Abstract
Recent studies indicate that neural cell development in the central nervous system (CNS) correlates with a reduction in acetylation of histone core proteins. Moreover, histone hypoacetylation is thought to be important to oligodendrocyte lineage development. The mechanisms mediating the reduction in acetylation during postnatal neural development remain to be defined. To begin to understand these mechanisms, we investigated the expression of histone deacetylase 11 (HDAC11), a newly identified HDAC, in mouse brain during postnatal development. We show that HDAC11 was widely expressed in the brain and that this expression gradually increased in a region-specific pattern between birth and 4 weeks of age. At the cellular level HDAC11 protein was predominately localized in the nuclei of mature oligodendrocytes but only minimally in astrocytes. Although dentate gyrus granule neurons abundantly expressed HDAC11, granule neuron precursors in the subgranule layer exhibited little HDAC11 immunoreactivity. Double-immunostaining of the corpus callosum and dentate gyrus demonstrated that HDAC11 and Ki67, a cell-proliferating marker, are rarely colocalized in same cells. Our data show that HDAC11 was expressed in the developing brain in a temporal and spatial pattern that correlates with the maturation of neural cells, including cells of the oligodendrocyte lineage. These findings support a role for HDAC11 in CNS histone deacetylation and the development of oligodendrocytes and neurons during postnatal development.
Keywords: HDAC, deacetylation, oligodendrocytes, hippocampus
Epigenetic modification of histone core proteins has been implicated in gene regulation and cellular development. Recent studies indicate that regulation of histone acetylation plays a pivotal role in the development of the central nervous system (CNS). Acetylation is a dynamic process that is regulated by two classes of enzymes: histone acetyltransferases (HATs) and histone deacetylases (HDACs). During postnatal development in rat corpus callosum and cortical neurons, acetylation of histones H3 and H4 gradually decreases with increasing age (Pina et al., 1988; Shen et al., 2005). Similarly, acetylation of H3 and H4 is reduced when cultured adult rat hippocampal neural precursor cells (NPCs) are induced to differentiate into mature oligodendrocytes and astrocytes by growth factors (Hsieh et al., 2004). Furthermore, global suppression of HDAC activity by inhibitors, such as vaproic acid and trichostatin A, blunts oligodendrocyte development and myelination in developing rats (Shen et al., 2005), in cultured rat adult hippocampi NPC (Hsieh et al., 2004), and in prenatal oligodendrocyte precursors (Liu et al., 2003).
The mechanisms by which acetylation of histone is regulated in the brain during postnatal development have not been elucidated. In a recent study of rat corpus callosum development, Shen et al. (2005) showed that the abundance of 2 HATs, p300 and CBP (cyclic AMP response element–binding protein), significantly increased in the first week of postnatal life before gradually declining in the ensuing 2 weeks. Although this decline in HAT activity could contribute to the post-natal decrease in CNS acetylation, available information on changes in HDAC mRNA and protein expression is not consistent with a decrease in acetylation. Corpus callosum HDAC activity and the number of CC1 oligodendrocytes that express HDACs (including HDAC2, HDAC3, HDAC5, and HDAC8) gradually decrease postnatally, with no significant change in the abundance of HDAC1–HDAC8 proteins in the corpus callosum (Shen et al., 2005). Our unpublished findings also demonstrate that HDAC2–HDAC9 mRNA expression either does not change or moderately decreases during maturation of an oligodendrocyte cell line (Liu et al., unpublished observations).
In the past 6 years three new HDACs—HDAC9, HDAC10, and HDAC11—have been identified (Zhou et al., 2001; Fischer et al., 2002; Gao et al., 2002; Guardiola and Yao, 2002; Kao et al., 2002; Mahlknecht et al., 2002; Petrie et al., 2003). Among them, HDAC11 is the lone known mammalian class IV HDAC (Gregoretti et al., 2004). In humans expression of HDAC11 is limited to only a few organs, including brain, heart, muscle, and kidney (Gao et al., 2002). This localized expression suggests that HDAC11 has functions important to these tissues and prompted us to examine its pattern of expression in mouse brain during postnatal development as a first step toward defining its role in the CNS. In this report we demonstrate that HDAC11 is predominately expressed in mature oligodendrocytes and neurons. The temporal and spatial pattern of HDAC11 expression in neural cells such as oligodendrocytes and neurons is negatively correlated with histone acetylation. Our data are consistent with a role for HDAC11 in CNS deacetylation and suggest that HDAC11 plays a role in the development of oligodendrocytes and some neurons including hippocampal granule neurons.
MATERIALS AND METHODS
Animals
C57BL/6 mice were originally purchased from the Charles River Laboratory (Wilmington, MA), and a breeding colony was established in the laboratory. At designated ages, the mice were sacrificed using either isofluoran or CO2. All procedures used were approved by the institutional review committees of the University of North Carolina at Chapel Hill.
