Abstract
Varicella–zoster virus (VZV) is a human herpesvirus that causes varicella (chicken pox) as a primary infection and, after a variable period of latency in trigeminal and dorsal root ganglia, reactivates to cause herpes zoster (shingles). Both of these conditions may be followed by a variety of neurological complications, especially in immunocompromised individuals such as those with human immunodeficiency virus (HIV) infection. There have been a number of conflicting reports regarding the cellular location of latent VZV within human ganglia. To address this controversy we examined fixed wax-embedded trigeminal ganglia from 30 individuals obtained at autopsy, including 11 with HIV infection, 2 neonates, and 17 immunocompetent individuals, for the presence of latent VZV. Polymerase chain reaction (PCR), in situ hybridization, and PCR in situ amplification techniques with oligonucleotide probes and primer sequences to VZV genes 18, 21, 29, and 63 were used. VZV DNA in ganglia was detected in 15 individuals by using PCR alone, and in 12 individuals (6 normal non-HIV and 6 positive HIV individuals, but not neonatal ganglia) by using PCR in situ amplification. When in situ hybridization alone was used, 5 HIV-positive individuals and only 1 non-HIV individual showed VZV nucleic acid signals in ganglia. In all of the VZV-positive ganglia examined, VZV nucleic acid was detected in neuronal nuclei. Only occasional nonneuronal cells contained VZV DNA. We conclude from these studies that the neuron is the predominant site of latent VZV in human trigeminal ganglia.
The herpesvirus varicella–zoster virus (VZV) is an important human pathogen, particularly in immunosuppressed individuals such as those with malignant disease or human immunodeficiency virus (HIV) infection. After a primary infection of chickenpox (varicella) (usually in childhood), VZV establishes a latent infection in the trigeminal and dorsal root ganglia (1, 2). After a variable latent interval the virus reactivates to produce shingles (herpes zoster). This reactivation may occur either spontaneously or after one or more of a number of triggering factors. Varicella and herpes zoster may be followed by a wide variety of neurological complications, including encephalitis, segmental motor weakness, myelitis, or arteritis, which may be fatal or followed by significant morbidity (2, 3). Furthermore, the syndrome of postherpetic neuralgia is a particularly significant complication of herpes zoster, particularly in the elderly, and can have a major impact in both human and financial terms (4).
Any attempt to modify the clinical course or prevent the incidence of VZV infection of the nervous system must be preceded by a clear understanding of the neuropathogenesis of the latent virus infection in human ganglia. Although knowledge of the site and mechanisms of herpes simplex virus (HSV) latency in human ganglia is now substantial (5), corresponding data for VZV latency have been slow to accrue, due in part to the scarcity of suitable human nervous tissues, and to the relative difficulty of experimental manipulation of VZV. A key question to be answered is the identity of the cell type in which latent VZV resides, but this has been the source of controversy for almost a decade. In 1983 initial application of molecular biological techniques to the problem by using in situ hybridization (ISH) with nick-translated probes indicated the neuron to be the site of latent VZV infection in human ganglia (6). Such findings were apparently confirmed in both human ganglia (7–9) and ganglia from animals experimentally infected with VZV (10) in subsequent studies by different groups. By contrast, in 1988, VZV was reported in perineuronal satellite cells in latently infected human trigeminal ganglia when ISH was used (11). In this study VZV was not detected in neurons, findings that were apparently confirmed in a subsequent study (12). Furthermore, a recent study reported the presence of VZV DNA in both neurons and satellite cells in latently infected human ganglia (13).
To resolve this issue unequivocally, we applied a range of molecular techniques to a large number of samples of human ganglionic tissue. We obtained trigeminal ganglia from different sources, and used polymerase chain reaction (PCR), ISH, and PCR in situ amplification technologies with several gene probes to determine the cell specificity of latent VZV. We report here that in our studies latent VZV was found to reside predominantly in the nuclei of neurons in trigeminal ganglia from both normal individuals and those infected with HIV.
