Abstract
Molecular mimicry, defined as similar structures shared by molecules from dissimilar genes or proteins, is a general strategy used by pathogens to infect host cells. Severe acute respiratory syndrome (SARS) is a new human respiratory infectious disease caused by SARS coronavirus (SARS-CoV). The spike (S) protein of SARS-CoV plays an important role in the virus entry into a cell. In this study, eleven synthetic peptides from the S protein were selected based on its sequence homology with human proteins. Two of the peptides D07 (residues 927–937) and D08 (residues 942–951) were recognized by the sera of SARS patients. Murine hyperimmune sera against these peptides bound to proteins of human lung epithelial cells A549. Another peptide D10 (residues 490–502) stimulated A549 to proliferate and secrete IL-8. The present results suggest that the selected S protein regions, which share sequence homology with human proteins, may play important roles in SARS-CoV infection.
1. INTRODUCTION
Severe acute respiratory syndrome (SARS) is a new emerging infectious disease, which was first reported in China in 2002 and was rapidly spreading all over the world in 2003 [1, 2]. The disease was transmitted by droplets and close contact. Patients develop persistent fever, dry cough, progressive radiographic changes of chest, and lymphopenia once infected. Despite treatment, about 10–15% of the patients would die due to the acute respiratory distress [3–6]. A novel coronavirus (SARS-CoV) was isolated from SARS patients [7–9]. SARS-CoV is a positive-stranded RNA virus with an envelop. The genome of SARS-CoV is around 29,727 nucleotides in length. The sequence was annotatedin silico [10]. Comparative genomic studies using the in silico annotated proteins have suggested that SARS virus belongs to a new group of coronavirus.
According to the genomic sequence of SARS-CoV, it is predicted that there are several structural proteins can be produced by SARS-CoV including spike (S), envelop (E), membrane (M), and nucleocapsid. Spike protein is very important in the binding and fusion of coronavirus to the host cells [11, 12]. The S protein of SARS-CoV has 1255 amino acids in length and 23 potential N-linked glycosylation sites. The amino terminus of the SARS-CoV S protein contains a short type 1 signal sequence composed of hydrophobic amino acids that are presumably removed during cotranslational transport through the endoplasmic reticulum. The carboxyl terminus consists of a transmembrane domain and a cytoplasmic tail rich in cysteine residues. The majority of protein (residues 12–1195) is outside the virus particle, which can be divided into amino-terminal S1 and carboxyl-terminal S2 domain. The S1 domain (residues 12–672) binds to the host cell receptor, angiotensin-converting enzyme 2 (ACE2), while the S2 domain is responsible for membrane fusion [13–15]. Monoclonal antibodies against S1 domain can block the receptor binding and contain potent neutralization activity against SARS-CoV [16]. However, peptides derived from S2 domain can also inhibit SARS-CoV infection [12].
Molecular mimicry, which is defined as similar structures shared by molecules from dissimilar genes or by their protein products, is a general strategy for pathogens to infect host cells and has been proposed as a pathogenic mechanism for autoimmune disease [17]. Therefore, identification of the molecular mimic regions of pathogen may be helpful to understand the disease induced by that pathogen. At present, it is unclear whether molecular mimicry occurs between SARS-CoV S proteins and human peptides. We have approached this question using computer to analyze the sequence of spike protein of SARS-CoV and select regions that share the sequence homology with human proteins. The criteria for the selection of potential regions include antigenic analysis and surface accessibility. In this study, we find that several regions of the S protein share sequence homology with human proteins. Synthetic peptides, which represent some of these regions, were synthesized to understand their roles in SARS-CoV infection.