RNA Isolation and Quantitative Real-Time PCR
After the mice were sacrificed and tissues were removed, brains were dissected into five regions: the cerebral cortex (CTX), hippocampus (HIP), brain stem (BS), diencephalon (DIE), and cerebellum (CB). Tissues were rapidly frozen in liquid N2 and stored at −80°C until use. Total RNA was isolated using Trizol (Invitrogen, Carlsbad, CA). Reverse transcription was performed using 1 μg of total RNA, random decamers as primers, and Superscript II reverse transcriptase (Invitrogen). The resultant cDNA was subjected to quantitative real-time PCR (qRT-PCR) analysis using Sybr Green PCR Master Mix (Qiagen, Valencia, CA), HDAC11 primers, and a LightCycler (Roche, Indianapolis, IN) or a Realplex cycler (Eppendorf, Westbury, NY). The sequences for the HDAC11 forward and reverse primers were 5′-AATGGGG CAAGGTGATCAAC-3′ and 5′-AGCCACCACCAACATT GATG-3′, respectively (Genebank Accession number NM_144919). PCR using the primer set produced a single HDAC11 cDNA amplicon with the expected size of 299 bp; and its identity was further confirmed by DNA sequencing. Melt temperature analysis and size fragmentation of PCR products were also used to confirm specificity of each qRT-PCR.
To quantify mRNA abundance, a standard curve for HDAC11 mRNA was generated from serial dilutions of cDNA derived from an adult mouse whole brain. A standard curve for 18S rRNA also was generated using a set of 18S rRNA–specific primers (Ambion, Austin, TX). The relative abundance of HDAC11 mRNA was estimated based on its standard curve and normalized against the abundance of 18S rRNA.
Western Immunoblot Analysis
To extract total soluble protein, tissues were lysed with a lysis buffer [20 mM Tris-HCl (pH 7.5), 1% NP-40, 137 mM NaCl, 1 mM CaCl2, 1 mM EDTA, and freshly added 0.5 mM PMSF and 2.5μg/mL each of aprotinin, leupeptin, and antipain]. After sonication, soluble proteins were collected by centrifugation at 5,000 rpm for 5 min at 4°C. Protein concentration was determined using a BCA protein kit (Pierce, Rockford, IL). Aliquots of proteins (50 μg) were separated on 4%–12.5% polyacrylamide gradient gels and transferred onto PVDF nylon membranes (Amersham, Arlington Heights, IL). After staining with Ponceau S solution (Sigma, St. Louis), the membranes were incubated with antibody specific for HDAC11 (1:3,000; Abcam, Cambridge, MA). Specific immunoreactivity was visualized using an ECL kit (Amersham). Images of specific protein bands on x-ray films were digitally scanned. After immunoblot analysis of HDAC11, the blots were stripped and reprobed with β-actin antibody (Sigma). Because the abundance of β-actin appears to decrease during development, the abundance of HDAC11 was normalized against the abundance of Ponceau S staining of an approximately 60-kDa protein band.
Immunohistochemical Staining
After the mice were sacrificed, brains were removed and split along the midsagittal line. Left brains were fresh-frozen in liquid N2, and sagittally sectioned (10-μm-thick sections) on a cryostat. After being fixed with 4% paraformaldehyde and washing with PBS, the sections were subjected to double-immunostaining with a rabbit antibody for HDAC11 (1:100, Abcam) and mouse monoclonal antibodies against CC1 (1:50; Oncogene Research Products, San Diego, CA), NeuN (1:200; Chemicon International, Temecula, CA), GFAP (1:200; Chemicon), or Ki67 (1:500; Victor Laboratories, Burlington, CA). Antibody–antigen complexes were detected by either Alexa Fluor 594–conjugated antirabbit antibody or Alexa Fluor 488–conjugated antimouse secondary antibody (Invitrogen). Cell nuclei were counterstained with the nuclear dye 4,6-diamidine-2′-phenylindole dihydrochloride (DAPI). Staining without primary antibodies served as negative controls. Fluorescent images were digitally captured and analyzed using a fluorescent microscope and a Spot Jr. digital camera (Diagnostic Instruments, Sterling Heights, MI). To quantify HDAC11-expressing oligodendrocytes, astrocytes, and neurons, immunolabeled cells in which nuclei were clearly observed (>300 cells) in the corpus callosum and dentate gyrus of P20 mice in digitally captured images were scored. The number of the CC1-, GFAP-, and NeuN-positive cells that also immunoreacted with HDAC11 antibody was calculated as the percentage of the total number of each neural cell type.
Statistics
One-way ANOVA was used to test statistical significance among the groups and was followed by comparison of each group mean using the Newman-Keuls Student test assisted with SigmaStat software for Windows (SPSS, Inc., Chicago, IL).
RESULTS
To determine HDAC11 changes in the CNS during postnatal development, developing brains were dissected, and their mRNA abundance was quantified using quantitative real-time PCR. When compared with that in whole brain at P0, the abundance of HDAC11 mRNA gradually increased during the first postnatal week in all five brain regions examined (CTX, HIP, BS, CB, and DIE). During the next 3 weeks HDAC11 mRNA increased more rapidly, reaching levels 3- to 14-fold higher than that in P0 whole brain from 40-day-old mice (Fig. 1). Among the brain regions examined, DIE exhibited the most abundant HDAC11 mRNA in 40-day-old mice (Fig. 1). HDAC11 mRNA was also highly expressed in kidney but only expressed at low levels in heart, liver, lung, and spleen (Fig. 2), observations in line with the study of human tissues by Gao et al. (2002). Consistent with the changes in mRNA expression, the abundance of HDAC11 protein also significantly increased during development when assessed in CTX and BS (Fig. 3).
Fig. 1.