METHODS
Tissue Specimens.
Human trigeminal ganglionic autopsy tissues were kindly donated by J. Bell (Edinburgh), S. Straus and R. Kost (Bethesda), D. Gilden (Denver), and D. Doyle (Glasgow). Data from trigeminal ganglia studied from 30 individuals are shown in Table 2. Eleven of these samples were HIV-infected. In addition, two neonatal trigeminal ganglia served as putative negative controls. Samples were fixed in formalin or paraformaldehyde prior to wax embedding. Control tissue included human heart, rat spinal cord, and VZV-infected and uninfected CV-1 (African green monkey) cells. All ganglia were studied by direct PCR in situ on at least two separate occasions (apart from individuals 20 and 23).
Table 2.
VZV detection in human ganglia
| Patient | Age | Sex | Diagnosis | PCR gene 29 | ISH gene 29 | ISH gene 63 | PCR in situ gene 21 | PCR in situ gene 29 | PCR in situ indirect gene 29 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 33 yr | M | AIDS | + | + | ND | + | + | + |
| 2 | 37 yr | M | AIDS | + | + | + | + | + | + |
| 3 | 28 yr | M | AIDS | + | + | + | + | + | ND |
| 4 | 50 yr | M | AIDS | + | + | + | ND | ND | ND |
| 5 | 30 yr | M | AIDS | + | − | − | ND | + | ND |
| 6 | 35 yr | M | Pre-AIDS | + | + | ND | + | + | ND |
| 7 | 27 yr | M | Normal (OD) | − | − | ND | ND | − | ND |
| 8 | 26 yr | M | Normal (OD) | − | − | ND | ND | + | ND |
| 9 | 26 yr | M | Normal (OD) | + | + | ND | ND | + | ND |
| 10 | 29 yr | M | Normal (Non-HIV) | − | − | ND | ND | − | ND |
| 11 | 36 yr | F | Normal (asthma) | − | − | − | ND | − | ND |
| 12 | 37 yr | M | Normal (gunshot) | − | − | − | ND | + | + |
| 13 | 30 yr | M | Normal (gunshot) | − | − | − | ND | − | ND |
| 14 | Full-term infant | M | Normal (respiratory failure) | − | − | ND | ND | − | − |
| 15 | 2-week infant | M | Normal (pulmonary failure) | − | − | ND | ND | − | − |
| 16* | 44 yr | F | Normal (Crohn’s) | + | ND | ND | ND | + | ND |
| 17 | Adult | ? | Normal (non-HIV) | + | − | − | ND | − | ND |
| 18 | Adult | ? | Normal (non-HIV) | + | ND | ND | ND | − | ND |
| 19 | 63 yr | M | Normal (myocardial infarction) | + | − | − | ND | + | ND |
| 20 | 34 yr | F | Normal (smoke inhalation) | − | − | ND | ND | + | + |
| 21 | 30 yr | M | AIDS | + | ND | ND | ND | − | ND |
| 22 | 24 yr | F | AIDS | − | − | ND | ND | + | ND |
| 23 | 46 yr | M | AIDS | + | − | ND | ND | − | ND |
| 24 | 26 yr | M | AIDS | + | − | ND | ND | − | ND |
| 25 | 29 yr | M | Pre-AIDS | + | ND | ND | ND | − | ND |
| 26 | Adult | ? | Normal (non-HIV) | − | − | ND | ND | − | ND |
| 27 | 22 yr | M | Normal (gunshot) | − | − | − | ND | − | ND |
| 28 | 9 yr | F | Rett syndome | ND | − | ND | ND | − | ND |
| 20 | 4 yr | F | Neurodegenerative disorder | ND | − | ND | ND | − | ND |
| 30 | 6-week infant | ? | Sudden infant death | − | − | ND | ND | − | − |
ND, not determined; OD, drug overdose; +, VZV-positive; −, VZV-negative; ?, unknown. ∗, PCR in situ for VZV gene 18 also positive for sample 16.