2. MATERIALS AND METHODS
2.1. Peptide prediction and synthesis
Publically available human and coronavirus genome sequences at the National Center for Biotechnology Information (Md, USA) were used for in silicoprediction. Algorithms predicting immunogenicity, second structure prediction, protein topology analysis, and hydrophobicity were conducted to design the tested peptides. Immunogenic viral peptides were calculated based on the algorithm developed by Kolaskar and Tongaonkar [18]. The algorithm is based on a table constructed from the occurrence of amino acid residues in experimentally known antigenic epitopes. The reported accuracy of the method is about 75% [18]. In silico secondary structural analyses of spike protein were performed based on PHD [19] and PREDATOR [20] algorithms. Protein topology prediction was based on the algorithm developed by TMHMM [21]. Hydrophobicity of the peptides was calculated based on the algorithm HMOMENT [22]. Similarity searches between S protein and human genome database were performed by using BLASTP [23] with BLOSUM 62. Extra amino acid residues were added at either N- or C-terminus to keep the hydrophobic amino acid content below 50%. Peptides with high hydrophobicity are difficult to be tested in biochemical experiments since most of in vitro assays are conducted in aqueous buffers. Also, on average, one charged residue is added for every five amino acids. Multiple antigen peptides were synthesized by CytoMol Corp (Mountain View, Calif, USA). In addition, bradykinin and angiotensin I (Ang I) were purchased from Sigma (St. Louis, Mo, USA).
2.2. SARS patient sera
SARS patient sera were collected by the Center for Disease Control, Department of Health (Taipie, Taiwan) from March to June, 2003. Diagnosis of SARS was based on the clinicalcriteria established by the World Health Organization (WHO). Patients with SARS-CoV were confirmed by laboratory methods, including viral antigen detection, RT-PCR, and serologic methods. Ten SARS patient sera collected at the convalescent stage (≥20 days after disease onset) were included in this study. Ten normal sera from healthy individuals were used as controls.
2.3. Enzyme-linked immunosorbent assay (ELISA)
Antibodies against peptides in human sera were detected by solid-phase capture technique using individual peptide-coated plates. ELISA plate was coated with or without 50 μL peptides (100 μg/mL) per well and blocked by 1% bovine serum albumin (BSA) in 0.05% Tween-20 in phosphate-buffered saline (PBS) for 1 hour at room temperature. Test serum samples were diluted and added to the plate for 2 hours at room temperature. After incubation, the ELISA plate was washed with 0.05% Tween-20 in PBS for three times. The bound antibodies were detected by horseradish peroxidase- (HRP-) conjugated antihuman immunoglobulin antibodies (Sigma Aldrich, St. Louis, Mo, USA) and peroxidase substrate, TMB (Promega, Madison, Wiss, USA). The absorbance was measured using the Vmax microplate reader (Molecular Devices Corporation, Sunnyvale, Calif, USA) at 450 nm. Antibodies against peptides in mouse sera were assayed by ELISA as in human sera except HRP-conjugated antimouse immunoglobulin antibodies (Sigma Aldrich) was used to detect bound antibodies.
2.4. Cell culture
Human lung adenocarcinoma cell line A549 and Vero cells were grown in DMEM supplemented with 10% heat-inactivated FCS, 2 mM L-glutamate, and 50 ng/mL gentamycin. Cells were incubated in CO2 incubator at 37°C with 5% CO2 in a humidified atmosphere. For immunofluorescent microscopy observation, monolayers of A549 cells were cultured on sterile glass slides before the experiment.
2.5. Mice immunization
Six- to eight-week-old female BALB/c mice were used in this study. These mice were originally purchased from Jackson Laboratory (Bar Harbor, Me, USA) and bred in the Laboratory Animal Center, National Cheng Kung University (Tainan, Taiwan). Synthetic peptides (1 mg/mL) were emulsified with complete Freund's adjuvant and injected intraperitoneally into BALB/c mice. Mice were boosted with the same peptide in PBS (50 μg/mouse) intraperitoneally two weeks after priming. Sera were collected from the axially plexus of the mice at different time intervals and tested for the presence of antibody against peptides by ELISA as mentioned above. Significant increase of antibody titer (greater than 4 folds) against immunized-peptide was found in mouse hyperimmune sera as compared to normal sera after boosting.
2.6. Immunofluorescent stain
Mouse hyperimmune sera against peptide D08 were incubated with A549 cells at 4°C for 1 hour. After washing three times with PBS, cells were incubated with 1 mL of 1 μg/mL FITC-conjugated antimouse IgG (Jackson ImmunoResearch Laboratories Inc., West Grove, Pa, USA) at 4°C for 1 hour and washed again with PBS. The immunofluorescent stain of cells was observed by fluorescent microscopy.