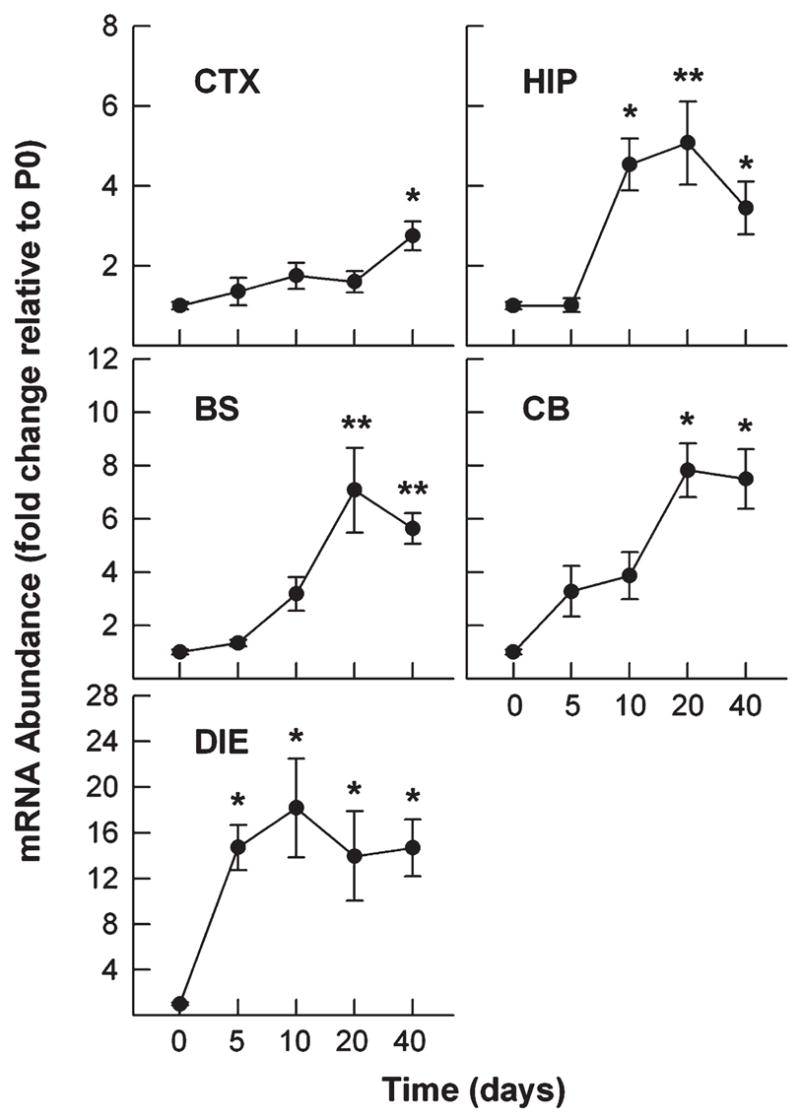
Expression of HDAC11 mRNA in mouse brain during development. Mice were sacrificed at the ages indicated. Total RNA was isolated from the brain regions indicated and subjected to qRT-PCR analysis. The abundance of HDAC11 mRNA was normalized to 18S rRNA abundance and is expressed as the magnitude of the change in abundance relative to that in P0 whole brains (CTX, cerebral cortex; HIP, hippocampus; BS, brain stem; CB, cerebellum; DIE, diencephalon). Results expressed as mean ±SE of measurements of 3 or 4 samples (*P < 0.05, **P < 0.01; compared to mRNA abundance in P0 whole brains).
Fig. 2.
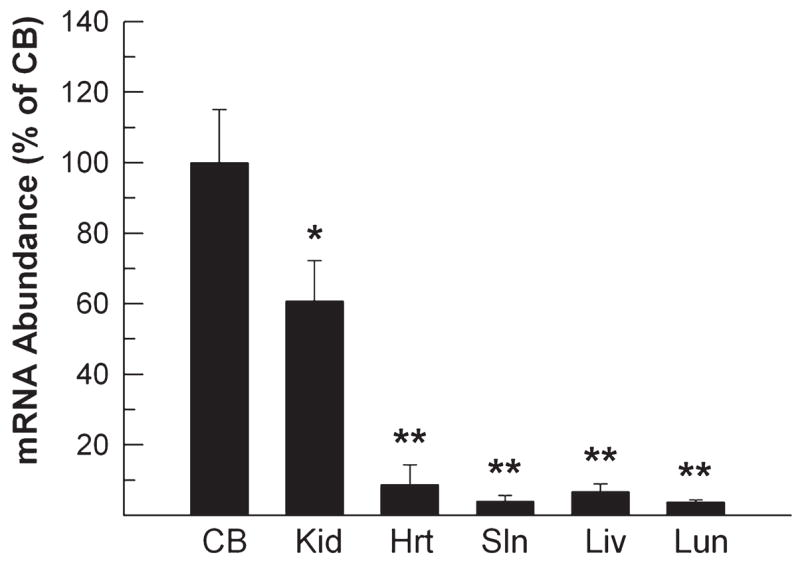
HDAC11 mRNA expression in non–nervous system organs. Mice were sacrificed on P40. Total RNA was isolated from kidney (Kid), heart (Hrt), spleen (Sln), liver (Liv), and lung (Lun) and subjected to qRT-PCR analysis. RNA from the cerebellum (CB) was used as a standard for comparison. The abundance of HDAC11 mRNA was normalized to 18S rRNA abundance and is expressed as that percentage of that in the CB. Results expressed as mean ±SE of the measurements of three samples (*P < 0.01, **P < 0.001, compared to mRNA abundance in CB).