Oligonucleotide Probes and PCR Primers.
The probes and PCR primers used in this study are shown in Table 1. They included oligonucleotide probes and primer sequences for VZV genes 18, 21, 29, and 63 and also human β-globin. All oligonucleotides were synthesized by Genosys UK.
Table 1.
Oligonucleotide sequences
| Gene | Sequence |
|---|---|
| VZV gene 18 | 5′-AATCGTTTATCACTGTGCCCGC-3′ |
| PCR primers | 5′-GATTCGGACTTTCCACTTGCA-3′ |
| VZV gene 21 | 5′-ACAAGGCAGCAGTTTCATTCG-3′ |
| PCR primers | 5′-GGTCACTCCCACTTGTATTCC-3′ |
| VZV gene 29 | 5′-AGAGACTTGGAGGAGTTACACG-3′ |
| PCR primers | 5′-ACGTTAGATGTGGTGTCATGGC-3′ |
| β-Globin (180 bp) | 5′-CTGTGGGGCAAGGTGAACG-3′ |
| PCR primers | 5′-CAAAGGACTCAAAGAACCTC-3′ |
| VZV gene 29 70-mer probe | 5′-TCATCTAGAATCTTTACTGCTTCTAG AGCGCCTTCTACGGTCCAGGGCGTTTCCAGGGTTTGGATAATC-3′ nt 54180–54248 |
| VZV gene 63 probe | 5′-CGCGCTCGAGCTCATGTTTTGCACCTCACCGGC-3′ |
PCR Studies.
Standard DNA PCR was performed on DNA isolated from 5-μm sections cut from blocks of fixed paraffin-embedded tissue. Paraffin was removed through xylene and ethanol washes. Samples were digested in proteinase K, and DNA was isolated by extraction with phenol/chloroform and precipitation with ethanol.
ISH.
Five-micrometer sections were cut onto slides coated with 3-aminopropyltriethoxysilane (APES), incubated overnight at 37°C, dewaxed by xylene, and rehydrated through graded ethanol to diethyl pyrocarbonate/H2O. Slides were treated with HCl and digested with proteinase K (for cultured cells the acid step was omitted). After inactivation of proteinase K by glycine/PBS, the sections were acetylated in triethanolamine/acetic anhydride and postfixed in 4% paraformaldehyde. Sections were then dehydrated and rehybridized in buffer containing 50% formamide (for the 70-mer probe), 2× SSC, 1× Denhardt’s solution, and denatured salmon sperm DNA. Samples were then hybridized overnight with the appropriate digoxigenin (DIG)-labeled probe in dextran sulfate prehybridization buffer. Hybrids were detected by using standard DIG reagents (Boehringer Mannheim). Slides were dipped in buffer 1 (0.1 M maleic acid/0.15 M NaCl, pH 7.5) then blocked for 30 min in buffer 2 (10% DIG blocking reagent). Antibody conjugated to alkaline phosphatase (AP) was diluted 1:200 in buffer 2 and left on sections for 60 min, and then slides were washed in buffer 1, twice for 10 min each. Slides were equilibrated in buffer 3 (100 mM Tris⋅HCl, pH 9.5/100 mM NaCl/50 mM MgCl2) and detection reagent [45 μl of nitro blue tetrazolium (NBT) and 35 μl of 5-bromo-4-chloro-3-indolyl phosphate (X-phosphate) in 10 ml of buffer 3] with 2.4 mg/ml levamisole applied to the slides. Samples were left in the dark for between 15 min and 2 hr until the purple color had developed. Slides from all in situ techniques were read without the reader knowing their identities, and to overcome the possibility of bias during the color development, several sections (including controls) were processed on the same slide.
PCR in Situ (Direct Visualization of Amplicons).