2.7. SDS-PAGE and western blot analysis
Proteins in the cell lysate of A549 were separated by 12% SDS-PAGE and transferred to nitrocellulose sheets as described previously [24]. Proteins recognized by normal or peptide D08 hyperimmune mice sera were detected using HRP-conjugated antimouse immunoglobulin antibodies (Sigma Aldrich) and substrate.
2.8. Cell proliferation
Vero E6 () and A549 cells () were incubated with different doses of synthetic peptides as indicated for 72 hours. Cell proliferation was detected using commercial XTT assay (Roche Diagnostics, Indianapolis, Ind, USA).
2.9. IL-8 assay
The IL-8 production was assessed by commercial ELISA kits (R&D systems, Minneapolis, Minn, USA) according to the manufacturer's instructions. Briefly, A549 cells () were cultured alone or with different doses of peptides for 48 hours. Culture supernatants were collected after incubation, added to precoated ELISA plates, and incubated for 2 hours at 37°C. Plates were washed four times with the washing buffer. The bound IL-8 was detected by HRP-conjugated antibodies and substrate. The developed color was read by the Vmax microplate reader (Molecular Devices, Calif, USA). The concentration of IL-8 was calculated according to the standard curve.
2.10. Statistical analyses
Data are expressed as mean ± standard deviation (SD). The levels of significance for the differences between groups were analyzed using Student's t-test. A value of P < .05 was considered to be significant.
3. RESULTS
3.1. Search for molecular mimic regions in S protein
The whole amino acid sequence of spike protein was analyzed to find out the potential immunogenic regions and the regions shared sequence homology with human proteins, which is defined as the pathogenic regions. As shown in Figure 1, there are 4 pathogenic regions. Region 1 (residues 199–254), region 2 (residues 658–715), region 3 (residues 893–951), and region 4 (residues 1127–1184) have shared sequence homology with hydroxyacid oxidase, human golgi autoantigen, Angrgm-52, and pallidin, respectively. Among these regions, region 3 with homology to human Angrgm-52 has the highest score (with 34% identities and 48% similarity of conservative substitutions). Its sequence comparison with angrgm-52 is shown in Figure 2(a). In addition, because des-Arg bradykinin and Ang I are the substrates for ACE2 [25], we also compared the sequence of S protein against bradykinin (RPPGFSPFR) and Ang I (DRVYIHPFHL) and found that residues 490–502 (GYQPYRVVVLSFEE) of S protein showed sequence homology with bradykinin as indicated by bold letters here and in Figure 2(b). The identity score is 27%; and the similarity score from conservative substitutions is 36%.
Figure 1.
Sequence analysis of S protein. Putative S protein amino acid sequence was analyzed to find immunogenic regions (yellow regions) and pathogenic regions (regions shared sequence homology with human proteins, blue regions). Purple regions are both immunogenic and pathogenic regions. The grey region is the leader sequence and the brown region is the transmembrane region.
Figure 2.
Sequence homology between S protein, (a) human Angrgm-52, and (b) bradykinin. Spike protein sequence (protein databank NP_828851) was compared with sequences from all nonredundant GenBank CDS translations, PDB, SwissProt, PIR, and PRF of human, by using blastp (NCBI, NIH, USA). For Panel (a), BLASTP was used to calculate the similarity between S protein and human Angrgm-52, with matrix set at default. For an identical residue, one letter symbol of amino is shown between the two sequences, and for a conservative substation, “+” is shown. For Panel (b), pairwise alignment was calculated based on Smith-Water local alignment with matrix set at BLOSUM 45. “” is annotated for identical residues; “:” and “.” are for similar residues.
3.2. Screen for peptides recognized by SARS patients' sera
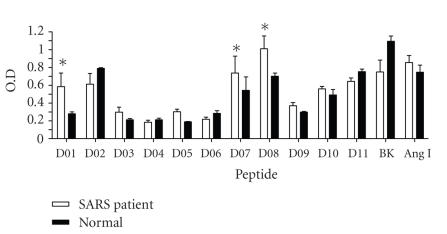
Eleven peptides (D01–D11, see Table 1), which represent those pathogenic regions were synthesized as well as bradykinin and Ang I were tested to see whether those peptides can be recognized by SARS patients' sera. The peptides were designed based on the algorithms predicting immunogenicity, second structure, protein topology, and hydrophobicity. Our goal is to select for peptides with high immunogenicity, with location on the protein surface, and with low hydrophobicity. The designed peptide sequences were synthesized and tested with clinical samples of SARS patient sera. A significant increase of SARS patients' sera binding to peptide D01, D07, and D08 was found as compared to the binding of normal sera (see Figure 3).