Fig. 3.
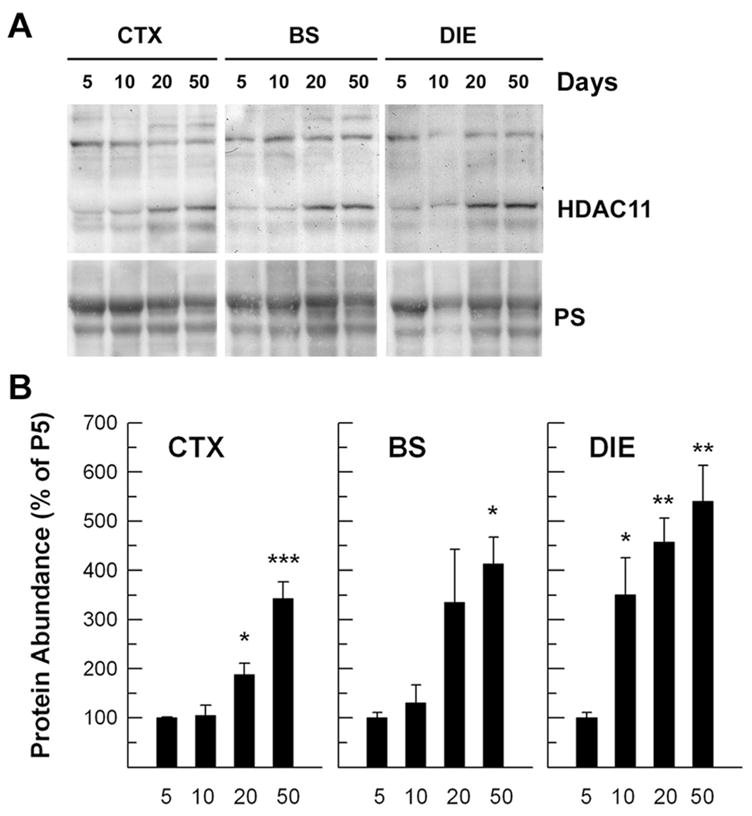
HDAC11 protein abundance in developing brain stem (BS), cerebral cortex (CTX), and diencephalon (DIE) during postnatal development. A: Representative Western immunoblots of HDAC11. Mice were sacrificed on P5, P10, P20, and P50, as indicated at the top. The bottom shows Ponceau S staining of about a 60-kDa protein band (PS). B: Quantification of HDAC11 protein abundance in brain regions during development. HDAC11 abundance was normalized to the abundance of the Ponceau S–stained 60-kDa protein band and is expressed as the percentage of that in P5 mice. Results expressed as mean ± SE of the measurements of three samples (*P < 0.05, **P < 0.01, ***P < 0.001, compared to mRNA abundance in CB).
Immunofluorescent staining with an antibody specific for HDAC11 was used to determine the cellular localization of HDAC11 in the developing brain. HDAC11 immunoreactivity was observed in most brain regions throughout postnatal development. As Figure 4 shows, HDAC11 protein was found predominantly in nuclei, including those of hippocampal granule neurons and glia-like cells in the corpus callosum. In contrast, sections of liver and lung from adult mice only weakly reacted with HDAC11 antibody (data not shown). These findings are consistent with our mRNA abundance data and with a previous report that showed nuclear localization of HDAC11 and low expression of HDAC11 in the liver and lung of human (Gao et al., 2002).
Fig. 4.
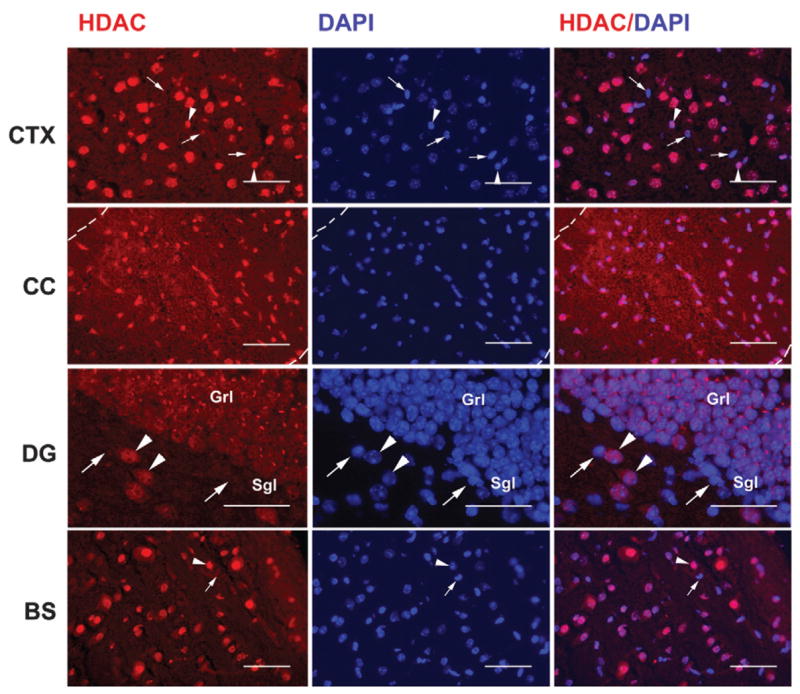
Representative photomicrographs of HDAC11 immunostaining in mouse brain. A brain from a P20 mouse was fresh-frozen, and near-midsagittal sections of cerebral cortex (CTX), corpus callosum (CC), dentate gyrus (DG), and brain stem (BS) were subjected to immunostaining. Images of HDAC11 red immunostaining and blue DAPI counterstaining were captured digitally and superimposed using a Spot Jr. camera and software. Dotted lines in the upper panels outline the CC (Grl, granule cell layer; Sgl, subgranule cell layer; Hil, hilus); arrowheads point to HDAC11-positive cells, and arrows point to HDAC11-negative cells. Scale bar = 50 μm.