Three 5-μm wax sections per APES-coated slide were incubated at 60°C for 24–48 hr. Sections were then dewaxed in xylene, washed, and rehydrated through graded ethanol solutions before treatment with 0.02 M HCl for 10 min at room temperature. Cells were cultured on coverslips that were subsequently glued (Glassbond, Loctite) onto slides. Slides were washed in H2O and PBS, then treated for 2 min in 0.01% Triton X-100 in PBS prior to 50 μg/ml proteinase K digestion at 37°C for 35 min (culture cells were digested for 3 min only). Samples were boiled for 3 min in 0.01 M citric acid, pH 6.0, fixed at 4°C in 20% acetic acid for 30 sec, and washed in PBS. Samples were then dehydrated through graded ethanol solutions and stored in 100% ethanol prior to use.
For PCR in situ the Perkin–Elmer Gene Amp system 1000 was used. PCR master mixes containing the following components were applied at 50 μl per section: 5 μl of 10× Perkin–Elmer buffer, 1 μl of each dNTP at 10 mM, 50 pmol of PCR primers, 9 μl of 25 mM MgCl2, 0.5 μl of Taq DNA polymerase (20 units/μl), 0.3 μl of DIG-11-dUTP (0.4 mM), and H2O to 50 μl. Slides were air-dried and 50 μl of the PCR mix was pipetted onto each section prior to amplification in the thermal cycler. Typical cycling parameters were 5 min at 94°C, 1.5 min at 55°C, followed by 30 cycles of 45 sec at 94°C and 55°C (the second temperature corresponding to the annealing temperature of the primers). Slides were cooled to 4°C and washed in 2× SSC, and color development was carried out as above. Slides were immersed in detection reagent for a maximum of 30 min because longer times increased background signal.
Indirect PCR in Situ.
This method combined the techniques described above but with the omission of the citric acid/acetic acid steps. For the indirect PCR in situ, no labeled dNTP was used during the amplification procedures and DIG was therefore not incorporated into the amplicons. Consequently, the amplicons were detected by using an in situ hybridization technique similar to that used for nonamplified sections.
RESULTS
Results are summarized in Table 2.
PCR.
To assess which cases were VZV-positive, we carried out PCR with primers to gene 29. VZV DNA was detected in 10 of the 11 HIV-positive patients, and 5 of the 17 non-HIV normal controls, and not in the two neonatal ganglia (patients 14 and 15). However, 4 of the PCR-negative patients (patients 8, 12, 20, and 22) were subsequently found to be positive by using PCR in situ (see below).
ISH.
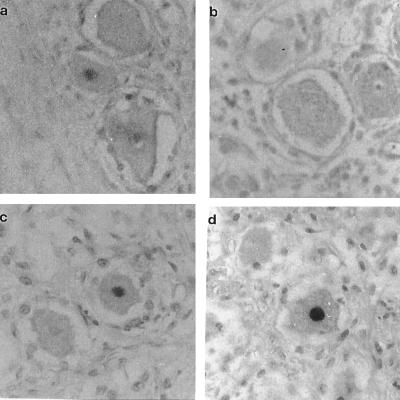
A VZV-positive signal was detected in 5 HIV-infected individuals but only 1 non-HIV individual. Approximately 2–5% of neurons were VZV-positive for genes 29 and/or 63 when the ISH technique was used, and this figure was confirmed by the other in situ techniques. A monoclonal mouse antibody to human neuron-specific enolase (NSE) was used on HIV and non-HIV ganglia to confirm the identity of the neurons. In addition, in several samples positive signals were seen in less than 0.1% of nonneuronal cells. No signal was detected in negative control tissues such as human heart, rat spinal cord, or uninfected CV-1 cells. Examples of ISH on trigeminal ganglia from non-HIV and HIV-infected individuals are shown in Fig. 1.
Figure 1.
