Abstract
One of the requirements for a federated information system is interoperability, the ability of one computer system to access and use the resources of another system. This feature is particularly important in biomedical research systems, which need to coordinate a variety of disparate types of data. In order to meet this need, the National Cancer Institute Center for Bioinformatics (NCICB) has created the cancer Common Ontologic Representation Environment (caCORE), an interoperability infrastructure based on Model Driven Architecture. The caCORE infrastructure provides a mechanism to create interoperable biomedical information systems. Systems built using the caCORE paradigm address both aspects of interoperability: the ability to access data (syntactic interoperability) and understand the data once retrieved (semantic interoperability). This infrastructure consists of an integrated set of three major components: a controlled terminology service (Enterprise Vocabulary Services), a standards-based metadata repository (the cancer Data Standards Repository) and an information system with an Application Programming Interface (API) based on Domain Model Driven Architecture. This infrastructure is being leveraged to create a Semantic Service Oriented Architecture (SSOA) for cancer research by the National Cancer Institute’s cancer Biomedical Informatics Grid (caBIG™).
Keywords: Semantic Interoperability, Model Driven Architecture, Metadata, Controlled Terminology, ISO 11179
Section 1 Introduction
The rise of high-throughput biomedical research tools has given scientists and clinicians data of unprecedented depth, timeliness and diversity to attack the problem of alleviating human cancer. However, this diversity brings with it the problem of integration and interoperation; that is, making disparate data sets created by a wide range of individuals available to others for the purposes of performing analyses that span multiple data types. For example, many researchers would like to correlate the results of high-throughput gene expression microarray experiments with clinical outcomes or toxicology results, or correlate loss of heterozygocity with tumor susceptibility to certain therapeutic compounds. Even though there are often multiple sources of this sort of data, they are often inaccessible to information systems at runtime (so-called data ‘stovepipes’), or the data is insufficiently annotated to determine if successful reuse is possible.
The solution to this problem is to build data systems utilizing an architecture that facilitates such interoperability. The IEEE Standard Computer Dictionary defines interoperability as the “ability of two or more systems or components to exchange information and to use the information that has been exchanged”[1]. From this definition it is possible to decompose interoperability into two distinct components: the ability to exchange information, and the ability to use the information once it has been received. The former process is denoted as ‘syntactic interoperability’ and the latter ‘semantic interoperability’. A small example suffices to demonstrate the importance of solving both problems. Consider two persons who do not share a common language. They can speak to one another and both individuals will recognize that data has been transferred (they can also probably parse out individual words, recognize the beginning and end of message units, etc.). Nevertheless, the meaning of the message will be mostly incomprehensible; they are syntactically but not semantically interoperable. Similarly, consider a person who is blind and one who is deaf, but who both utilize a single language. They can attempt to exchange information, one by speaking and one by writing, but since they are incapable of receiving the messages, they are semantically but not syntactically interoperable.
The creation of an interoperable data system, therefore, requires several elements, including a convenient mechanism that provides a clear and consistent interface into a data repository and a source of terminology whose meaning is clear and unambiguous to those who would record and use the data maintained in that repository. Object Oriented Application Programming Interfaces (APIs) built using the Model Driven Architecture (MDA) paradigm can begin to address the first problem, and controlled terminology available at runtime can help with the latter. There is however, a third piece to this problem; a mechanism to bind the controlled terminology to a model driven, object-oriented data system. Such descriptive information, commonly referred to as semantic oriented metadata, provides a formal description of the meaning of the types of data that are supplied by the individual classes and attributes that are part of the data system as well as what constitutes a valid value for each attribute of the classes.
The creation of an infrastructure that supports such a ‘semantically annotated’ data system is the central innovation of caCORE version 3. The caCORE infrastructure provides a runtime-accessible terminology service and a standards-based, runtime accessible metadata repository that is used to bind the terminology to a domain model-based information system. This infrastructure has been successfully used to create a reference implementation, cancer Bioinformatics Infrastructure Objects (caBIO), but is fully extensible to any arbitrary data system, regardless of subject.