To determine the identity of HDAC11-positive cells, double-immunostaining was performed using antibodies specific for HDAC11 and CC1 (a mature oligodendrocyte marker), glial fibrillary acidic protein (GFAP, an astrocyte-specific marker), or NeuN (a mature neuron marker). As shown in Figure 5, numerous CC1-positive oligodendrocytes and NeuN-positive neurons strongly reacted with HDAC11 antibody. In contrast, most GFAP-positive astrocytes showed no HDAC11 immunoreactivity.
Fig. 5.
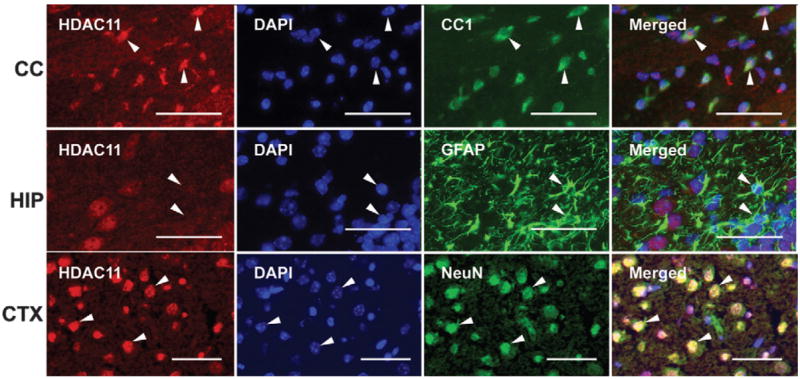
Representative photomicrographs of HDAC11 immunostaining in CC1-positive oligodendrocytes, GFAP-positive astrocytes, and NeuN-positive neurons. Sections from a P20 brain were double-immunostained with antibodies to HDAC11 (red) and CC1, GFAP, or NeuN (green). Nuclei were counterstained with DAPI (blue). Arrowheads point to CC1-positive/HDAC11-positive oligodendrocytes and NeuN-positive/HDAC11-positive neurons in the upper and lower panels, respectively, whereas arrowheads point to a GFAP-positive/HDAC11-negative astrocyte in the middle panels (CC, corpus callosum; HIP, hippocampus; CTX, cerebral cortex). Scale bar = 50 μm.
As expected during development, the number of CC1-positive mature oligodendrocytes in all brain regions rapidly increased between P5 and P40, especially in oligodendrocyte-rich regions such as the corpus callosum and the brain stem. Similarly, HDAC11-positive cells were significantly increased during the same developmental period. Double-immunostaining demonstrated that most CC1-positive cells also were positive for HDAC11. Representative photomicrographs of CC1-and HDAC11-double-immunolabeled cells in the corpus callosum are shown in Figure 6.
Fig. 6.
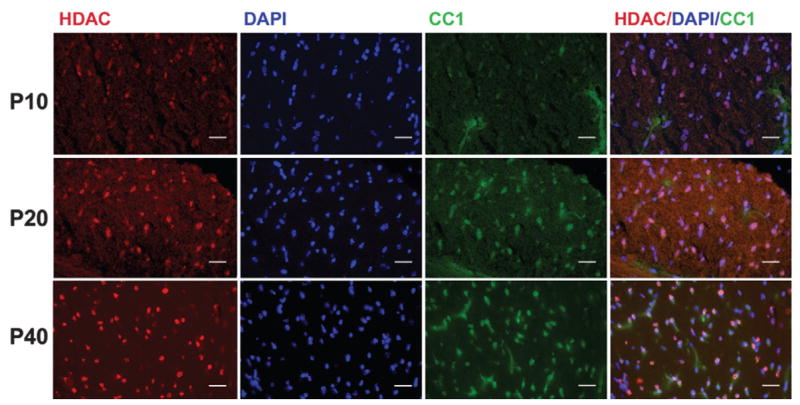
HDAC11 expression in oligodendrocytes during development. Representative photomicrographs of HDAC11 immunostaining in CC1-positive oligodendrocytes. Sections of corpus callosum (CC) from P10, P20, or P40 mice were double-immunostained with antibodies to HDAC11 (red) and CC1 (green). Nuclei were counter-stained with DAPI (blue). Note that with increased age, the number of CC1-positive oligodendrocytes (green) and the number of cells positive for both CC1 and HDAC11 (pink) increased. Scale bar = 50 μm.