In situ hybridization of human trigeminal ganglia with DIG-labeled probes. (a) Sample 9 (normal) hybridized with VZV gene 29 probe. (b) Control, ganglion hybridized with non-VZV plasmid pGEM 3 DNA probe. (c and d) Sample 2 (HIV-positive) hybridized with VZV gene 63 probe and with VZV gene 29 probe respectively. Positive signals in neurons can be seen in a, c, and d, but not in b. (×450.)
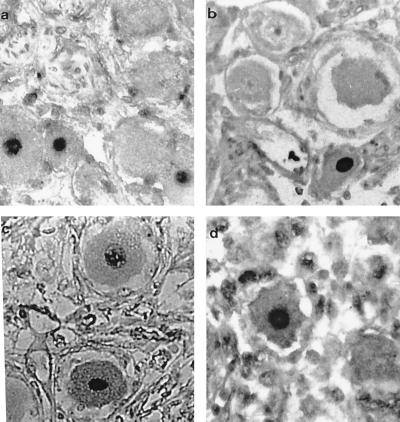
PCR in Situ.
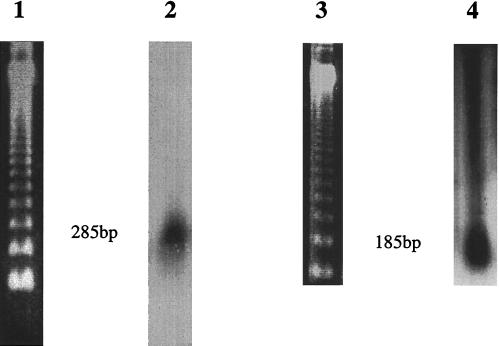
Six ganglia from normal non-HIV-infected individuals were found to be positive by direct PCR in situ with oligonucleotide primers to VZV genes 21 and/or 29. PCR-positive signals were predominantly located within neuronal nuclei (Fig. 2). Overall, approximately 2–5% of the neurons in the tissue sections were positive; only occasional nonneuronal (including satellite) cells (less than 0.1%) harbored the virus. Similarly, positive signals were detected almost exclusively within the neuronal nuclei of ganglia from 6 of the HIV-infected individuals. Four of these positive ganglia had been negative by DNA PCR alone (Table 2). Both of the neonatal trigeminal ganglia were consistently negative for VZV signals. No signal was detected in the negative control tissues or in experiments in which PCR primers had been omitted from the reaction mix. In addition, “nonsense” primers including combinations of Borna disease virus (BDV), enterovirus, and rat mitochondrial deletion primers were used and gave no signal. Also, one BDV primer with one or other VZV gene 29 primer gave no signal. To help exclude the possibility that this method could give rise to nonspecific results we determined whether the direct PCR in situ amplification signals had been obtained from discrete-sized amplicons and not from nonspecific primer extension. Nucleic acid was isolated by scraping cells from eight pooled PCR in situ sections and purified by digestion with proteinase K and extraction with phenol/chloroform. Samples were electrophoresed, blotted onto nylon membranes (GeneScreenPlus, DuPont), and hybridized with the 285-bp or 185-bp random primed 32P-labeled VZV gene 21 or 29 PCR product probe where appropriate. In Fig. 3 it can be seen that discrete hybridization signals of the expected sizes were detected, indicating that the direct PCR in situ procedure did not give rise to an artifact caused by nonspecific primer extension or DNA repair.
Figure 2.
PCR in situ (direct and indirect) of normal human trigeminal ganglia with VZV gene primers. (a) Sample 16 amplified with primers to VZV gene 18 (direct PCR in situ). (b and c) Sample 12 and sample 19 amplified with primers to VZV gene 29 (direct PCR in situ). (d) Sample 12 amplified with primers to VZV gene 29 and the section subsequently hybridized to the DIG-labeled internal 70-mer oligonucleotide probe (indirect PCR in situ). Positive signals in neurons can be seen in all panels. (×450.)
Figure 3.
