Abstract
A previous genetic analysis of a reporter gene carrying a 375bp region from a dpp intron (dppMX-lacZ) revealed that the Wingless and Dpp pathways are required to activate dpp expression in posterior spiracle formation. Here we report that within the dppMX region there is an enhancer with binding sites for TCF and Mad that are essential for activating dppMX expression in posterior spiracles. There is also a binding site for Brinker likely employed to repress dppMX expression. This combinatorial enhancer may be the first identified with the ability to integrate temporally distinct positive (TCF and Mad) and negative (Brinker) inputs in the same cells. Cuticle studies on a unique dpp mutant lacking this enhancer showed that it is required for viability and that the Filzkorper are U-shaped rather than straight. Together with gene expression data from these mutants and from brk mutants, our results suggest that there are two rounds of Dpp signaling in posterior spiracle development. The first round is associated with dorsal-ventral patterning and is necessary for designating the posterior spiracle field. The second is governed by the combinatorial enhancer and begins during germ band retraction. The second round appears necessary for proper spiracle internal morphology and fusion with the remainder of the tracheal system. Intriguingly, several aspects of dpp posterior spiracle expression and function are similar to demonstrated roles for Wnt and BMP signaling in proximal-distal outgrowth of the mammalian embryonic lung.
Keywords: Drosophila, Dpp, Mad, TCF, Brinker, combinatorial signaling, posterior spiracle enhancer, gene regulation
Introduction
Secreted proteins in the Transforming Growth Factorβ (TGFβ) and Wingless/Int-1 (Wnt) families have important roles in many species. In Drosophila, the TGFβ family member decapentaplegic (dpp) influences numerous developmental events (e.g., Ashe et al., 2000; Waltzer and Bienz, 1999). Typically, the transcription factor Mad is responsible for Dpp-dependent gene expression (e.g., Massagué et al., 2005). The Drosophila Wnt family member wingless (wg) also influences many developmental decisions (e.g., Cordero et al., 2004; Hatini et al., 2005). In canonical Wg signal transduction the transcription factor TCF is largely responsible for Wg-dependent gene expression (e.g., Willert and Jones, 2006).
In a genetic analysis we demonstrated that combinatorial signaling by the Wg and Dpp pathways regulates dpp expression in the posterior dorsal ectoderm. At stage 11, the dpp intron-derived reporter gene dppMX-lacZ is expressed in two bilaterally symmetrical clusters of dorsal ectoderm cells in the eighth abdominal segment. At stage 17, dppMX expression is present in posterior regions of the tracheal system: 1) in posterior portions of the dorsal trunk branches that connect the anterior and posterior spiracles, 2) in the spiracular branches that connect the spiracular chambers to the dorsal trunk branches and 3) the spiracular chambers of the posterior spiracles (Takaesu et al., 2002a). Interestingly, the posterior spiracles are the only functional tracheal opening at hatching and for the first larval instar only the spiracular branches and the spiracular chambers participate in gas exchange (Manning and Krasnow, 1993).
Substantial genetic and fate-map data shows that the development of the posterior spiracles is separable from the remainder of the tracheal system (Martinez-Arias and Lawrence, 1985; Jurgens, 1987). Consistent with these studies the spiracular branches and posterior spiracles are not detected with reagents commonly employed to study tracheal development such as trachealess or breathless (Takaesu et al., 2002a). In addition to dpp, gene expression studies revealed that the transcription factors spalt, cut and esg are also expressed in posterior spiracle cells. An analysis of their mutant phenotypes suggests that these genes are required for cell fate choices in the spiracles (Hu and Castelli-Gair, 1999; Merabet et al., 2005). Further, developmental studies of the external morphology of the posterior spiracles revealed a role for Rho signaling in invagination and formation of the spiracular lumen (Simoes et al., 2006). Here we report that dpp posterior spiracle activity does not influence cell fate or external morphology but instead regulates spiracle internal morphology.
Viability and cuticle studies of a unique mutant we created showed that the MX intronic region of the dpp locus is required for posterior spiracle development but not for dorsal-ventral patterning. This result contrasts with the prevailing wisdom that considers all dpp posterior spiracle defects simply downstream consequences of dorsal-ventral patterning defects. Instead, our data suggests that there are two rounds of Dpp signaling in posterior spiracle development. The first round is necessary for setting up the posterior spiracle field in association with dorsal-ventral patterning of the blastoderm stage embryo. The second begins during germ band retraction, is regulated by an enhancer in the MX region and appears to regulate fusion of the posterior spiracles with the dorsal trunk branches late in embryogenesis. In addition, the enhancer within the dppMX region contains binding sites recognized by TCF and Mad that are essential for activating dpp expression. There is also a binding site recognized by Brinker that appears to be employed to repress dppMX expression late in development. To our knowledge, this enhancer is the first one known that provides cells with the ability to respond to sequential positive and negative signals from three transcription factors.
Materials and Methods
Molecular biology
To create the dpp-ΔKX rescue construct we began with a NotI to PstI clone from the dpp chromosome walk (St. Johnston et al., 1990). This is a subclone from the 8kb EcoRI fragment that constitutes the dpp rescue construct (Padgett et al., 1993). A 100bp deletion from Kas1 to XbaI (ΔKX) was made in the subclone. An SphI to EcoRI subclone from the 8kb EcoRI fragment was also generated. The SphI to PstI fragment of the SphI to EcoRI subclone was replaced with the SphI to PstI fragment from the ΔKX version of the NotI to PstI subclone. An XhoI (from the MCS) to SphI fragment from the 8kb EcoRI clone was then inserted upstream of the ΔKX version of the SphI to PstI fragment. The 8kb EcoRI fragment was recreated minus the 100bp KasI to XbaI fragment and utilized to generate the dpp-ΔKX rescue construct in Casper4. To create the dpp-ΔKX reporter gene a Bluescript clone of the dppMX reporter gene was digested with Kas1 and XbaI, the ends were polished with T4 ligase and reclosed. To create the dppMX-MadM1+2 and dppMX-TCFM1+2 reporter genes oligos bearing mutations that match those shown in Supplemental Table 1 were incorporated into a dppMX subclone with Stratagene’s QuickchangeII kit (La Jolla, CA). Then each fragment was excised and inserted into the HZR-lacZ transformation vector as described (Takaesu et al., 2002a).
Drosophila genetics
PB{Gal4}43 is as described (Horn et al., 2003), PS{Gal4}8B4B is as described (Takaesu et al., 2002b), P{UAS-pan.TCF.□N}4, P{UAS-wg.H.T:HA1}6C and P{UAS.Brk}2.2 are as described (Flybase, 2007), P{UAS.Dpp}5 is as described (Staehling-Hampton and Hoffmann, 1994). In Gal4-UAS crosses where a transgene was not homozygous viable experimental embryos were positively identified by the absence of blue-balancer or GFP-balancer chromosomes. dppHin46, dppHin47 and dppHin61 are Haploinsufficient alleles as described in St. Johnston et al., (1990). The CyO.23 balancer carrying the dpp rescue construct is as described (Padgett et al., 1993). Strains homozygous for the dpp rescue or dpp-ΔKX rescue construct on chromosome III and a dppHin allele on chromosome II over In(2LR)Gla were generated via standard schemes. Lethality tests of these strains were conducted as described (Hoffman and Goodman, 1987). Cuticles were prepared as described (Wharton et al., 1993). brkF124 and brkM68 are null or nearly null allele as described (Jazwinska et al,. 1999, Lammel et al., 2000, Saller et al., 2002). The P[lacW] insertion insertions esgB7-2-22 and brk37 are as described (Flybase, 2007).
Biochemistry
Expression of the Histadine-tagged HMG box of TCF-A in pET15b (Novagen) was induced according to van de Wetering et al., (1997). Protein was purified using Ni2+-coated resin (New England Biolabs). Oligos were labeled with [γ-32P]ATP and purified by PAGE. Binding reactions were conducted according to Xu et al., (1998). Expression of the MH1-domain of Mad fused to GST in pGEX (Amersham) was induced according to Kim et al., (1997). Protein was purified with a GSTtrap column (Amersham). Oligos were end labeled with [γ-32P]dCTP and purified with QIAquick Nucleotide Removal kit (QIAGEN). Binding reactions were conducted according to Kim et al., (1997). Expression of full-length Brk protein (Minami et al., 1999) was conducted with the TNT Rabbit Reticulocyte Coupled Transcription Translation System (Promega). Oligos were labeled and purified as described for Mad-MH1. Binding reactions were conducted according to Sivasankaran et al., (2000). Bound and unbound oligos were separated using 5% native PAGE in 0.5X TBE buffer followed by autoradiography. All oligo sequences are shown in Supplemental Table 1.