Numerous cells positive for both NeuN and HDAC11 were found in the CTX, HIP, BS, and CB. Hippocampal granule neurons, cortical neurons, and large brain-stem neurons showed strong HDAC11 immunoreactivity. However, internal granule cells and Purkinje cells in the CB only weakly reacted with HDAC11 antibody (data not shown). In the dentate gyrus, the outer layer of mature granule neurons strongly reacted with antibodies specific for both HDAC11 and NeuN. Cells in and near the subgranule layer, however, did not react or only weakly reacted with these antibodies (Fig. 7). Similarly, in the cerebellum of P4 mice, the external granule layer cells weakly reacted with HDAC11 (data not shown). Quantification of cells in P20 corpus callosum and hippocampus, two well-defined regions, showed that most CC1-positive oligodendrocytes (~93%) and NeuN-positive neurons (~96%) were HDAC11 positive, whereas only about 6.5% of GFAP-positive astrocytes were HDAC11 reactive (Fig. 8).
Fig. 7.
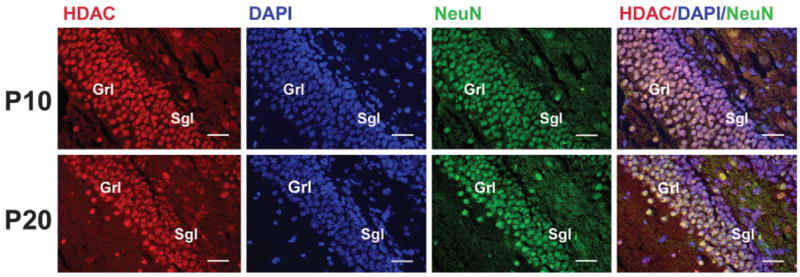
Expression of HDAC11 in dentate gyrus during development. Sections of dentate gyrus from P10 and P20 mice were double-immunostained with antibodies to HDAC11 (red) and NeuN (green) and counterstained with DAPI (blue). Note that a few cells in the sub-granule layer are positive for either NeuN or HDAC11 and that nearly all granule neurons are positive for both NeuN and HDAC11 (very light pink to white); Grl, granule cell layer; Sgl, subgranule cell layer. Scale bar = 50 μm.
Fig. 8.
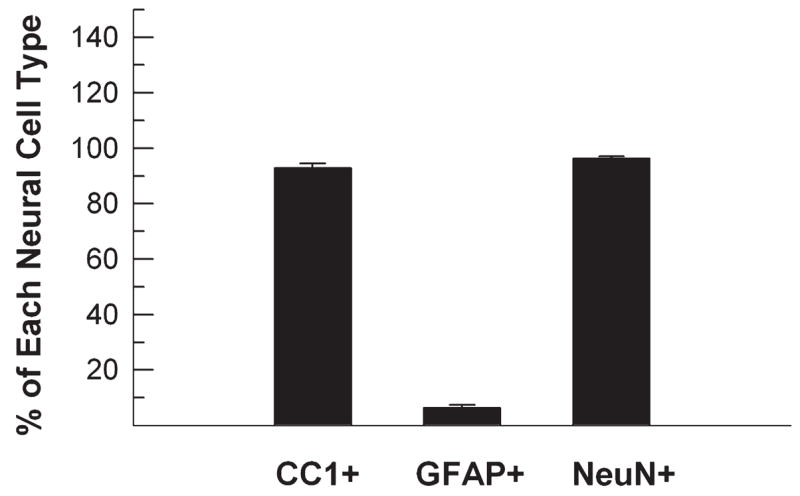
Quantification of oligodendrocytes, astrocytes, and neurons expressing HDAC11. Sections of corpus callosum and dentate gyrus from P20 mice were double-immunostained with HDAC11 and CC1, GFAP, or NeuN antibodies. The number of double-immuno-positive neural cells was counted and is expressed as the percentage of each neural cell type. Results expressed as mean ±SE of measurements of three animals.
The lack of HDAC11 immunostaining in the sub-granule layer in the dentate gyrus and CB external granule layer, two brain regions that primarily contain neural precursors during postnatal development, led us to speculate that HDAC11 is either minimally expressed or not expressed in neural precursors. To address this possibility, proliferating neural precursor cells were identified by immunostaining for Ki67, an antigen that is expressed in the G1, S, G2, and M phases of the cell cycle, and then double-immunostained for HDAC11. In both the corpus callosum and dentate gyrus of 5- and 10-day-old mice, ages when neural precursors in these regions proliferate actively, few Ki67-positive proliferating neural precursors were positive for HDAC11. Similarly, few HDAC11-positive cells reacted with Ki67 antibody (Fig. 9). This mutual exclusion of immunoreactivity for HDAC11 and Ki67 strongly indicates that HDAC11 is predominately expressed in mature cells and minimally or not expressed in proliferating neural precursors.
Fig. 9.
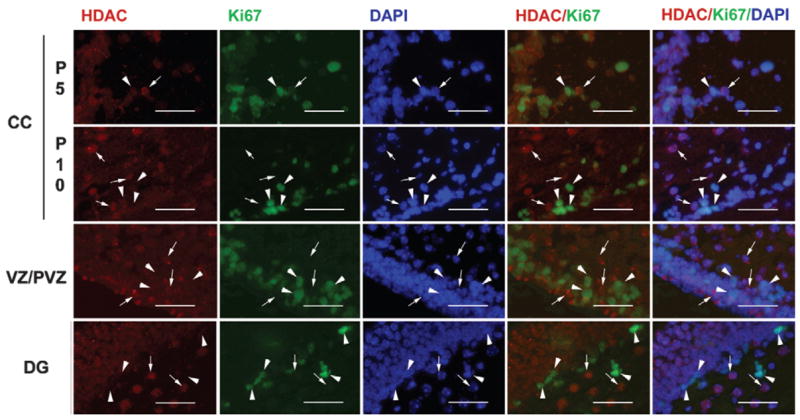
Representative microphotographs of HDAC11 and Ki67 double immunostaining in corpus callosum (CC), ventricular/subventricular zones (VZ/SVZ), and dentate gyrus (DG) during development. Sections of P5 and P10 CC and P10 DG and VZ/SVZ were double-immunostained with antibodies to HDAC11 (red) and Ki67 (green) and counterstained with DAPI (blue). Arrowheads point to Ki67-positive proliferating cells, whereas arrows show HDAC11-positive cells. Note that Ki67-positive cells and HDAC11-positive cells are mutually exclusive. Scale bar = 50 μm.