Southern blot analysis of amplicons from in situ PCR sections. Lane 1, BRL 123-bp DNA ladder; lane 2, DNA isolated from 8 5-μm sections of ganglia that had undergone direct PCR in situ with primers to VZV gene 21; lane 3, BRL 123-bp DNA ladder; lane 4, DNA isolated from 8 5-μm sections of ganglia that had undergone direct PCR in situ with primers to VZV gene 29. DNA was electrophoresed through 2% agarose gels, blotted onto GeneScreenPlus (DuPont), and hybridized with either the 285-bp gene 21 (lane 2) or the 185-bp gene 29 (lane 4) random primed 32P-labeled VZV PCR product probe.
Indirect PCR in Situ.
To further exclude the possibility that the PCR in situ results may have been because of nonspecific primer extension, we carried out PCR in situ using primers to VZV gene 29 and unlabeled dNTPs. Sections were then hybridized with a DIG-labeled 70-mer oligonucleotide probe (Table 1). Both of the HIV-positive individuals analyzed and 2 of the 4 non-HIV individuals showed positive signals in neuronal nuclei when this method was used. Neither of the two neonatal ganglia (patients 14 and 15) were positive (Table 2).
DISCUSSION
The data reported in this study of 30 human trigeminal ganglia show that latent VZV DNA resides predominantly in neuronal nuclei. In addition, in some cases as well as detecting VZV DNA in 2–5% of neurons, we also noted viral signal in less than 0.1% of nonneuronal cells. These observations were consistently made in both normal individuals and those who had been infected with HIV. Further, VZV could be readily detected in 5 of 11 HIV-positive ganglia by ISH alone. However, in only one of the 19 non-HIV patients (patient 9, Fig. 1a) was the ISH technique sufficiently sensitive to demonstrate VZV in neurons. Similar observations were made in dorsal root ganglia that we examined from 12 of the patients (1–9, 23, 24, and 25; data not shown). These findings suggest that VZV is present in slightly higher copy numbers in HIV-infected individuals, which would be consistent with the immunosuppression characteristic of this infection. These findings are also consistent with, and may partially explain, the increased incidence of clinical VZV infection in AIDS patients, including herpes zoster eruptions and other more generalized VZV infections of the central nervous system (2, 3).
It is recognized that any technique involving PCR amplification may be prone to artifacts due to contamination, nonspecific binding of oligonucleotide primers, or primer oligomerization, any of which may give rise to a false positive signal, especially in fixed embedded tissues. In this study we have examined tissue from four different sources, fixed and embedded under different conditions, and we have maximized our efforts with appropriate controls to reduce these various possibilities to a minimum. It is possible that variations in tissue collection, fixation, and experimental techniques, including the use of diverse probes, are likely to be important factors in explaining the inconsistent reports of the cell specificity of latent VZV that have appeared over the last 9 years. It is noteworthy that the percentage of ganglionic neurons harboring VZV DNA in our study was considerably lower than that recently reported by Lungu et al. (13), but the precise reasons for this are unclear. In the current study we have helped to settle the question of which cell harbors latent VZV through the use of sensitive molecular techniques, including direct and indirect PCR in situ amplification of VZV signals in the ganglia. To help exclude the possibility that nonspecific primer extension or DNA repair had occurred, giving rise to a false-positive DIG signal, Southern blot analysis of DNA isolated from PCR in situ sections demonstrated the specific amplification of VZV genes 21 and 29. In addition, the demonstration of VZV in neurons was confirmed by ISH and also by indirect PCR in situ, providing further strong evidence to support the validity of our findings. It was of interest that four ganglia were negative when standard PCR was used, but were then found to be positive by using PCR in situ; this difference may reflect the focal nature of latent VZV or the difficulty of extracting intact nucleic acids of a suitable quality for PCR from fixed tissue.