Developmental biology
mRNA in situ hybridization to embryos with a digoxigenin labeled dpp cDNA was conducted as described (Takaesu et al., 2002a) and with a fluorescent labeled cDNA as described (Kosman et al,. 2004). Immunohistochemistry was performed as described (Johnson et al., 2003). The following primary antibodies were utilized: rabbit α-Spalt (Kuhnlein et al., 1994), rabbit α-phospho-Smad1 (Persson et al., 1998), rabbit α-lacZ (Organon Teknika) and mouse monoclonal 2B10 α-Cut (Jack et al., 1991). Secondary antibodies include: biotinylated goat α-rabbit and α-mouse (Vector Laboratories), Alexa Fluor 488- and 633-conjugated goat α-rabbit and α-mouse and horseradish peroxidase conjugated goat α-rabbit and α-mouse (Molecular Probes). The Vectastain Elite kit (Vector Laboratories) was employed to detect biotinylated antibodies and the TSA Amplification kit (Molecular Probes) was utilized to detect HRP conjugated antibodies.
Results
TCF, Mad and Brk recognize a combinatorial dpp posterior spiracle enhancer
Previously we showed that the dppMX reporter gene is expressed in posterior spiracles in a pattern that matches dpp transcript accumulation (Takaesu et al., 2002a). An analysis utilizing loss of function mutations suggested that combinatorial signaling by the Wg and Dpp pathways activates dppMX posterior spiracle expression. Subsequently, additional results supporting this hypothesis were obtained in overexpression studies with Dpp and Wg pathway components driven by two non-overlapping posterior spiracle specific Gal4 lines. In these studies we employed PB{Gal4}43 inserted in larp (Horn et al., 20003) and PS{Gal4}8B4B derived from the dppMX region (Takaesu et al., 2002b). Here the cell autonomous, dominant negative version of TCF was lethal when driven by PS{Gal4}8B4B (in dpp expressing posterior spiracle cells) but inconsequential when expressed with PB{Gal4}43. Alternatively, the nonautonomous Dpp and Wg proteins were lethal when expressed with either Gal4 line.
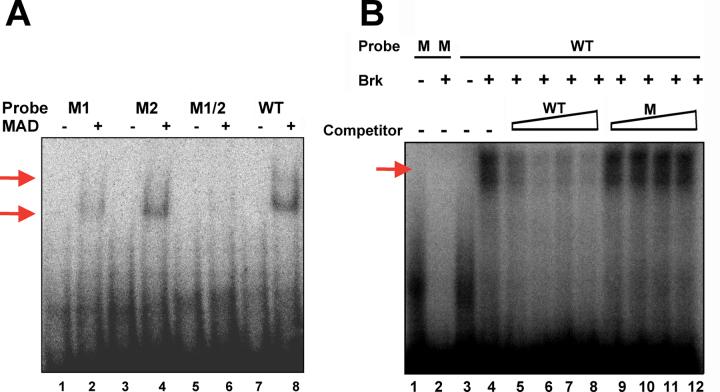
We then identified within the dppMX region a 54bp enhancer that is highly conserved and contains features appropriate for an enhancer responsive to Dpp and Wg signaling (Fig. 1A). This enhancer contains two bi-partite binding sites potentially recognized by both TCF and Mad. There is also a site recognizable by Brinker, a protein with the ability to antagonize Dpp (Jazwinska et al., 1999) and Wg signaling (Saller et al., 2002). Both TCF sites contain the core CTTT of the consensus shown in Tetsu and McCormick (1999). The Mad sites roughly match the sequence GCCGNCGC from Kim et al., (1997). The Brk site perfectly matches the sequence GGCGCC shown in Sivasankaran et al., (2000). Interestingly, the Brk site is contained within one of the Mad sites suggesting the possibility of competition between Mad and Brk as shown for enhancers of zerknüllt (Rushlow et al., 2001) and Ultrabithorax (Saller et al., 2002).
Fig. 1.
TCF efficiently and specifically binds the dpp posterior spiracle enhancer. (A) Combinatorial signaling enhancer within the dppMX reporter. I. An alignment of dpp intron sequences from D. melanogaster, D. simulans, D. pseudoobscura and D. virilis is shown. The D. melanogaster sequence begins 796 nucleotides downstream of the dpp exon2 splice donor. Identities are shown as dots and gaps as dashes. The D. melanogaster sequence is 94% identical to D. pseudoobscura and 84% identical to D. virilis. The sequences removed in the ΔKX deletion mutant are shown. II. Both strands of the D. melanogaster sequence are shown with putative binding sites for TCF, Mad and Brk. (B) Gelshift with TCF-HMG domain protein. Lane 8 in red serves as a positive control for two sets of experiments run on the same gel. Lanes 1-8 demonstrate the efficiency of TCF enhancer binding to both sites (shift and supershift indicated with red arrows). Lanes 1, 3, 5, and 7 show the mobility of each oligo alone. Mobility of the WT oligo is significantly impeded in the presence of TCF (Lane 8). Under identical conditions reduced TCF binding to M1 (Lane 2), M2 (Lane 4), and M1+2 (Lane 6) is seen. Binding of M1+2 is significantly less than WT. Lanes 8-24 demonstrate the specificity of TCF enhancer binding. Lanes 9-24 reveal the affects of adding increasing amounts of cold competitor oligo (50X, 100X, 200X and 300X) to the WT reaction in Lane 8. Adding cold WT (Lanes 9-12) significantly reduces binding at 50X and essentially eliminates binding at 200X. Adding cold M1 (Lanes 13-16) shows a similar reduction at 50X. Slightly more binding is seen at 200X and 300X than in WT indicating that this oligo cannot compete as efficiently as WT. Adding cold M2 (Lanes 17-20) does not reduce binding as efficiently as WT or M1 - at 50X and 100X greater binding is seen. Adding cold M1+2 (Lanes 21-24) has only modest effects on binding at 50X or 100X. Even at 300X, TCF binding to the WT oligo is not eliminated with M1+2 (Lane 24).
First we tested if the Wg pathway effector TCF could bind the enhancer in gel shift studies. We analyzed the specificity of TCF binding by utilizing mutants with CCC replacing TTT as in Yang et al., (1997). We also analyzed the efficiency TCF binding by utilizing 50-300 fold excess unlabeled wild type or mutant competitor oligos in binding reactions. Results of these studies are shown in Fig. 1B. The presence of shifts and supershifts in these studies demonstrate that TCF binds to both sites in the enhancer. This is consistent with our data that dpp expression in posterior spiracles is absent in Wg mutants (Takaesu et al., 2002a).
Then we tested if the Dpp pathway effector Mad could bind the enhancer. We analyzed the specificity of Mad binding by utilizing mutants with AT replacing CG as in Kim et al., (1997). Results of these studies are shown in Fig. 2A and the presence of shifts and supershifts demonstrate that Mad binds both sites in the enhancer. This is consistent with our data that dpp expression in posterior spiracles is reduced in Medea zygotic mutants (Takaesu et al., 2002a).
Fig. 2.
Mad and Brk efficiently and specifically bind the dpp posterior spiracle enhancer. (A) Gelshift with Mad-MH1 domain protein. Lanes 1-8 demonstrate the efficiency of Mad enhancer binding to both sites (shift and supershift indicated with red arrows). Lanes 1, 3, 5, and 7 show the mobility of each oligo alone. Mobility of the WT oligo is significantly impeded in the presence of Mad (Lane 8). Under identical conditions reduced Mad binding to M1 (Lane 2) and M2 (Lane 4) is seen. No binding of Mad to M1+2 is evident. (B) Gelshift with Brk protein. Lanes 1-4 demonstrate the efficiency of Brk enhancer binding (shift indicated with red arrow). Lanes 1 and 3 show the mobility of each oligo alone. The WT oligo is significantly impeded by Brk binding (Lane 4) but the mutant oligo is not (Lane 2). Lanes 4-12 demonstrate the effect of adding increasing amounts of cold competitor oligo (10X, 50X, 100X and 200X) to the WT reaction in Lane 4. Adding cold WT (Lanes 5-8) significantly reduces probe binding at 10X and essentially eliminates binding at 50X. Adding cold M1 (Lanes 9-12) has no effect.
Overall, the gel shift data for TCF and Mad is consistent with the genetic data implicating combinatorial signaling by Wg and Dpp pathways in the activation of dpp expression in posterior spiracle development. Alternatively, the identification of an exact match to the Brinker (Brk) binding site reported by Sivasankaran et al., (2000) was unexpected. However, when we examined reports of brk mRNA expression we found that it is present in the posterior spiracles beginning at stage 14 (Jazwinska et al., 1999). The expression of brk in posterior spiracles and the presence of a Brk binding site in the dpp enhancer suggested to us that Brk might repress dpp expression at some point in posterior spiracle development. This idea is consistent with a report (Jazwinska et al., 1999) that ubiquitous brk expression prevents posterior spiracle formation.
We tested the ability of Brk to bind to the combinatorial enhancer in gel shift studies. We analyzed the specificity of Brk binding utilizing mutants with AT replacing CG as in Sivasankaran et al., (2000). We also analyzed the efficiency of Brk binding by utilizing 10-200 fold excess unlabeled wild type or mutant competitor oligos in the binding reaction. Results of these studies are shown in Fig. 2B. The data demonstrate that Brk binds to the enhancer supporting the hypothesis that Brk may repress dpp expression in posterior spiracle development.