DISCUSSION
In this study we have demonstrated that brain expression of HDAC11, a newly identified HDAC, gradually increases during development of the murine brain in a temporal and spatial pattern that correlates positively with neural cell maturation and negatively with histone acetylation. Furthermore, we have shown that HDAC11 is predominately expressed in the nuclei of oligodendrocytes and some neurons but only very weakly expressed in astrocytes, if expressed at all. Our data suggest that HDAC11 plays a role in the regulation of brain histone modification and development and that HDAC11 function is largely restricted to oligodendrocytes and some neurons during postnatal CNS development.
Our data show that HDAC11 is abundantly expressed in the mouse brain in a distinctive temporal-spatial pattern. Specifically, HDAC11 is abundantly expressed in mature oligodendrocytes and many neuron populations and modestly in CB granule cells and Pur-kinje cells. HDAC11, however, does not appear to be expressed in astrocytes and neural precursors. Furthermore, regional expression of HDAC11 in the brain significantly increases postnatally and peaks between P10 and P20. In general, the temporal pattern of HDAC11 expression is coincident with increases in the expression of oligodendrocyte/myelin-protein genes and the maturation of oligodendrocytes. Because neurogenesis is largely completed before birth in most brain regions, the increased number of mature oligodendrocytes exhibiting abundant HDAC11 is likely to significantly contribute to the increase in brain HDAC11 expression postnatally. Neuronal cells, however, also show increased HDAC11 expression postnatally.
Although histone acetylation is often linked to gene activation (Jenuwein and Allis, 2001), several studies have indicated that histone deacetylation is associated with neural cell development in the CNS (Pina et al., 1988; Liu et al., 2003; Hsieh et al., 2004; Shen et al., 2005), and accumulating evidence suggests that HDAC activity plays a pivotal role in gene expression (see review by Nusinzon and Horvath, 2005, 2006; Siebzehnrubl et al., 2007). Acetylation of histone H3 and H4 has been shown to gradually decrease with maturation of cortical neurons (Pina et al., 1988) and in the corpus callosum, an oligodendrocyte-rich region, during rat postnatal development (Shen et al., 2005). Although HDAC1 has been shown to be important to the development of zebra fish oligodendrocytes (Cunliffe and Casaccia-Bonnefil, 2006), no significant developmental changes have been observed in the abundance of HDAC1–HDAC8 in rat corpus callosum (Shen et al., 2005). This is despite the finding that the activity of both HAT and HDAC is apparently reduced (Shen et al., 2005). Although the importance of HDAC1 in mammalian cells remains to be elucidated, the data mentioned above suggest that the reduction in histone acetylation is not a result of increased abundance and/or activity of HDACs (HDAC1–HDAC8); rather, other factors likely contribute to histone deacetylation. In this study we have demonstrated that HDAC11 protein is preferentially localized in the nuclei of oligodendrocytes, as judged by double-immunostaining. Furthermore, its abundance, as well as that of its mRNA, is significantly increased in mouse brain during a developmental period when histone H3 and H4 acetylation is significantly decreased. Our data suggest, therefore, that increased HDAC11 abundance in developing oligodendrocytes contributes to the reduction in histone acetylation. This speculation is supported by the data of Gao et al. (2002), who showed that HDAC11 was capable of removing acetyl groups from histone core proteins, and by our unpublished data showing that suppressing HDAC11 expression by RNA interference increases histone H3 acetylation in a cultured oligodendroglial cell line (manuscript is in preparation).
There have been few studies on developmental regulation of histone acetylation in neurons, either in vivo or in vitro. Furthermore, reports on the role of acetylation, as well as that of HDAC functions, in neuronal development have not been consistent. In rat cortical neurons, acetylated histone H4 gradually decreased with maturation (Pina et al., 1988), and the expression of HDAC5, HDAC6, HDAC7, and HDAC9 increased in cultured hippocampal neural precursors during their maturation (Ajamian et al., 2003). Each of these studies suggests a role for HDACs in the reduction of histone acetylation. Similarly, our study has demonstrated increased expression of HDAC11 in the hippocampus during hippocampal postnatal development, in line with a role for HDAC11 in these neurons. In contrast, however, administration of HDAC inhibitors or small interfering RNA appears to promote neuronal differentiation in cultured adult rat forebrain (Siebzehnrubl et al., 2007) and hippocampus precursor cells (Hsieh et al., 2004), as well as the survival of CB granule neurons (Bolger and Yao, 2005). The differences in these experimental data may be a result of the diversity in the culture conditions used, the types of neurons studied, and/or the age of animals from which neural cells were obtained. In addition, HDAC inhibitors may promote growth by mechanisms independent of histone acetylation (Hao et al., 2004).