Table 1.
Amino acid sequence of the eleven synthetic peptides *Extra amino acid residues which were indicated by italic letters were added at either N- or C-terminus to decrease the hydrophobicity.
| Peptide | Amino acid positions | Amino acid sequence | No. of amino acids |
|---|---|---|---|
| D01 | 199–210 | GYQPIDVVRDLG | 12 |
| D02 | 658–669 | ASYHTVSLLRSTSQK | 15 |
| D03 | 733–744 | EEGNLLLQYGSFCTQ | 15 |
| D04 | 745–753 | EELNRALSGIAAGQ | 13 |
| D05 | 763–770 | VFAQVKQM | 8 |
| D06 | 911–919 | KAISQIQESLTTE | 13 |
| D07 | 927–937 | GLGKLQDVVNQNGE | 14 |
| D08 | 942–951 | ALNTLVKQLSSN | 12 |
| D09 | 1154–1162 | INASVVNIQK | 10 |
| D10 | 490–502 | GYQPYRVVVLSFEE | 14 |
| D11 | 306–317 | GFRVVPSGDVVRF | 13 |
Figure 3.
Antibody binding activity of SARS patients' sera to different peptides. Sera of SARS patients at convalescent stage as well as normal controls were collected as described in Section 2. Antibodies binding to different peptides were detected by ELISA as described in Section 2. BK represents bradykinin. “∗” indicates P < .05.
3.3. Hyperimmune sera against D08 crossreacted with A549 cells
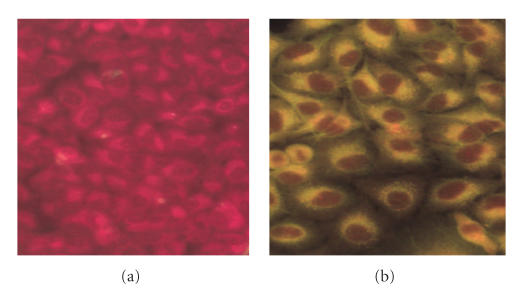
To test whether synthetic peptides D01, D07, and D08 can induce antibodies crossreacted with human proteins, we immunized mice with these peptides to generate hyperimmune sera against these peptides. We found hyperimmune sera against D08 can bind to the cytoplasmic region of human A549 cells as demonstrated by immunofluorescent stain (see Figure 4). Using Western blot analysis, hyperimmune sera against D08 could recognize more bands in A549 cell lysate as compared to normal mice sera (see Figure 5). In addition, hyperimmune sera against D07, but not D01, showed similar crossreactivity to A549 cells as hyperimmune sera against D08 did (data not shown).
Figure 4.
Immunofluorescent staining of mouse hyperimmune sera against A549 cells. A549 cells were grown on the slides and stained with secondary antibody alone (a) or mouse hyperimmune sera against D08 peptide (b) as described in Section 2.
Figure 5.