We then generated two transgenic strains carrying dppMX reporter genes with mutations that match those analyzed in our gel shift studies. One reporter has the dppMX region with both Mad sites mutated to mimic the MadM1+2 oligo. The second reporter has the dppMX region with both TCF sites mutated to mimic the TCFM1+2 oligo. LacZ staining of embryos from multiple transgenic strains for each reporter revealed that both of these mutation pairs completely eliminate dppMX-lacZ expression (data not shown).
The dpp combinatorial enhancer is required for posterior spiracle development
The combinatorial signaling enhancer is located in a dpp intron within the Haploinsufficient (Hin) region of the locus. This intron falls between the two protein coding exons (St. Johnston et al., 1990) and contains enhancer sequences regulating dpp blastoderm expression necessary for embryonic dorsal-ventral patterning (Hoffmann and Goodman, 1987). Embryos containing only a single functional copy of the dpp Hin region are inviable due to severe defects along the dorsal-ventral axis (Hoffmann and Goodman, 1987). A dpp rescue construct (Hoffmann and Goodman, 1987; Padgett et al., 1993) contains an 8kb EcoRI fragment encompassing the entire Hin region that alleviates dorsal-ventral patterning defects in an embryo carrying a dppHin mutation. dppHin mutations are often deletions removing a dpp protein coding exon (Hoffmann and Goodman, 1987).
To test the hypothesis that the combinatorial signaling enhancer is required for dpp function in posterior spiracle development we generated a unique dpp mutant. We utilized restriction sites in the dppMX reporter to delete a portion of the enhancer thereby creating the ΔKX deletion mutant (Fig. 1A). This deletion completely removes both Mad binding sites, the Brinker site, one of the TCF sites and ends immediately adjacent to the second TCF site possibly disrupting its context. We then replaced sequences corresponding to the dppMX region in the 8kb dpp rescue construct with the ΔKX deletion mutant creating the dpp-Δ□□ Multiple transgenic strains were the generated that carry this construct.
We conducted two viability studies on strains bearing the dpp-ΔKX rescue construct that also carried homozygous or heterozygous combinations of dppHin alleles. In these studies, dpp null embryos with two copies of the dpp-ΔKX rescue construct are still genetically null for the combinatorial enhancer. In the first study we learned that the original dpp rescue construct and our dpp-ΔKX rescue construct fully rescue embryonic viability in dpp Haploinsufficient embryos carrying one copy of a dppHin allele (Table 1A). In the second study we learned that two copies of the dpp rescue construct fully rescue embryonic viability in dpp null embryos (those carrying two dppHin alleles) but that two copies of the dpp-ΔKX rescue construct cannot rescue these embryos (Table 1B). Overall, these studies suggest that the combinatorial enhancer is required for embryonic viability but, as discussed below, it is not required for dorsal-ventral patterning.
Table 1.
Rescue of dpp Haploinsufficient and null genotypes
| A. % of expected Haploinsufficient adults rescued from a cross of dppHinX / CyO.23 females and the indicated males (>200 scored) | |||
|---|---|---|---|
| male genotype | |||
| y w | 0 | 0 | 0 |
| 1 copy dpp rescue construct (Padgett et al. 1993) | 106 | 108 | not done |
| 1 copy dpp rescue construct (Newfeld lab) | 98 | 113 | 97 |
| 1 copy dpp-ΔKX rescue construct | 103 | 109 | 87 |
| B. % of expected null embryos rescued from a cross of dppHin47/Gla females and dppHin61/Gla males both homozygous for the dpp rescue construct or for the dpp-ΔKX rescue construct (>200 scored) | |
|---|---|
| experimental embryos carrying these constructs | |
| 2 copies dpp rescue construct | 96 |
| 2 copies dpp-ΔKX rescue construct | 4 |
Note: CyO.23 carries the Padgett et al. (1993) dpp rescue construct
To provide evidence for the effect of the KX deletion on the expression of the combinatorial enhancer we generated a transgenic strain deleted for the KX region within the dppMX reporter. LacZ staining of embryos from several transgenic strains revealed that this deletion completely eliminates dppMX-lacZ expression (data not shown). Taken together the viability and reporter results suggest the hypothesis that dpp’s role in posterior spiracle development is genetically distinct from its role in dorsal-ventral patterning rather than a downstream effect. The hypothesis makes two predictions: 1) that the dpp-ΔKX rescue construct will act just like the dpp rescue construct in dorsal-ventral patterning but not in posterior spiracle development and 2) that posterior spiracle defects are the source of the lethality in dpp null embryos carrying two copies of the dpp-ΔKX rescue construct.
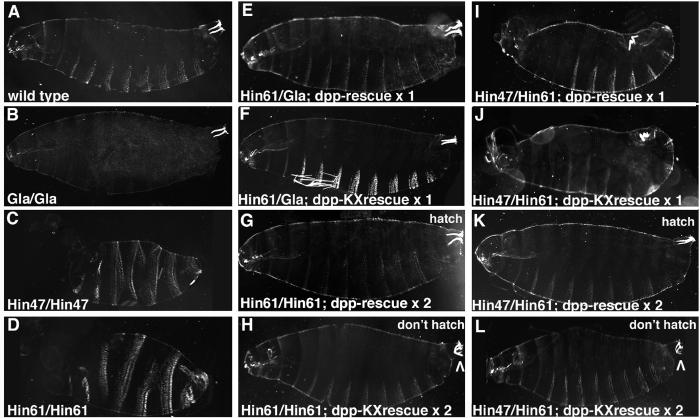
To investigate these predictions we examined cuticles from dpp mutant embryos generated in our viability studies (Fig. 3). First, we found normal cuticles on dpp Haploinsufficient embryos carrying one copy of either construct (Fig. 3E, F) reflecting the rescue of dorsal-ventral patterning defects. Second, dpp null embryos (full ventralization of the cuticle, Fig. 3C, D) were rescued to a Haploinsufficient phenotype (partial ventralization, Fig. 3I, J) by one copy of either construct. Third, dpp null embryos carrying two copies of either construct have normal dorsal-ventral patterning (Fig. 3G, H and 3K, L) but those with the dpp-ΔKX rescue construct have obvious posterior spiracle defects (Fig. 3H, L). dpp null embryos with two copies of the dpp-ΔKX rescue construct have U-shaped Filzkorper that appear unconnected to the remainder of the tracheal system.
Fig. 3.
Deleting the dpp posterior spiracle enhancer leads to Filzkorper defects without dorsal-ventral patterning defects. Darkfield images of embryonic cuticles in lateral view. The broad, white denticle belts on the ventral surface (bottom) and narrow, white Filzkorper in the posterior spiracles (upper right corner) are easily visible. (A) Wild type with two Filzkorper pointing straight into the embryo. (B) Homozygous Gla embryo with no denticle belts but normal Filzkorper. (C, D) Homozygous dppHin47 and dppHin61 embryos. In these dpp null genotypes denticles encircle the embryo and Filzkorper are missing. (E, F) Heterozygous dppHin61 embryos with one copy of the dpp rescue construct or one copy of the dpp-ΔKX rescue construct. This Haploinsufficient genotype is fully rescued by both constructs. (G, H) Homozygous dppHin61 embryos with two copies of the dpp rescue construct or two copies of the dpp-ΔKX rescue construct. This null genotype is rescued to hatching only by the dpp rescue construct. Both embryos have roughly normal denticle patterns but the dpp-ΔKX embryo has U-shaped Filzkorper (arrowhead) not likely connected to the remainder of the tracheal system. (I, J) dppHin47 and dppHin61 heterozygous embryos with one copy of the dpp rescue construct or one copy of the dpp-ΔKX rescue construct. This null genotype is rescued to a Haploinsufficient phenotype by both constructs. (K, L) dppHin47 and dppHin61 heterozygous embryos with two copies of the dpp rescue construct or two copies of the dpp-ΔKX rescue construct. This dpp null genotype is rescued to hatching only by the dpp rescue construct. Both embryos have roughly normal denticle patterns but again the dpp-ΔKX embryo has U-shaped Filzkorper (arrowhead).
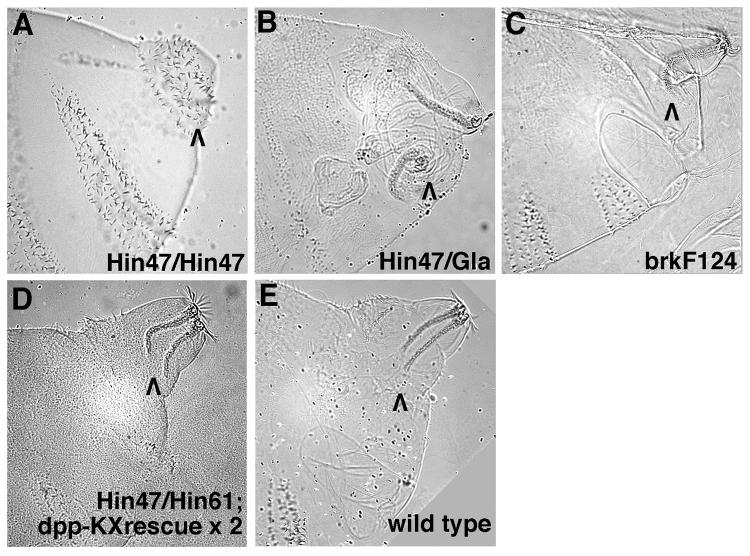
Comparisons of the posterior spiracles in cuticles from these dpp mutants suggested that there are two rounds of Dpp signaling in posterior spiracle development. Comparison of dpp null embryos (no Filzkorper, Fig. 4A) and dpp Haploinsufficient embryos (Filzkorper present but highly disorganized, Fig. 4B) identifies the first round. This round of Dpp signaling occurs in conjunction with dorsal-ventral patterning of the blastoderm stage embryo and specifies posterior spiracle primordia. Suppression of this round of Dpp signaling with maternally provided brk results in failure to form spiracles (Jazwinska et al. 1999). Comparison of dpp Haploinsufficient embryos (Fig. 4B) with dpp null embryos carrying two copies of the dpp-ΔKX rescue construct (Filzkorper present but U-shaped, Fig. 4D) identifies the second round. This round is regulated by the dppMX region enhancer and begins during germ band retraction. The second round of Dpp signaling appears to be required for refining the internal morphology of the posterior spiracles. Specifically, the second round of Dpp signaling seems to facilitate fusion of the posterior spiracles to the dorsal trunk branches of the tracheal system late in embryogenesis.