Introduction to Components of caCORE
Version 3 of caCORE consists of three major parts: 1) a primary technology stack encompassing three major components depicted in the center of Figure 1; 2) two major enabling technology components; and 3) one supporting technology. This is diagrammed in Figure 1. At the top of the primary technology stack are the cancer Biomedical Informatics Objects, caBIO, the interoperable data system system1; at the bottom of the stack is the Enterprise Vocabulary Services or EVS, supplying the controlled terminology that is leveraged to provide semantics for caBIO (or any other data system that utilizes the caCORE met methodology). Between these two components is the cancer Data Standards Repository or caDSR, a system for storing semantic metadata, that acts as the glue between the object oriented data system and the controlled terminology. The supporting technology component is a Common Security Module or CSM, which is designed to be readily integrated into systems designed along caCORE lines. The CSM contains a user provisioning tool for managing rights given to users within the system. Finally, there are two pieces of enabling technology, 1) the caCORE Software Development Kit [2] that is used to generate ‘caCORE-like’ systems, and 2) the Semantic Integration Workbench, an end-user application with a graphical user interface (GUI) that assists in creating the semantic metadata that is stored in the caDSR. This manuscript focuses primarily on the integration of the three components of the primary technology stack to enable semantic interoperability. The caCORE infrastructure is distributed under a non-viral open source software license which allows for any commercial or non-commercial reuse of the software, its components, or source code. Each component is described in further detail in upcoming sections.
Figure 1.

The major components of caCORE version 3. The primary technology stack contains a model driven, object oriented data system (caBIO in this example) and the metadata and controlled terminology services required to achieve semantic interoperability. Supporting this stack is a set of enabling technologies that simplifies the process of creating a ‘caCORE-like’ system and a supporting technology stack that includes a Common Security Module (CSM) that can be readily implemented through the caCORE SDK.
Section 2 Creating an interoperable, service oriented architecture
Model Driven Architecture
All caCORE components utilize a software development strategy known as Model Driven Architecture or MDA, that is developed by the Object Management Group (OMG http://www.omg.org). The MDA approach posits that prior to creating an object-oriented software system, the designer should create a graphical model of the functions, components, and behavior of the system that is independent of the computer language that the system will ultimately be created in. This model is then used to create the implemented software system, sometimes using programs called code generators to create either a skeleton of or the complete software system. Within the MDA paradigm, a Platform Independent Model (PIM) is captured using the Unified Modeling Language (UML). As a result, all caCORE systems are modeled in UML prior to implementation.
caCORE software systems utilize two UML artifacts: use cases and class diagrams. Use case diagrams capture requirements the system must be able to fulfill and certain high-level system behaviors related to fulfilling the requirement. Class diagrams describe the level objects that will be implemented in the system and the relationships between these objects. Figure 2 shows a portion of the UML model for the caBIO system, centered around a class that captures information about genes. Each box represents a class; each class has a series of attributes listed below the top line. Lines between classes represent associations, which are traversable linkages between the classes in the underlying system.
Figure 2.

An example of a Unified Modeling Language (UML) model used in the generation of caCORE-like data systems. This particular example is a portion of the caBIO system.
This particular model, and in fact all caCORE models, are specializations of a UML model known as a domain information model or a domain model for short. Domain models are meant not only to describe software systems, but to also be a representation of current understanding of knowledge of how the entity being modeled exists and works in the real world. Thus, in the example shown in Figure 2, classes corresponding to entities in the world of molecular biology research (i.e. Gene, Protein, Nucleic Acid Sequence, Protein, Taxon, etc.) display with associations describing the relationships between these entities (for example Genes are associated with Proteins). By utilizing domain models, interoperability is enhanced, because groups that are providing data are likely to create information models that are based on a common universe of scientific entities.
n-tier Architecture and multiple APIs
Data systems in caCORE generally provide three or four interfaces to client systems. These APIs include Java Bean and Web Services (always), http interfaces (usually) and PERL interfaces (caBIO only). The Java Bean and Web Services APIs are generated (see below) but the http and PERL interfaces currently require manual coding. Wherever possible, these APIs are built on top of each other (i.e. the Web Services API translates requests into Java Beans) so that maximum API consistency can be maintained.
