:4410. doi: 10.1093/nar/17.11.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curran J, Stewart C R. Virology. 1985;142:78–97. doi: 10.1016/0042-6822(85)90424-6. [DOI] [PubMed] [Google Scholar]
- 20.Harwood C R, Cutting S M. Molecular Biological Methods of Bacillus. New York: Wiley; 1990. [Google Scholar]
- 21.Harry E J, Pogliano K, Losick R. J Bacteriol. 1995;177:3386–3393. doi: 10.1128/jb.177.12.3386-3393.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barak I, Behari J, Olmedo G, Guzman P, Brown D, Castro E, Walker D, Westpheling J, Youngman P. Mol Microbiol. 1996;19:1047–1060. doi: 10.1046/j.1365-2958.1996.433963.x. [DOI] [PubMed] [Google Scholar]
- 23.Huang W M. Annu Rev Genet. 1996;30:79–107. doi: 10.1146/annurev.genet.30.1.79. [DOI] [PubMed] [Google Scholar]
- 24.Schmid M. J Bacteriol. 1990;172:5416–5424. doi: 10.1128/jb.172.9.5416-5424.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutkenhaus J. Mol Microbiol. 1993;9:404–409. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- 26.Kato J, Nishimura Y, Suzuki H. Mol Gen Genet. 1989;217:178–181. doi: 10.1007/BF00330959. [DOI] [PubMed] [Google Scholar]
- 27.Hussain K, Elliott E J, Salmond G. Mol Microbiol. 1987;1:259–273. doi: 10.1111/j.1365-2958.1987.tb01932.x. [DOI] [PubMed] [Google Scholar]
- 28.Thornton M, Armitage M, Maxwell A, Dosanjh B, Howells A J, Norris V, Sigee D C. Microbiology. 1994;140:2371–2382. doi: 10.1099/13500872-140-9-2371. [DOI] [PubMed] [Google Scholar]
- 29.Warburton P E, Earnshaw W C. BioEssays. 1997;19:97–99. doi: 10.1002/bies.950190203. [DOI] [PubMed] [Google Scholar]
- 30.Kato J, Suzuki H, Ikeda H. J Biol Chem. 1992;267:25676–25684. [PubMed] [Google Scholar]
- 31.Kusano T, Steinmetz D, Hendrickson W, Murchie J, King M, Benson A, Schaechter M. J Bacteriol. 1984;158:313–316. doi: 10.1128/jb.158.1.313-316.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niki H, Imamura R, Kitaoka M, Yamanaka K, Ogura T, Hiraga S. EMBO J. 1992;11:5101–5109. doi: 10.1002/j.1460-2075.1992.tb05617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiraga S, Niki H, Imamura R, Ogura T, Ichinose C, Mori H, Ezaki B, Jaffe A. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaffe A, Vinella D, D’Ari R. J Bacteriol. 1997;179:3494–3499. doi: 10.1128/jb.179.11.3494-3499.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]