Fig. 4.
Filzkorper defects in dpp and brk mutant embryos suggest that two rounds of Dpp signaling occur during posterior spiracle development. High magnification Nomarski images of embryonic cuticles seen in lateral view. The focus is on abdominal segments 8-11 with the posterior spiracle region of segment 8 indicated by a black arrowhead. (A) dpp null embryos (dppHin47 homozygotes) have an ectopic denticle belt in place of their posterior spiracles. (B) dpp Haploinsufficient embryos (dppHin61 / Gla) have highly disorganized posterior spiracle regions characterized by misshapen Filzkorper that are displaced from their normal location. (C) brkF124 embryos have essentially normal posterior spiracles except that their Filzkorper are U-shaped and do not appear to be connected to the remainder of the tracheal system. (D) dppHin47 / dppHin61 embryos with two copies of the dpp-ΔKX rescue construct also have U-shaped Filzkorper (this view is slightly ventral from true lateral). Wild type embryos have straight Filzkorper that are attached to the spiracular branches (the connection is visible just above the arrowhead).
Given our gel shift data for Brk we examined cuticles from brkF124 embryos (Fig. 4C). These embryos display a phenotype with clear similarity to that of dpp null embryos carrying two copies of the dpp-ΔKX rescue construct (Filzkorper present but U-shaped, Fig. 3H, 3L and 4D). This similarity was at first surprising as the loss of brk and the loss of the dppMX region enhancer would be predicted to have opposite effects on dpp posterior spiracle expression. However, upon reflection it occurred to us that if Dpp posterior spiracle signaling very tightly regulated then too much or too little Dpp could lead to the same phenotype. From this perspective, the brkF124 phenotype supports our hypothesis that Brk influences dpp expression in posterior spiracle development. We should note that though the brkF124 phenotype shown in Fig. 4C was the most frequent spiracle phenotype, spiracle morphology was variable (as might be expected for a gene involved in both rounds of Dpp posterior spiracle signaling). The same results were seen in cuticles from brkM68 embryos.
dpp mRNA and pMad are specifically expressed in posterior spiracles and dorsal trunk branches
To fully document dpp activity in posterior spiracle development we examined dpp mRNA accumulation and the expression of phosphorylated Mad (pMad - an indicator of active Dpp signal transduction) in wild type embryos. At stage 13, dpp mRNA expression (Fig. 5A, B) is visible in the spiracular chamber of the posterior spiracles. At this stage pMad expression (Fig. 5C) is consistent with dpp mRNA expression and is also visible in the spiracular chamber. A subset of spiracular chamber cells will eventually secrete dense cuticular extensions that form the tracheal filter known as the Filzkorper (Hu and Castelli-Gair, 1999). These expression patterns are consistent with the cuticle phenotype of dpp null embryos carrying two copies of the dpp-ΔKX rescue construct.
Fig. 5.
dpp mRNA and pMad are strongly expressed in the spiracular chambers, spiracular branches and dorsal trunk branches of the tracheal system. Staged wild type embryos in dorsal view with anterior to the left: digoxigenin labeled for dpp mRNA (A, D, G, J, M), fluorescently labeled for dpp mRNA (B, E, H, K, N) or fluorescently labeled for pMad (C, F, I, L, O). Embryos digoxigenin labeled for dpp mRNA are shown at low and high magnifications while fluorescently labeled embryos are shown only at high magnification. In all panels, expression in the bilayered spiracular chamber that contains the Filzkorper is indicated with a red arrowhead. dpp and pMad expression in the hindgut, illustrated with a white arrowhead in stage 13 embryos, was utilized for stage matching. (A, B, C) Stage 13. dpp posterior spiracle expression is visible in the outer layer of cells in the spiracular chamber. pMad expression is present in both layers in the spiracular chamber. (D, E, F) Stage 14. dpp and pMad expression persists in the spiracular chamber and expression is now visible in the posterior-most portion of the dorsal trunk branches. (G, H, I) Stage 15. dpp and pMad remain visible in spiracular chamber while pMad expression begins to clear from the lumen. dpp mRNA and pMad expression is now visible in the spiracular branches and throughout the length of the dorsal trunk branches. Note that in fluorescent images of dpp mRNA (Fig. 5E, H, K, N), expression in the spiracular branches is outside the plane of focus. (J, K, L) Stage 16. All aspects of dpp mRNA and pMad expression continue. (M, N, O) Stage 17. dpp expression is absent from the spiracular chamber but remains strong throughout the length of the dorsal trunk branches. In contrast, pMad expression remains strong in the spiracular chamber, the spiracular branches and the dorsal trunk branches.
At stage 14, dpp mRNA and pMad expression (Fig. 5D, E, F) is strong in the spiracular chamber and is emerging in the spiracular branches and posterior portions of the dorsal trunk branches. At stages 15 and 16, dpp mRNA and pMad expression (5G, H, I and J, K, L) remain strong in the spiracular chamber and pMad begins to clear from the lumen. Both are now strongly expressed throughout the length of the spiracular branches and the dorsal trunk branches. Note that in high magnification fluorescent images of dpp mRNA expression (Fig. 5E, H, K, N), the spiracular branches are outside the focal plane. At stage 17, dpp mRNA expression (Fig. 5M, N) is unexpectedly absent from the spiracular chamber but remains visible in the spiracular branches and the dorsal trunk branches. In contrast, pMad expression (Fig. 5O) appears strong in the spiracular chamber, the spiracular branches and the dorsal trunk branches.
Taken together our genetic, cuticle and expression data are all consistent with the hypothesis that the second round of Dpp posterior spiracle signaling modulates the internal morphology of the posterior spiracles to facilitate fusion with the dorsal trunk branches.
Brk represses dpp expression in the spiracular chamber late in development
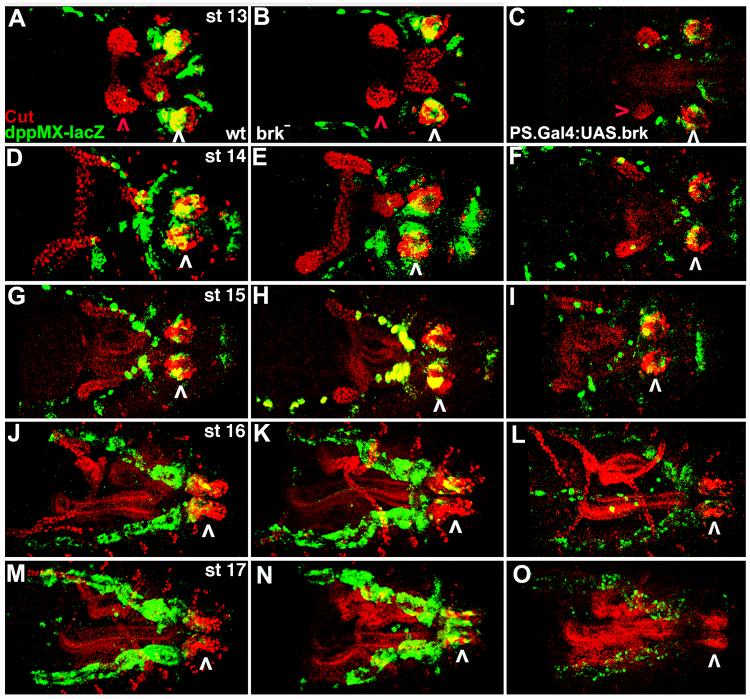
The unexpected finding that dpp mRNA expression is absent from the spiracular chamber of stage 17 embryos suggested that Brk repression of dpp expression occurs at this time. In our first test of this hypothesis we conducted co-localization studies for dppMX-lacZ and Brk (Campbell and Tomlinson, 1999). We found (as confirmed by others) that this antibody does not effectively detect Brk in embryos. Therefore we conducted side-by-side studies of dppMX-lacZ and brk-lacZ (utilizing the enhancer trap line brk37) and counterstained both sets of embryos with Cut. Cut is a transcription factor expressed in the spiracular chamber (Hu and Castelli-Gair, 1999). Cut expression in the nearby Malphigian tubules is highly dynamic and was utilized as a marker for stage matching of embryos.