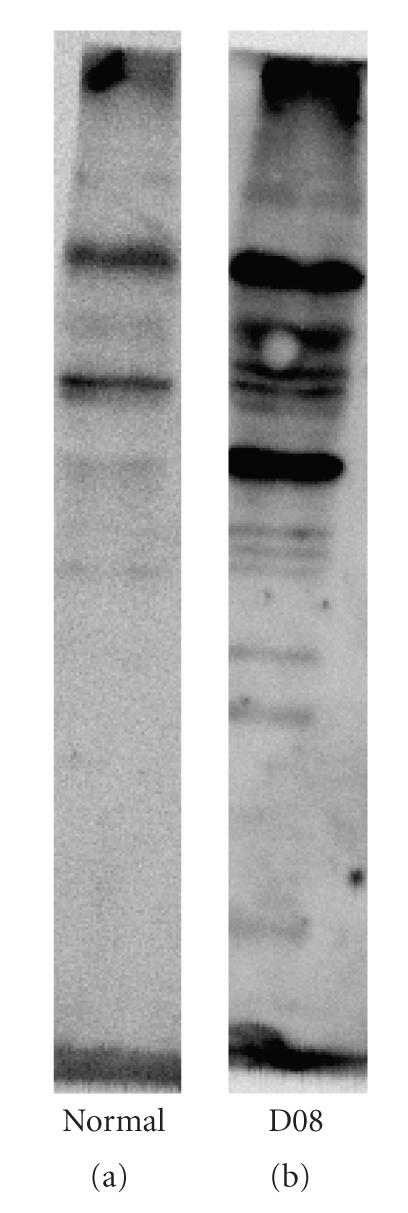
Mouse hyperimmune sera against D08 peptide recognize proteins in the A549 cell lysate. Proteins in the cell lysate of A549 cells were separated by SDS-PAGE and transferred to membrane as described in Section 2. Western blots against this membrane using normal mouse sera (a) or hyperimmune sera against D08 (b) are shown.
3.4. Hyperimmune sera against D10 crossreacted with bradykinin
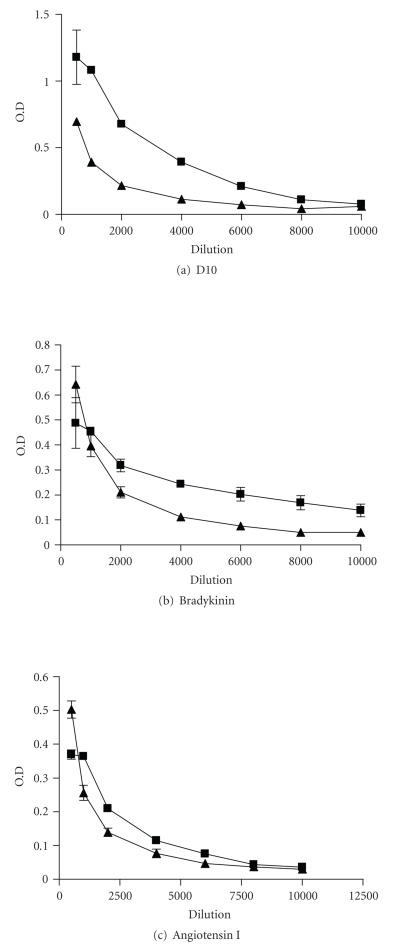
To test whether synthetic peptides D10, indeed, can induce antibodies crossreactive with bradykinin and Ang I, we immunized mice with D10 peptides to generate hyperimmune sera against this peptide. Significant increase of antibodies against D10 was found in D10 hyperimmune sera, which could crossreact with bradykinin, but not with Ang I-coated plates (see Figure 6).
Figure 6.
The crossreactivity of D10 antibody with bradykinin and Ang I. Hyperimmune sera from D10 immunized mice () or normal mice sera () were diluted as indicated and reacted with D10-, bradykinin-, or Ang I-coated ELISA plates as indicated. Bound antibodies were detected as described in Section 2. Data represents the mean ± SD of triplicates.