Systems constructed as part of the caCORE infrastructure are generally created as n-tier data systems with multiple APIs. This is illustrated in Figure 3. An n-tier architecture means that a client system does not directly access the stored data (the data or persistence tier). Instead, clients use one or more interfaces to access domain objects that are provided by a middleware layer. In the middleware layer, domain objects are translated into data access objects that are used to retrieve information from the data layer where the actual information is stored (or persisted). The advantage to such as system is that it shields clients from the implementation details of a software system. System owners can make changes to their data tier (change hardware or database type or design) without effecting a change in the API or interface used by clients.
Figure 3.
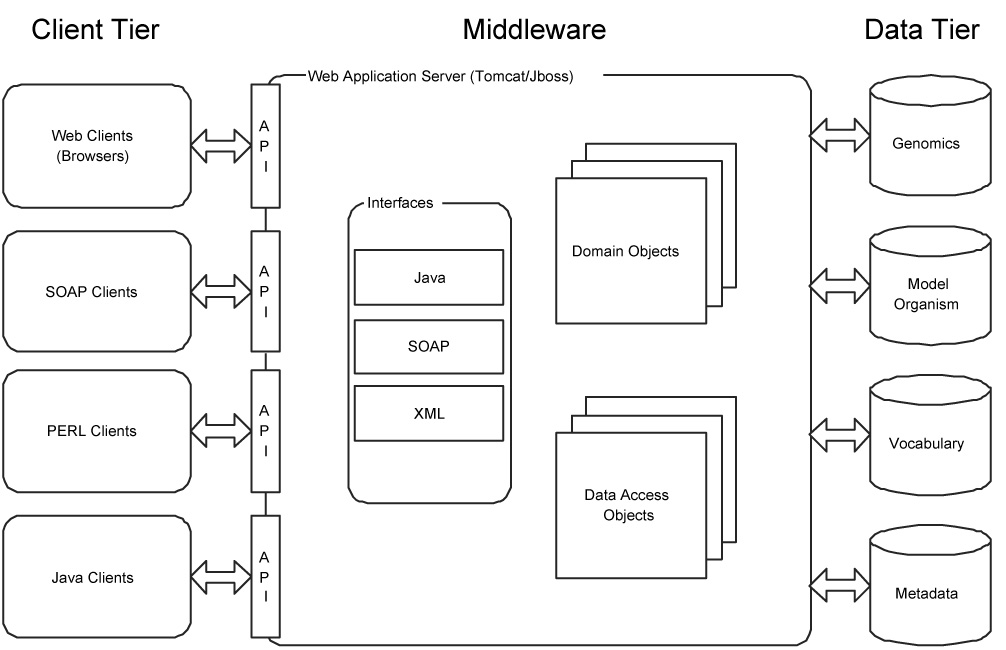
High level view of the architecture of the caCORE architecture. The architecture is split into three tiers: a client tier that a consuming application would use, a persistence or data tier that contains the data persisted in a relational database management system (RDBMS) and a middle tier that converts queries created in the form of domain objects into queries that can be understood by the persistence tier.
The mapping between the objects present in the middleware and API and the underlying data stored in a Relational Database Management System (RDBMS) is described in the UML model. The implementation of this mapping and the generation of the code that converts an object query into the Structured Query Language (SQL) used by the underlying RDBMS are created using the Hibernate Object to Relational Mapping (ORM) framework (http://www.hibernate.org). Hibernate is an open source framework that provides ORM for a number of different RDBMS systems.