In conclusion, the data reported here unambiguously show that latent VZV DNA resides in the nuclei of human ganglionic neurons. It is recognized that further work will be required to determine the sequence of events that result in the initiation and subsequent reactivation of the virus. The presence of VZV genome in a cell, however, does not necessarily indicate its transcription in that particular cell. Immunosuppression clearly plays a role in the reactivation process in some instances, but exactly how reactivated VZV produces an eruption in an entire sensory dermatome rather than in a single nerve distribution remains to be explained.
Acknowledgments
We thank our colleagues mentioned in the text for their generous gifts of ganglionic tissues, in particular Dr. R. Kost, to whom we are indebted. Tissue used in this study (courtesy of Dr. J. Bell) was also provided by the Medical Research Council HIV Brain and Tissue Bank in Edinburgh, and this support is gratefully acknowledged. This work was financially supported by a grant from The Wellcome Trust, and the support of the Greater Glasgow Health Board HIV financial allocation is also acknowledged.
ABBREVIATIONS
- VZV
varicella–zoster virus
- ISH
in situ hybridization
- DIG
digoxigenin
References
- 1.Meier J L, Straus S E. J Infect Dis. 1992;166:S13–S23. doi: 10.1093/infdis/166.supplement_1.s13. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy P G E. Infections of the Nervous System. London: Butterworth; 1987. pp. 117–208. [Google Scholar]
- 3.Amlie-Lefond C, Kleinschmidt-DeMasters B K, Mahalingam R, Davis L E, Gilden D H. Ann Neurol. 1995;37:784–790. doi: 10.1002/ana.410370612. [DOI] [PubMed] [Google Scholar]
- 4.Weller T H. J Infect Dis. 1992;166:S1–S6. [PubMed] [Google Scholar]
- 5.Steiner I, Kennedy P G E. Mol Neurobiol. 1993;7:137–159. doi: 10.1007/BF02935640. [DOI] [PubMed] [Google Scholar]
- 6.Hyman R W, Ecker J R, Tenser R B. Lancet. 1983;ii:814–816. doi: 10.1016/s0140-6736(83)90736-5. [DOI] [PubMed] [Google Scholar]
- 7.Gilden D H, Rozenman Y, Murray R, Devlin M, Vafai A. Ann Neurol. 1987;22:377–380. doi: 10.1002/ana.410220315. [DOI] [PubMed] [Google Scholar]
- 8.Dueland A N, Ranneberg-Nilsen T, Degre M. Arch Virol. 1995;140:2055–2066. doi: 10.1007/BF01322692. [DOI] [PubMed] [Google Scholar]
- 9.Mahalingam R, Wellish M, Cohrs R, Debrus S, Piette J, Rentier B, Gilden D H. Proc Natl Acad Sci USA. 1996;93:2122–2124. doi: 10.1073/pnas.93.5.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadzot-Delvaux C, Merville-Louis M-P, Delree P, Marc P, Piette J, Moonen G, Rentier B. J Neurosci Res. 1990;68:7900–7908. doi: 10.1002/jnr.490260110. [DOI] [PubMed] [Google Scholar]
- 11.Croen K D, Ostrove J M, Dragovic L J, Straus S E. Proc Natl Acad Sci USA. 1988;85:9773–9777. doi: 10.1073/pnas.85.24.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meier J L, Holman R P, Croen K D, Smialek J E, Straus S E. Virology. 1993;193:193–200. doi: 10.1006/viro.1993.1115. [DOI] [PubMed] [Google Scholar]
- 13.Lungu O, Annunziato P, Gershon A, Staugaitias S M, Josefson D, LaRussa P, Silverstein S J. Proc Natl Acad Sci USA. 1995;92:10980–10984. doi: 10.1073/pnas.92.24.10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohrs R J, Srock K, Barbour M B, Owens G, Mahalingam R, Devlin M E, Wellish M, Gilden D H. J Virol. 1994;68:7900–7908. doi: 10.1128/jvi.68.12.7900-7908.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohrs R J, Barbour M, Gilden D H. J Virol. 1996;70:2789–2796. doi: 10.1128/jvi.70.5.2789-2796.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]