Throughout embryogenesis dppMX-lacZ expression closely tracks dpp mRNA expression in posterior spiracles. dppMX-lacZ expression is strong in the spiracular chamber coincident with Cut expression, at stage 13 and is present in this tissue through stage 16. (Fig. 6B, E, H, K). By stage 17 no dppMX-lacZ expression is visible in the spiracular chamber (Fig. 6N). Alternatively, in the spiracular chamber, brk-lacZ is expressed in pattern that is complementary to dppMX-lacZ. At stage 13 when dppMX-lacZ expression in the spiracular chamber is at its height brk-lacZ is not present (Fig. 6C). During stages 14 through 16 brk-lacZ is strengthening in the spiracular chamber where it is also coincident with Cut expression (Fig. 6F, I, L). Finally, at stage 17, when dppMX-lacZ expression is absent from the spiracular chamber brk-lacZ is strongly expressed in this tissue (Fig. 6O).
Fig. 6.
dppMX-lacZ spiracular chamber expression is absent when brk-lacZ expression is strongest. Staged embryos in dorsal view with anterior to the left. Wild type embryos were single-labeled for Cut and detected with diaminobenzidine (A, D, G, J, M) or fluorescently double-labeled for Cut in red and dppMX-lacZ in green (B, E, H, K, N) or double-labeled for Cut in red and brk-lacZ (brk37) in green (C, F, I, L, O). Cut expression in the spiracular chamber is not a target of Dpp posterior spiracle signaling (see Fig. 8). Cut expression in the Malphigian tubules, illustrated with a red arrowhead in stage 13 embryos, was utilized for stage matching. Expression of Cut and/or lacZ in the spiracular chamber is indicated with a black (single labeled) or a white (double-labeled) arrowhead. (A, B, C) Stage 13. dppMX-lacZ is strongly expressed in the spiracular chamber coincident with Cut (yellow cells). brk-lacZ is not present in the spiracular chamber. (D, E, F) Stage 14. dppMX-lacZ expression remains strong in the spiracular chamber. brk-lacZ is now weakly present in the spiracular chamber coincident with Cut (yellow cells). (G, H, I) Stage 15. dppMX-lacZ expression weakens in the spiracular chamber and strengthens in the dorsal trunk branches. brk-lacZ remains visible in the spiracular chamber. (J, K, L) Stage 16. dppMX-lacZ expression continues to weaken in the spiracular chamber but is now very strong in the dorsal trunk branches. brk-lacZ remains at the same level in the spiracular chamber. (M, N, O) Stage 17. dppMX expression is absent from the spiracular chamber but remains very strong in the dorsal trunk branches. brk-lacZ is now strongly expressed in the spiracular chamber. brk-lacZ expression is also visible in the hindgut.
In the spiracular and dorsal trunk branches the absence of brk-lacZ expression at any stage contrasts with strong dppMX-lacZ expression in these tissues through stage 17. Thus, in the spiracular chamber at stage 17 when brk-lacZ expression is greatest we see the extinction of dppMX-lacZ expression but in the spiracular and dorsal trunk branches where brk-lacZ is absent we see significant levels of dppMX-lacZ expression (Fig. 6N, O).
In more rigorous tests of our hypothesis that Brk represses dpp posterior spiracle expression during stage 17 we examined dppMX-lacZ expression in brkM68 and UAS.Brk overexpression embryos. We found that overexpression of Brk in the dppMX pattern effectively repressed dppMX-lacZ expression in the spiracular chamber, spiracular branches and the dorsal trunk branches at all stages (Fig. 7C, F, I, L, O). This shows that Brk can antagonize dppMX-lacZ expression whenever and wherever Brk is strongly expressed. Consistent with this result, in brk mutants we see the opposite effect on dppMX-lacZ expression (Fig. 7B, E, H, K). In the absence of brk at stage 17 (when brk spiracular chamber expression is normally strongest) we see ectopic dppMX-lacZ expression (Fig. 7M, N). In the spiracular branches and dorsal trunk branches where brk is not expressed we see no difference between dppMX-lacZ expression in wild type and brk mutants (Fig. 7M, N).
Fig. 7.
Increasing or decreasing brk levels in the spiracular chamber engenders the opposite effects on dppMX-lacZ expression. Staged embryos in dorsal view with anterior to the left. Wild type embryos (A, D, G, J, M), brkM68 embryos (B, E, H, K, N) and embryos with UAS.Brk driven by PS{Gal4}8B4B (C, F, I, L, O) were double-labeled for Cut in red and dppMX-lacZ in green. PS{Gal4}8B4B driving UAS.Brk leads to overexpression of Brk in the dpp posterior spiracle pattern. Cut expression in the Malphigian tubules, illustrated with a red arrowhead in stage 13 embryos, was utilized for stage matching. Expression of Cut and lacZ in the spiracular chamber is indicated with a white arrowhead. (A, B, C) Stage 13. In wild type dppMX-lacZ is strongly expressed in the spiracular chamber. In brk mutants dppMX-lacZ spiracular chamber expression is also present but it is barely visible in UAS.Brk embryos. (D, E, F) Stage 14. In wild type dppMX-lacZ expression weakens in the spiracular chamber. In brk mutants dppMX expression is present and is still faint in UAS.Brk embryos. (G, H, I) Stage 15. In wild type dppMX-lacZ expression continues to weaken in the spiracular chamber but is now present in the dorsal trunk branches. In brk mutants dppMX-lacZ spiracular chamber and dorsal trunk branch expression are roughly equal to wild type. dppMX-lacZ spiracular chamber expression remains faint in UAS.Brk embryos and is below wild type in the dorsal trunk branches. (J, K, L) Stage 16. In wild type dppMX expression is weakly visible in the spiracular chamber but is now strong in the dorsal trunk branches. In brk mutants dppMX-lacZ spiracular chamber and dorsal trunk branch expression remains roughly equal to wild type levels. dppMX-lacZ expression is absent in the spiracular chamber and very faint in the dorsal trunk branches in UAS.Brk embryos. (M, N, O) Stage 17. In wild type dppMX-lacZ expression is absent from the spiracular chamber and very strong in the dorsal trunk branches. In brk mutants dppMX-lacZ ectopic spiracular chamber expression is present while dorsal trunk branch expression remains roughly equal to wild type. dppMX-lacZ expression is again absent in the spiracular chamber and very faint in the dorsal trunk branches in UAS.Brk embryos.
Taken together our data plus that of (Takaesu et al. 2002a) support the hypothesis that combinatorial signaling by the Dpp and Wg pathways turn on the dpp posterior spiracle enhancer at stage 13 in the spiracular chamber and slightly later in the spiracular branches and dorsal trunk branches. Brk then turns off the dpp posterior spiracle enhancer at stage 17 in the spiracular chamber.
Neither spalt nor cut nor esg-lacZ is a target of Dpp posterior spiracle signaling
Returning to the issue of Dpp’s role in late stage posterior spiracle development we viewed three proteins (Spalt, Cut and Escargot; Hu and Castelli-Gair, 1999; Merabet et al., 2005) expressed in the posterior spiracle as candidate targets for Dpp signaling. Spalt is a well-known Dpp target gene in wing development (de Celis et al., 1996) that is expressed in the stigmatophore - the epidermal layer that surrounds the spiracular chamber where Dpp is expressed. Cut is expressed in the spiracular chamber coincident with dppMX-lacZ expression. Escargot is expressed in a subset of Cut expressing cells (Whiteley et al., 1992).
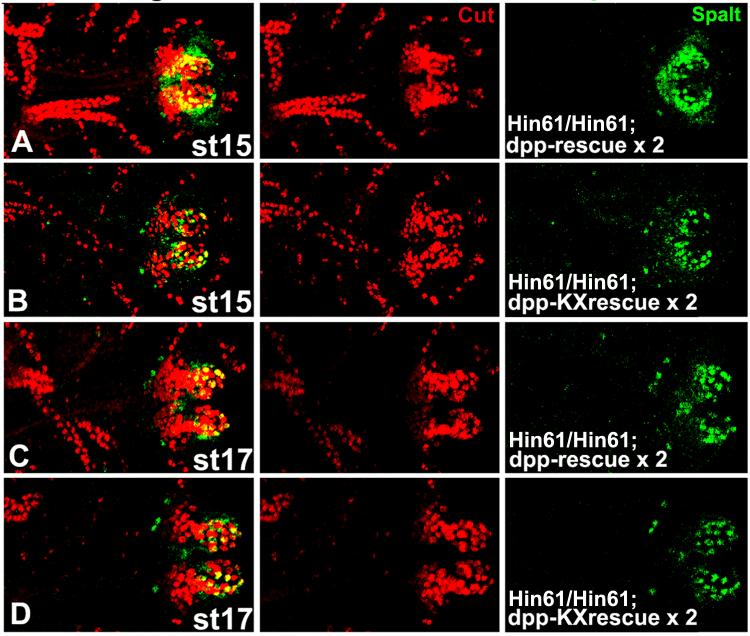
We examined the expression of these three genes in two sets of experiments. First, we employed the unique dpp posterior spiracle mutant analyzed in our cuticle study. As shown in Fig. 8, dpp null embryos (homozygous for dppHin61) that carry two copies of the dpp-rescue construct or two copies of the dpp-ΔKX rescue construct were examined for Cut and Spalt expression. We found that both Cut and Spalt are visible in a wild type pattern in dpp posterior spiracle mutants. This suggests that neither Cut nor Spalt are targets of Dpp posterior spiracle signaling and is consistent with the demonstration that Cut is unaffected in brk mutants or in UAS.Brk overexpression genotypes (Fig. 7).