API Generation
The Java Bean API for most caCORE components is generated using the caCORE SDK [3]. The Castor framework (http://www.castor.org) is utilized to create the Web Services API. The caCORE Web Services APIs are WS-I compliant (http://www.ws-i.org), as specified by the Web Services Interoperability Organization. Although the Hibernate mapping files are initially generated by the SDK, they are subsequently manually edited to enhance certain types of query performance, and to account for the fact that the data is maintained in more than one database schema. In addition, certain modifications to the generated code are made for the EVS APIs and provenance model (see below).
For the Java API, an application that wanted to make use of the data in a caCORE component would incorporate a client JAR file that is one of the artifacts of the caCORE SDK. This client JAR contains the relevant code to communicate with the middleware layer. Prior to caCORE 3.0, this communication was via Java Remote Method Invocation (RMI). Beginning with caCORE 3.0, RMI has been replaced with HTTP tunneling. Since this uses port 80, which is open to allow conventional Web protocols, it is more reliable than the older RMI method.
Service Layers
Domain objects in caCORE software systems have setter and getter methods associated with each attribute and traversable association. Other types of methods are generally not incorporated into the classes in caCORE components. Instead, convenience methods are encapsulated into a service layer. As an example, a molecular weight computation method would not be coded into a Protein class, but would be implemented as a method in an application service layer that takes a Protein object. The decision to remove most methods from the objects themselves was made to improve maintainability. Custom methods implemented in the classes would have to be added back to the code base each time a new version was generated. By moving these met methods to the service layer, the code base can be regenerated without losing the custom methods.
Semantic Interoperability
As defined earlier, semantic interoperability is the ability to use information that has been extracted from an information resource. At its most fundamental level, it involves providing a mechanism to obtain and interpret the meaning of the classes and attributes of a software system and for the instance values that are recorded as data. Model Driven Architecture (MDA) provides a framework for creation of semantically interoperable systems through the organization of data into classes that represent entities in the knowledge space; however, it is not complete in that it is dependent upon an implied common understanding of specific terms. In other words, MDA systems would be fully interoperable if there was unambiguous understanding of the names of the classes and attributes. Unfortunately, this is never the case. Consider the example of a class named ‘Agent’ with two properties ‘name’ and ‘NSCCode’. It is not a priori possible to determine if this refers to a drug with a name and a code from the Nomenclature Standards Committee (NSC) of the Food and Drug Administration (FDA) or a spy with a name and a code number from the National Security Council (NSC). The problem, therefore, is to find a way to bind unambiguous meaning to data systems.
The components of caCORE provide a mechanism for such ‘semantic annotation’. By binding formal semantics (i.e. unambiguous meanings) to classes and attributes in MDA-based systems, it becomes possible to create a semantically integrated and interoperable based data system. The semantic interoperability solution created as part of caCORE 3 utilizes a variety of pre-existing technologies, including terminologies/ontologies, terminology servers, and a metadata repository based on the ISO11179 metamodel, however, it combines them in a unique manner to supply unambiguous semantic metadata.
ISO 11179
The ISO 11179 Edition 2 metadata repository standard provides standard grammar and syntax for describing data element metadata that (with extensions) result in unambiguous data representation and interpretation. The formalism behind the caDSR implementation of the ISO 11179 metamodel is shown in a simplified form with a hypothetical example in Figure 42. A Data Element (often called a Common Data Element or CDE at the NCI), indicated by the shaded box, consists of two parts, a Data Element Concept (DEC) and a Value Domain (VD). The DEC is a formal description of the thing about which we are recording a data value, in this case, the name of an Agent; a Value Domain consists of the properties that constitute a valid response for the thing that we are recording. Value Domains can be either enumerated (i.e. there exists a finite list of valid responses), or constraints can be defined around the valid values (a number, a string, an integer between 0 and 120, etc.).
Figure 4.