3.5. Peptide D10 induced IL-8 secretion and cell proliferation of A549 cells
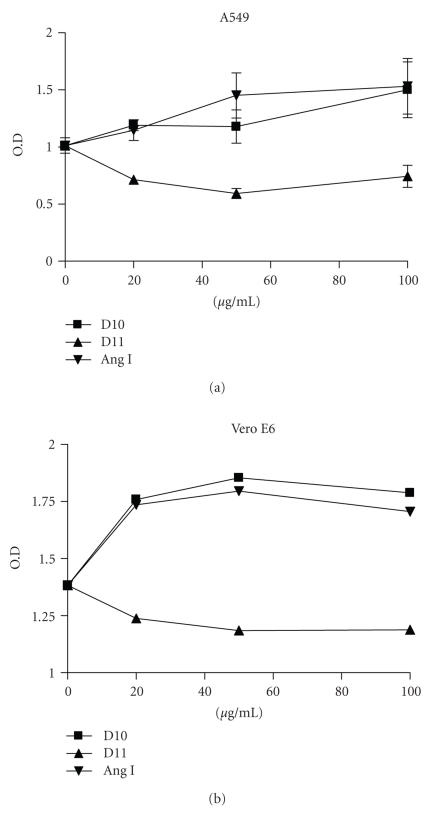
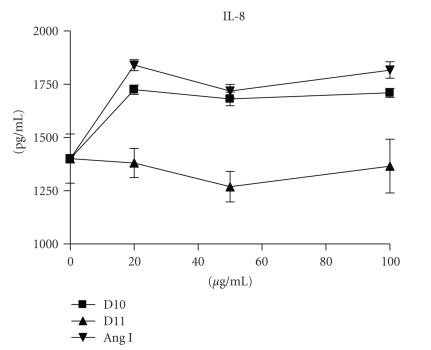
To understand whether D10 has similar biological activity as Ang I, we incubated Vero cells and lung epithelial A549 cells with D10, Ang 1, or control peptide D11. Both Vero and A549 cells were induced to proliferate in the presence of D10 and Ang I but not the control peptide (see Figure 7). In addition, D10 and Ang I could also induce chemokine IL-8 production of A549 cells (see Figure 8).
Figure 7.
Cell proliferation induced by D10 peptide and Ang I. Vero and A549 cells were incubated with different doses of peptides as indicated. Cell proliferation was detected after 72 hours of incubation by XTT assay as described in Section 2. Data represents the mean ± SD of triplicates.
Figure 8.
IL-8 production of A549 cells induced by D10 peptide and Ang I. A549 cells () were incubated with or without peptides for 48 hours. The levels of IL-8 in the culture supernatants were assayed as described in Section 2. Data represents the mean ± SD of triplicates.
4. DISCUSSION
In this study, we have identified four pathogenic regions of SARS-CoV S protein which share sequence homology with different human proteins. Among them, pathogenic region 3 (residues 893–941), which shares sequence homology with Angrgm-52 (GenBank accession no. AAL62340), a novel gene upregulated in human mesangial cells stimulated by angiotensin II, may deserve further investigation. Peptides D07 and D08 of this region were recognized by the sera of SARS patient indicating that this region is immunogenic and can be recognized by the immune system during SARS-CoV infection. Murine hyperimmune sera against peptides D07 or D08 were able to bind to recombinant S2 but not S1 domain of S protein (data not shown). In addition, hyperimmune sera against D07 or D08 also bounded to the cytoplasmic region of A549 cells and recognized several proteins in the A549 cell lysate. These results indicate that regions represented by D07 and D08 are immunogenic and may induce autoantibodies. However, further study is required to understand the biological function of these regions and the role of their antibodies in the pathogenesis of SARS-CoV infection.
In addition to D07 and D08 peptides, we also noticed that D10 peptide which represents residues 490–502 of S1 domain contained some interesting activities. The D10 peptide, which shared sequence homology with bradykinin, was able to generate antibodies crossreactive with bradykinin. In addition, D10 peptide could stimulate A549 to produce IL-8 and proliferation as Ang I did. These results suggest that the region of D10 in S protein may bind to Ang I receptor, ACE2, and may be involved in the binding of SARS-CoV to ACE2. This is consistent with the previous report, which indicates that residues 318–510 of S1 domain can bind to ACE2 [25] and is similar to the receptor binding domain of the HCoV-229E, which is within a fragment containing residues 407 to 547 [26]. Therefore, region 490–502 of S1 domain may be involved in the receptor binding domain of SARS-CoV.
In summary, our results suggest that molecular mimicry occurs between SARS-CoV and host proteins. Motifs shared sequence homology with host proteins of SARS-COV may be involved in the binding and fusion of SARS-CoV to host cells. Antibody against these motifs may contain neutralization activity against SARS-CoV infection or participate in the immunopathogenesis induced by SARS-CoV. As reported previously, SARS-CoV, like influenza, can inhibit the host's corticosteroid stress response via a molecular mimicry strategy [27]. Our studies on the mimicry motifs of S protein, which is involved in the virus, entry may provide alternative approaches to disrupt the infection of SARS-CoV, similar to the previous studies on the virus entry [28, 29].