Fig. 8.
Cut and Spalt are not targets of dpp posterior spiracle activity in rescue experiments. dpp null embryos (homozygous for dppHin61) from the indicated stages that carry two copies of the dpp-rescue construct (A, C) or two copies of the dpp-ΔKXrescue construct (B, D) are shown with anterior to the left. Embryos were double labeled for Cut and Spalt expression with the merged image shown in the left column. Cut expression in the Malphigian tubules was utilized for stage matching. Cut spiracular chamber expression is comparable in both genotypes and at both stages. The reduction in Spalt stigmatophore expression at stage 15 in dpp-ΔKX rescue embryos is transient and by stage 17 Spalt expression is the same in both genotypes.
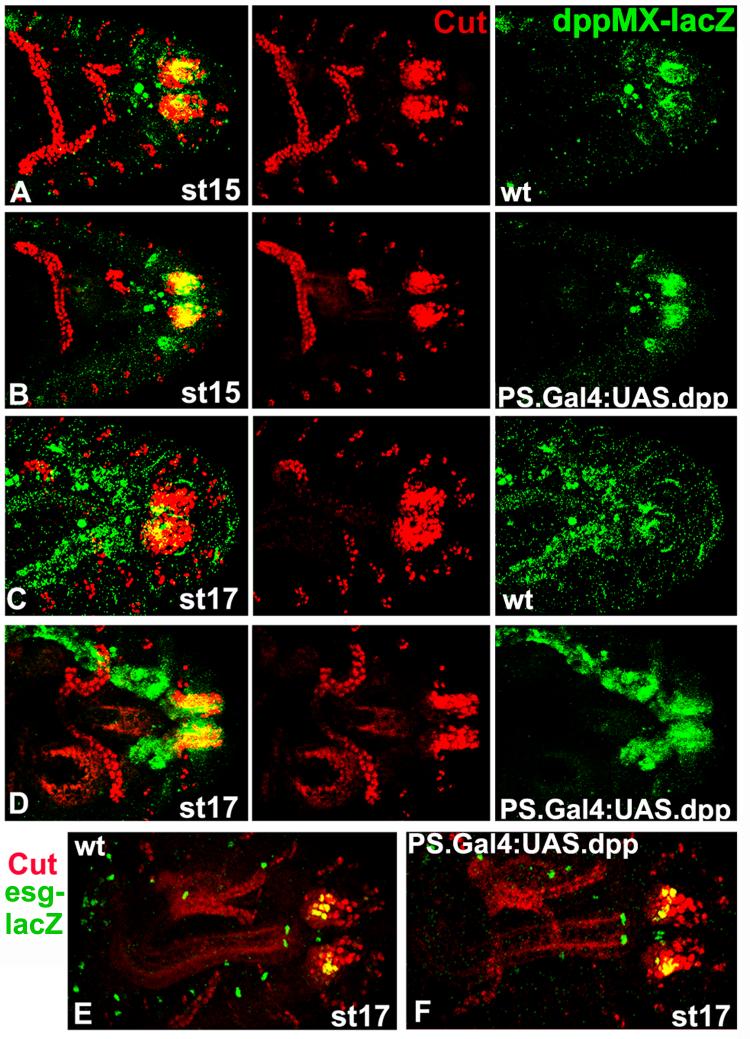
Next we examined the expression of Cut and esg-lacZ (esgB7-2-22) in embryos with UAS.Dpp driven by our posterior spiracle Gal4 line (Takaesu et al., 2002b). These embryos have overexpression of Dpp in its native posterior spiracle pattern. In the Cut experiment, as a positive control we included the dppMX reporter gene and monitored its expression. As shown in Fig. 9A-D, notwithstanding the increase in dppMX-lacZ expression in the UAS.Dpp embryos we noted no changes in Cut expression. Similarly (though in this case neither embryo carried dppMX-lacZ) we see no changes in esg-lacZ expression. Thus, esg-lacZ is also not a target of Dpp posterior spiracle signaling.
Fig. 9.
Cut and esg-lacZ are not targets of Dpp posterior spiracle signaling in overexpression experiments. Staged wild type embryos (A, C) and embryos with UAS.Dpp driven by PS{Gal4}8B4B that express dppMX-lacZ (B, D)are shown with anterior to the left. PS{Gal4}8B4B driving UAS.Dpp generates overexpression of Dpp in its native posterior spiracle pattern. Cut expression in the Malphigian tubules was utilized for stage matching. (A-D) Embryos were double labeled for Cut and dppMX-lacZ with the merged image in the left column. Cut spiracular chamber expression is comparable in both genotypes and at both stages. The overexpression of Dpp in the PS{Gal4}8B4B embryos is visible as increased dppMX-lacZ expression at both stages (right column). (E) A staged wild type embryo and (F) an embryo with UAS.Dpp driven by PS{Gal4}8B4B that both express lacZ from the esgB7-2-22 enhancer trap are shown with anterior to the left. The embryos were double labeled for Cut and lacZ. esg-lacZ expression in the spiracular chamber, coincident with Cut is largely the same in both genotypes.
Discussion
Our previous genetic analysis of the dppMX reporter gene revealed that combinatorial signaling by the Wg and Dpp pathways is required to activate dpp expression in posterior spiracles. This study advances our understanding of the regulation of dpp posterior spiracle expression and suggests a possible function for Dpp signaling in posterior spiracle development.
A novel combinatorial enhancer regulates dpp posterior spiracle expression
Our data shows that within the dppMX region there is a combinatorial enhancer that contains binding sites recognized by TCF and Mad that are essential for activating dpp expression in the spiracular chambers, spiracular branches and in the dorsal trunk branches. There is also a binding site recognized by Brinker that is likely employed to repress dpp expression late in spiracle development. To our knowledge, this is the first enhancer with the ability to sequentially integrate two positive and one negative factor in the same cells.
What makes this enhancer different from other enhancers in Drosophila also capable of integrating three inputs in the same cells (e.g., Flores et al., 2000; Halfon et al., 2000; Xu et al., 2000)? These enhancers integrate only positive signals. In all cases PointedP2 binding displaces the Yan repressor that is constitutively bound to the enhancer in the absence of PointedP2. The difference is that the dppMX enhancer is actively repressed by Brk binding after being stimulated by positive input from the Dpp and Wg pathways. What makes this enhancer different from other enhancers in Drosophila that integrate positive and negative signals such as the enhancer of Ultrabithorax where positive input from TCF is associated with a competition between Mad (positive) and Brk (negative) inputs (Saller et al. 2002)? The difference is that in the same cells the dppMX enhancer responds sequentially to positive combinatorial input from TCF and Mad and then to negative input from Brinker. The Ultrabithorax enhancer responds simultaneously to positive input from TCF and Mad in parasegment seven and to negative input from Brinker in the adjacent cells of paraegment 8.
If combinatorial signaling by the Dpp and Wg pathways, via TCF and Mad, turn on the dppMX enhancer in posterior spiracle primordia of the dorsal ectoderm at stage 13, then where do the Dpp signals originate? One possibility is that Dpp signals derive from the adjacent region of the dorsal ectoderm - leading edge cells located just anterior to the posterior spiracle primordia. In leading edge cells of the dorsal ectoderm dpp expression is activated at stage 8. We and others have shown that dpp leading edge expression is activated by enhancers distinct from the dppMX enhancer (e.g., Newfeld and Takaesu, 2002) and that the leading edge enhancers are themselves stimulated, in part, by dpp blastoderm expression that sets up the embryonic dorsal/ventral axis. In this scenario, the activation of the dppMX enhancer in posterior spiracles by Dpp leading edge signaling represents the last step in a cascade, covering nearly all of embryogenesis, of increasingly spatially restricted rounds of Dpp dorsal ectoderm signaling.
The most likely the source of the Wg signal is a small group of cells in the spiracular chamber (Mirabet et al., 2005). wg expression in the spiracular chamber becomes visible at stage 11 and is present through the remainder of embryogenesis (van den Heuvel et al., 1989). This group of Wg expressing cells is required for the maintenance of Cut and Spalt expression (Mirabet et al. 2005), genes shown here to be independent of Dpp signaling. The involvement of Wg in spiracle cell fate determination and dpp activation results in more severe spiracle defects in wg mutants (Mirabet et al. 2005) than in brkF124 embryos or dpp null embryos with two copies of the dpp-ΔKX rescue construct.
The source of the signal that activates brk in the posterior spiracles is less easy to identify. However, one possibility is suggested by the mutant phenotype generated by ubiquitous expression of unpaired (a ligand of the Jak/Stat pathway with a role in posterior spiracle formation, Brown et al., 2003). These embryos display a U-shaped Filzkorper similar to brkF124 embryos and dpp null embryos with two copies of the dpp-ΔKX rescue construct.