In summary, we have shown that HDAC11 is predominately expressed in oligodendrocytes and some neurons and that its expression exhibits a temporal and spatial pattern consistent with histone deacetylation and maturation in oligodendrocyte lineage cells. The exact function or functions of HDAC11 in developing oligodendrocytes and neurons remain to be determined.
Acknowledgments
The authors thank Dr. Billie Moats-Staats for her critical reading of this manuscript.
Contract grant sponsor: NIH; Contract grant numbers: NS038891 and NS048868.
References
- Ajamian F, Suuronen T, Salminen A, Reeben M. Upregulation of class II histone deacetylases mRNA during neural differentiation of cultured rat hippocampal progenitor cells. Neurosci Lett. 2003;346:57–60. doi: 10.1016/s0304-3940(03)00545-7. [DOI] [PubMed] [Google Scholar]
- Bolger TA, Yao TP. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J Neurosci. 2005;25:9544–9553. doi: 10.1523/JNEUROSCI.1826-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe VT, Casaccia-Bonnefil P. Histone deacetylase 1 is essential for oligodendrocyte specification in the zebrafish CNS. Mech Dev. 2006;123:24–30. doi: 10.1016/j.mod.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Fischer DD, Cai R, Bhatia U, Asselbergs FA, Song C, Terry R, Trogani N, Widmer R, Atadja P, Cohen D. Isolation and characterization of a novel class II histone deacetylase, HDAC10. J Biol Chem. 2002;277:6656–6666. doi: 10.1074/jbc.M108055200. [DOI] [PubMed] [Google Scholar]
- Hao Y, Creson T, Zhang L, Li P, Du F, Yuan P, Gould TD, Manji HK, Chen G. Mood stabilizer valproate promotes ERK pathway-dependent cortical neuronal growth and neurogenesis. J Neurosci. 2004;24:6590–6599. doi: 10.1523/JNEUROSCI.5747-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Cueto MA, Asselbergs F, Atadja P. Cloning and functional characterization of HDAC11, a novel member of the human histone deacetylase family. J Biol Chem. 2002;277:25748–25755. doi: 10.1074/jbc.M111871200. [DOI] [PubMed] [Google Scholar]
- Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Guardiola AR, Yao TP. Molecular cloning and characterization of a novel histone deacetylase HDAC10. J Biol Chem. 2002;277:3350–3356. doi: 10.1074/jbc.M109861200. [DOI] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1079. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kao HY, Lee CH, Komarov A, Han CC, Evans RM. Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J Biol Chem. 2002;277:187–193. doi: 10.1074/jbc.M108931200. [DOI] [PubMed] [Google Scholar]
- Liu A, Muggironi M, Marin-Husstege M, Casaccia-Bonnefil P. Oligodendrocyte process outgrowth in vitro is modulated by epigenetic regulation of cytoskeletal severing proteins. Glia. 2003;44:264–274. doi: 10.1002/glia.10290. [DOI] [PubMed] [Google Scholar]
- Mahlknecht U, Schnittger S, Will J, Cicek N, Hoelzer D. Chromosomal organization and localization of the human histone deacetylase 9 gene (HDAC9) Biochem Biophys Res Commun. 2002;293:182–191. doi: 10.1016/S0006-291X(02)00193-6. [DOI] [PubMed] [Google Scholar]
- Nusinzon I, Horvath CM. Histone deacetylases as transcriptional activators? Role reversal in inducible gene regulation. Sci STKE 2005. 2005:re11. doi: 10.1126/stke.2962005re11. [DOI] [PubMed] [Google Scholar]
- Nusinzon I, Horvath CM. Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation. Mol Cell Biol. 2006;26:3106–3113. doi: 10.1128/MCB.26.8.3106-3113.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie K, Guidez F, Howell L, Healy L, Waxman S, Greaves M, Zelent A. The histone deacetylase 9 gene encodes multiple protein isoforms. J Biol Chem. 2003;278:16059–16072. doi: 10.1074/jbc.M212935200. [DOI] [PubMed] [Google Scholar]
- Pina B, Martinez P, Suau P. Differential acetylation of core histones in rat cerebral cortex neurons during development and aging. Eur J Biochem. 1988;174:311–315. doi: 10.1111/j.1432-1033.1988.tb14099.x. [DOI] [PubMed] [Google Scholar]
- Siebzehnrubl FA, Buslei R, Eyupoglu IY, Seufert S, Hahnen E, Blumcke I. Histone deacetylase inhibitors increase neuronal differentiation in adult forebrain precursor cells. Exp Brain Res. 2007;176:672–678. doi: 10.1007/s00221-006-0831-x. [DOI] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebzehnrubl FA, Buslei R, Eyupoglu IY, Seufert S, Hahnen E, Blumcke I. Histone deacetylase inhibitors increase neuronal differentiation in adult forebrain precursor cells. Exp Brain Res. 2007;176:672–678. doi: 10.1007/s00221-006-0831-x. [DOI] [PubMed] [Google Scholar]
- Zhou X, Marks PA, Rifkind RA, Richon VM. Cloning and characterization of a histone deacetylase, HDAC9. Proc Natl Acad Sci USA. 2001;98:10572–10577. doi: 10.1073/pnas.191375098. [DOI] [PMC free article] [PubMed] [Google Scholar]