ACKNOWLEDGMENT
This work was supported by Grant no.NSC92-2751-B006-Y from the National Science Council, Taipei, Taiwan.
References
- 1.Christian MD, Poutanen SM, Loutfy MR, Muller MP, Low DE. Severe acute respiratory syndrome. Clinical Infectious Diseases. 2004;38:1420–1427. doi: 10.1086/420743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuiken T, Fouchier RAM, Schutten M, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee N, Hui D, Wu A, et al. A major outbreak of severe acute respiratory syndrome in Hong Kong. New England Journal of Medicine. 2003;348(20):1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 4.Poutanen SM, Low DE, Henry B, et al. Identification of severe acute respiratory syndrome in Canada. New England Journal of Medicine. 2003;348(20):1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 5.Tsang KW, Ho PL, Ooi GC, et al. A cluster of cases of severe acute respiratory syndrome in Hong Kong. New England Journal of Medicine. 2003;348(20):1977–1985. doi: 10.1056/NEJMoa030666. [DOI] [PubMed] [Google Scholar]
- 6.Wenzel RP, Edmond MB. Managing SARS amidst uncertainty. New England Journal of Medicine. 2003;348(20):1947–1948. doi: 10.1056/NEJMp030072. [DOI] [PubMed] [Google Scholar]
- 7.Drosten C, Günther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New England Journal of Medicine. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 8.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. New England Journal of Medicine. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 9.Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 10.Marra MA, Jones SJM, Astell CR, et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 11.Bonavia A, Zelus BD, Wentworth DE, Talbot PJ, Holmes KV. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. Journal of Virology. 2003;77(4):2530–2538. doi: 10.1128/JVI.77.4.2530-2538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosch BJ, Martina BE, van der Zee R, et al. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(22):8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He Y, Zhou Y, Wu H, et al. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. Journal of Immunology. 2004;173(6):4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- 14.Li W, Moore MJ, Vasllieva N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong SK, Li W, Moore MJ, Choe H, Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. Journal of Biological Chemistry. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sui J, Li W, Murakami A, et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(8):2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oldstone MBA. Molecular mimicry and immune-mediated diseases. The FASEB Journal. 1998;12(13):1255–1265. doi: 10.1096/fasebj.12.13.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Letters. 1990;276(1-2):172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- 19.Rost B, Yachdav G, Liu J. The predict protein server. Nucleic Acids Research. 2004;32, Web Server Issue:321–326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frishman D, Argos P. Incorporation of long-distance interactions into a secondary structure prediction algorithm. Protein Engineering. 1996;9(2):133–142. doi: 10.1093/protein/9.2.133. [DOI] [PubMed] [Google Scholar]
- 21.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 22.Eisenberg D, Weiss RM, Terwilliger TC. The hydrophobic moment detects periodicity in protein hydrophobicity. Proceedings of the National Academy of Sciences of the United States of America. 1984;81(1):140–144. doi: 10.1073/pnas.81.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gish W, States DJ. Identification of protein coding regions by database similarity search. Nature Genetics. 1993;3(3):266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 24.Yeh TM, Chang HC, Liang CC, Wu JJ, Liu MF. Deoxyribonuclease-inhibtory antibodies in systemic lupus erythematosus. Journal of Biomedical Science. 2003;10(5):544–551. doi: 10.1159/000072382. [DOI] [PubMed] [Google Scholar]
- 25.Wong SK, Li W, Moore MJ, Choe H, Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. Journal of Biological Chemistry. 2004;279(5):3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breslin JJ, Mork I, Smith MK, et al. Human coronavirus 229E: receptor binding domain and neutralization by soluble receptor at C. Journal of Virology. 2003;77(7):4435–4438. doi: 10.1128/JVI.77.7.4435-4438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wheatland R. Molecular mimicry of ACTH in SARS—implications for corticosteroid treatment and prophylaxis. Medical Hypotheses. 2004;63(5):855–862. doi: 10.1016/j.mehy.2004.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kao RY, Tsui WH, Lee TS, et al. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetic. Chemistry & Biology. 2004;11(9):1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han DP, Penn-Nicholson A, Cho MW. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350(1):15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]