Dpp signaling may regulate posterior spiracle outgrowth and dorsal trunk branch fusion
Our data advances our understanding of posterior spiracle development and the role that Dpp signaling plays in this process in three areas: 1) that dpp activity in dorsal/ventral patterning is genetically separable, in part, from its activity in posterior spiracle development, 2) that dpp signaling does not appear to influence posterior spiracle cell fate or external morphology but instead regulates spiracle internal morphology and 3) that a functioning posterior spiracle is necessary for viability prior to hatching.
Regarding the separability of dpp dorsal/ventral patterning and posterior spiracle functions, this view contrasts with the prevailing wisdom that all dpp posterior spiracle defects are downstream consequences of dorsal-ventral patterning defects. Instead, our results suggest that there are two rounds of Dpp signaling in posterior spiracle development. The first round is necessary for setting up the posterior spiracle field in association with dorsal-ventral patterning at the blastoderm stage. The second begins during germ band retraction and appears to regulate the internal morphology of the spiracles. In our view, one possible explanation for why these distinct aspects of dpp function were connected in the conventional wisdom is that the dppMX enhancer is located in an intron alongside dorsal/ventral patterning enhancers and is deleted in several widely studied dppHin alleles (e.g., dppHin61; St. Johnston et al., 1990).
This two-round model for dpp signaling in posterior spiracle development fits well with our analysis of Dpp signaling in heart development. Here we discovered that there is a second round of Dpp dorsal ectoderm to mesoderm signaling late in development that maintains the boundary between pericardial cells and the adjacent dorsal muscle cells (Johnson et al., 2007). The second round of Dpp signaling in heart development is autoactivated by Dpp signals that also likely derive from dpp leading edge expression. Thus, in heart development there is also evidence of a multi-step cascade of increasingly spatially restricted rounds of Dpp dorsal ectoderm signaling.
Regarding the function of the second round of Dpp signaling in posterior spiracle development, our data shows that the expression of three transcription factors essential for cell fate determination in the spiracles are independent of Dpp signaling. In addition, our pMad data shows that the lumen of the spiracular chamber forms normally suggesting that spiracle external morphology and invagination, under the control of Rho signaling is also independent of Dpp.
Our cuticle data indicates that the primary defect in dpp posterior spiracle mutants is fully differentiated but U-shaped Filzkorper that do not appear to connect to the dorsal trunk branches. This phenotype plus the fact that dpp mRNA and pMad expression normally span the spiracular chamber, spiracular branches and dorsal trunk branches suggest the hypothesis that Dpp regulates the internal morphology of the spiracles. Given the mutant phenotype and gene expression patterns it is tempting to speculate that Dpp signaling via pMad directs the anterior outgrowth of the spiracles, the posterior outgrowth of the dorsal trunk branches and their eventual fusion into a coherent tracheal system.
Regarding posterior spiracle function in embryos, the fact that our dpp posterior spiracle mutants do not hatch suggests that gas exchange through the posterior spiracles and the spiracular branches begins, and is required to sustain the individual, prior to hatching. This is an advance in our understanding of Drosophila embryonic and larval respiration.
Conservation of Dpp posterior spiracle signaling in mammalian lung development
Numerous studies have shown that there is strong functional conservation for components of Dpp/BMP4 signaling pathways across species. Most robust is the conservation of the signaling mechanism (e.g., pathway dedication of receptor-associated Smads; Marquez et al., 2001). In addition, there are numerous instances where the role of Dpp/BMP4 signaling in development is conserved (e.g., induction of cardiogenic mesoderm; Cripps and Olson, 2002).
Several aspects of dpp posterior expression and function are consistent with recent studies of morphogenesis in mammalian lung epithelia. Shu et al. (2005) showed that Wnt signals from the proximal epithelium activates BMP4 expression in distal epithelium and that this interaction is required for proper proximal-distal outgrowth of the lung during late stages of mouse development (E16.5 to birth). Their experiments also implicate TCF and TCF binding sites in the BMP4 promoter in this interaction. The similarity between this inductive event and Wg activation of Dpp in late stage spiracle formation is intriguing.
Additional similarities are found in a study of mice with a conditional knockout in the lung epithelium of the BMPR1a type I receptor (Alk3 homologous to Thickveins - the Dpp type 1 receptor; Newfeld et al., 1999). This study reports two very interesting parallels with dpp posterior spiracle signaling (Eblaghie et al., 2006). First, autocrine signaling by BMP4 in lung epithelial cells during late stages of development (E16.5 to birth) is necessary for growth of the distal lung epithelium just as dpp autoactivation is necessary for posterior spiracle development. Second, the mutant phenotypes for BMP4 loss of function and BMP4 overexpression in the distal lung epithelium are the same, just as dpp loss of function (dpp null embryos carrying two copies of the dpp-ΔKX rescue construct) and dpp overexpression (brkF124 embryos) both generate U-shaped Filzkorper. Viewed together with our data these studies suggest that combinatorial activation of dpp/BMP4 expression by Wnt and BMP family members is essential for late stage airway development in both species.
Supplementary Material
Acknowledgements
Adela Syed, Alan Laughon and Tetsuya Tabata provided expression plasmids. We thank Adela Syed, Rolf Bodmer, Ernst Wimmer, Mike Hoffmann and the Bloomington Stock Center for flies. The Developmental Studies Hybridoma Bank, Calle Heldin and Bertrand Mollereau provided antibodies. Alan Laughon, Laural Raftery, Adela Syed and two anonymous reviewers provided valuable comments. We thank Duke University Model Systems Genomics for fly injections. Joel Frandsen and Charlotte Konikoff assisted with embryo staining. Mike Stinchfield helped with fly pushing. This study was supported by grants from NSF (IBN-0196059 to TVO) and NIH (CA095875 to SJN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashe H, Mannervik M, Levine M. Dpp signaling thresholds in the dorsal ectoderm of the Drosophila embryo. Development. 2000;127:3305–3312. doi: 10.1242/dev.127.15.3305. [DOI] [PubMed] [Google Scholar]
- Brown S, Hu N, Hombria J. Novel level of signaling control in the Jak/Stat pathway revealed by in situ visualization of protein-protein interaction during Drosophila development. Development. 2003;130:3077–3084. doi: 10.1242/dev.00535. [DOI] [PubMed] [Google Scholar]
- Campbell G, Tomlinson A. Transducing the Dpp morphogen gradient in the wing of Drosophila: regulation of Dpp targets by brinker. Cell. 1999;96:553–562. doi: 10.1016/s0092-8674(00)80659-5. [DOI] [PubMed] [Google Scholar]
- Cordero J, Jassim O, Bao S, Cagan R. A role for wingless in a pupal cell death event that contributes to patterning the Drosophila eye. Mech. Dev. 2004;121:1523–1530. doi: 10.1016/j.mod.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Cripps R, Olson E. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev. Biol. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- de Celis J, Barrio R, Kafatos F. A gene complex acting downstream of Dpp in Drosophila wing morphogenesis. Nature. 1996;381:421–424. doi: 10.1038/381421a0. [DOI] [PubMed] [Google Scholar]
- Eblaghie M, Reedy M, Oliver T, Mishina Y, Hogan B. Evidence that autocrine signaling through BMPR1a regulates the proliferation, survival and morphogenetic behavior of distal lung epithelial cells. Dev. Biol. 2006;291:67–82. doi: 10.1016/j.ydbio.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Flores G, Duan H, Yan H, Nagaraj R, Fu W, Zou Y, Noll M, Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75–85. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- Flybase 2007 http://flybase.bio.indiana.edu/
- Halfon M, Carmena A, Gisselbrecht S, Sackerson C, Jimenez F, Baylies M, Michelson A. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Jan Y. Studying Drosophila embryogenesis with P-lacZ enhancer trap lines. Roux Arch. Dev. Biol. 1992;201:194–220. doi: 10.1007/BF00188752. [DOI] [PubMed] [Google Scholar]
- Hatini V, Green R, Lengyel J, Bray S, Dinardo S. The Drumstick/Lines/Bowl regulatory pathway links antagonistic Hedgehog and Wingless signaling inputs to epidermal cell differentiation. Genes Dev. 2005;19:709–718. doi: 10.1101/gad.1268005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann F, Goodman W. Identification in transgenic animals of the Drosophila dpp sequences required for embryonic dorsal pattern formation. Genes Dev. 1987;1:615–625. doi: 10.1101/gad.1.6.615. [DOI] [PubMed] [Google Scholar]
- Horn C, Offen N, Nystedt S, Hacker U, Wimmer E. piggyBac-based insertional mutagenesis and enhancer detection as a tool for functional insect genomics. Genetics. 2003;163:647–661. doi: 10.1093/genetics/163.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N, Castelli-Gair J. Study of the posterior spiracles of Drosophila as a model to understand the genetic and cellular mechanisms controlling morphogenesis. Dev. Biol. 1999;214:197–210. doi: 10.1006/dbio.1999.9391. [DOI] [PubMed] [Google Scholar]
- Jack J, Dorsett D, Delotto Y, Liu S. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development. 1991;113:735–747. doi: 10.1242/dev.113.3.735. [DOI] [PubMed] [Google Scholar]
- Jazwinska A, Rushlow C, Roth S. The role of Brinker in mediating the graded response to Dpp in early Drosophila embryos. Development. 1999;126:3323–3334. doi: 10.1242/dev.126.15.3323. [DOI] [PubMed] [Google Scholar]
- Johnson A, Bergman C, Kreitman M, Newfeld S. Embryonic enhancers in the dpp disk region regulate a second round of Dpp signaling from the dorsal ectoderm to the mesoderm that represses Zfh-1 expression in a subset of pericardial cells. Dev. Biol. 2003;262:137–151. doi: 10.1016/s0012-1606(03)00350-6. [DOI] [PubMed] [Google Scholar]
- Johnson A, Burnett L, Sellin J, Paululat A, Newfeld S. Defective Decapentaplegic signaling results in heart overgrowth and reduced cardiac output in Drosophila. Genetics. 2007;176:1609–1624. doi: 10.1534/genetics.107.073569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G. Segmental organization of the tail region in the embryo of Drosophila. A blastoderm fate map of cuticle structures of the larval tail. Roux Arch. Dev. Biol. 1987;196:141–157. doi: 10.1007/BF00376308. [DOI] [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen H, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Dpp. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Kosman D, Mizutani C, Lemons D, Cox W, McGinnis W, Bier E. Multiplex detection of RNA Expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- Kuhnlein R, Frommer G, Friedrich M, Gonzalez-Gaitan M, Weber A, Wagner-Bernholz J, Gehring W, Jackle H, Schuh R. spalt encodes an evolutionarily conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. EMBO J. 1994;13:168–179. doi: 10.1002/j.1460-2075.1994.tb06246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamka M, Boulet A, Sakonju S. Ectopic expression of UBX and ABD-B proteins during Drosophila embryogenesis: competition, not a functional hierarchy, explains phenotypic suppression. Development. 1992;116:841–854. doi: 10.1242/dev.116.4.841. [DOI] [PubMed] [Google Scholar]
- Lammel U, Meadows L, Saumweber H. Analysis of Drosophila salivary gland, epidermis and CNS development suggests an additional function of brinker in anterior-posterior cell fate specification. Mech. Dev. 2000;92:179–191. doi: 10.1016/s0925-4773(99)00337-8. [DOI] [PubMed] [Google Scholar]
- Manning G, Krasnow M. Development of the Drosophila tracheal system. In: Bate, Martinez-Arias, editors. The Development of Drosophila melanogaster. CSHL Press; Cold Spring Harbor, NY: 1993. pp. 609–685. [Google Scholar]
- Marquez R, Singer M, Takaesu N, Waldrip W, Kraytsberg Y, Newfeld S. Transgenic analysis of the Smad family of TGF-β signal transducers in Drosophila suggests new roles and new interactions between family members. Genetics. 2001;157:1639–1648. doi: 10.1093/genetics/157.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Arias A, Lawrence P. Parasegments and compartments in the Drosophila embryo. Nature. 1985;313:639–642. doi: 10.1038/313639a0. [DOI] [PubMed] [Google Scholar]
- Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Merabet S, Hombria J, Hu N, Pradel J, Graba Y. Hox-controlled reorganisation of intrasegmental patterning cues underlies Drosophila posterior spiracle organogenesis. Development. 2005;132:3093–3102. doi: 10.1242/dev.01889. [DOI] [PubMed] [Google Scholar]
- Minami M, Kinoshita N, Kamoshida Y, Tanimoto H, Tabata T. brinker is a target of Dpp in Drosophila that negatively regulates Dpp-dependent genes. Nature. 1999;398:242–246. doi: 10.1038/18451. [DOI] [PubMed] [Google Scholar]
- Newfeld S, Wisotzkey R, Kumar S. Molecular evolution of a developmental pathway: Phylogenetic analyses of TGF-β family ligands, receptors and Smad signal transducers. Genetics. 1999;152:783–795. doi: 10.1093/genetics/152.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newfeld S, Takaesu N. An analysis using the hobo genetic system reveals that combinatorial signaling by the Dpp and Wg pathways regulates dpp expression in leading edge cells of the dorsal ectoderm in Drosophila. Genetics. 2002;161:685–692. doi: 10.1093/genetics/161.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R, Wozney J, Gelbart W. Human BMP sequences confer normal dorsal-ventral patterning in the Drosophila embryo. Proc. Natl. Acad. Sci. USA. 1993;90:2905–2909. doi: 10.1073/pnas.90.7.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson U, Izumi H, Souchelnytskyi S, Itoh S, Grimsby S, Engstrom U, Heldin C, Funa J, ten Dijke P. The L45 loop in type I receptors for TGF-β family members is critical in specifying Smad isoform activation. FEBS Lett. 1998;434:83–87. doi: 10.1016/s0014-5793(98)00954-5. [DOI] [PubMed] [Google Scholar]
- Rushlow C, Colosimo P, Lin M, Xu M, Kirov N. Transcriptional regulation of Drosophila gene zen by competing Smad and Brinker inputs. Genes Dev. 2001;15:340–351. doi: 10.1101/gad.861401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saller E, Kelley A, Bienz M. The transcriptional repressor Brinker antagonizes Wingless signaling. Genes Dev. 2002;16:1828–1838. doi: 10.1101/gad.230002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W, Guttentag S, Wang Z, Andl T, Ballard P, Lu M, Piccolo S, Birchmeier W, Whitsett J, Millar S, Morrisey E. Wnt/β-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev. Biol. 2005;283:226–239. doi: 10.1016/j.ydbio.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Simões S, Denholm B, Azevedo D, Sotillos S, Martin P, Skaer H, Castelli-Gair J, Jacinto A. Compartmentalisation of Rho regulators directs cell invagination during tissue morphogenesis. Development. 2006;133:4257–4267. doi: 10.1242/dev.02588. [DOI] [PubMed] [Google Scholar]
- Sivasankaran R, Vigano M, Muller B, Affolter M, Basler K. Direct control of the Dpp target omb by the DNA binding protein Brinker. EMBO J. 2000;19:6162–6172. doi: 10.1093/emboj/19.22.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Johnston R, Hoffmann F, Blackman R, Segal D, Grimaila R, Padgett R, Irick H, Gelbart W. Molecular organization of the decapentaplegic gene in Drosophila. Genes Dev. 1990;4:1114–1127. doi: 10.1101/gad.4.7.1114. [DOI] [PubMed] [Google Scholar]
- Staehling-Hampton K, Hoffmann F. Ectopic Dpp in the Drosophila midgut alters the expression of five homeotic genes dpp and wg causing specific morphological defects. Dev. Biol. 1994;164:502–512. doi: 10.1006/dbio.1994.1219. [DOI] [PubMed] [Google Scholar]
- Takaesu N, Sultani O, Johnson A, Newfeld S. Combinatorial signaling by an unconventional Wg pathway and the Dpp pathway requires Nejire (CBP/p300) to regulate dpp expression in posterior tracheal branches. Dev. Biol. 2002a;247:225–236. doi: 10.1006/dbio.2002.0693. [DOI] [PubMed] [Google Scholar]
- Takaesu N, Johnson A, Newfeld S. Posterior spiracle specific GAL4 lines: new reagents for developmental biology and respiratory physiology. genesis. 2002b;34:16–18. doi: 10.1002/gene.10109. [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene TCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- van den Heuvel M, Nusse R, Johnston P, Lawrence P. Distribution of the wingless gene product in Drosophila embryos: a protein involved in cell-cell communication. Cell. 1989;59:739–749. doi: 10.1016/0092-8674(89)90020-2. [DOI] [PubMed] [Google Scholar]
- Waltzer L, Bienz M. A function of CBP as a transcriptional co-activator during Dpp signaling. EMBO J. 1999;18:1630–1641. doi: 10.1093/emboj/18.6.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K, Ray R, Gelbart W. An activity gradient of dpp is necessary for the specification of dorsal pattern elements in the Drosophila embryo. Development. 1993;117:807–822. doi: 10.1242/dev.117.2.807. [DOI] [PubMed] [Google Scholar]
- Whiteley M, Noguchi P, Sensabaugh S, Odenwald W, Kassis J. The Drosophila gene escargot encodes a zinc finger motif found in snail-related genes. Mech. Dev. 1992;36:117–127. doi: 10.1016/0925-4773(92)90063-p. [DOI] [PubMed] [Google Scholar]
- Xu X, Yin Z, Hudson J, Ferguson E, Frasch M. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev. 1998;12:2354–2370. doi: 10.1101/gad.12.15.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Kauffmann R, Zhang J, Kladny S, Carthew R. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell. 2000;103:87–97. doi: 10.1016/s0092-8674(00)00107-0. [DOI] [PubMed] [Google Scholar]
- Yang X, van Beest M, Clevers H, Jones T, Hursh D, Mortin M. dpp is a direct target of TCF repression in the Drosophila visceral mesoderm. Development. 2000;127:3695–3702. doi: 10.1242/dev.127.17.3695. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.