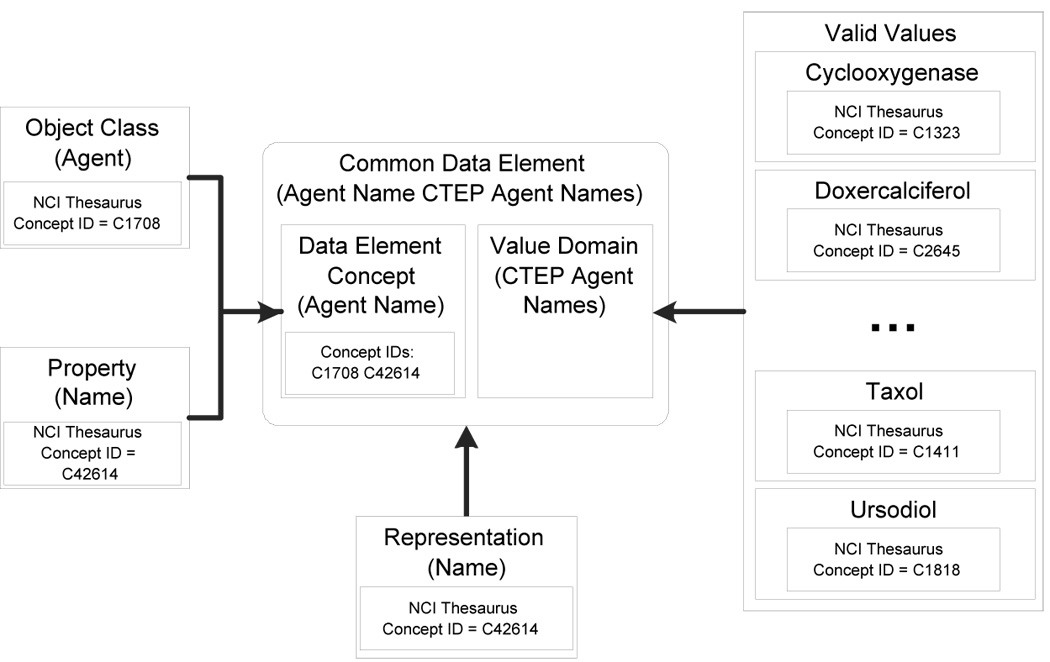
Simplified view of a Common Data Element (CDE) in the caDSR implementation of the ISO 11179 metamodel. This example is for a CDE that describes the name of an agent (i.e. a drug compound) that is constrained to an enumerated list of values provided by the Cancer Therapeutics Evaluation Program (CTEP) of the NCI.
Data Element Concepts are further refined into two subcomponents, Object Classes and Properties. An Object Class is the entity that is being described by the data element, while the Property is a specific attribute of the entity whose value is being recorded. In the example in figure 4, the entity being described is an Agent, and the property (the characteristic being used to distinguish instances of one Agent from another) is its name. If the Data Element was recording the temperature of a patient during a clinical trial, the Object Class would be Patient (or perhaps Patient Person) and the Property would be body temperature. Data Elements also have other associated components, including a Representation which describes the nature of the data that is being recorded (code, text, number) and a Conceptual Domain, which is a means of classifying CDE components (such as Data Element Concepts and Value Domains) for easier search and identification.
The ISO 11179 model has been implemented at the National Cancer Institute in the cancer Data Standards Repository or caDSR, an enterprise class system for storing and maintaining metadata about questions on clinical case report forms and object-oriented data systems regardless of their data domain. This system provides both tools for viewing and editing metadata content and object oriented APIs that can be accessed by other software systems to retrieve semantic metadata at runtime (see section 6).
Binding of Controlled Terminology
As designed, the ISO 11179 metamodel does not provide a completely unambiguous definition of a data element, because it uses words or phrases to name and represent the meaning of most of the components of a data element (Object Classes being the notable exception). Consider the hypothetical example in Figure 4, Agent Name. As described before, it is impossible for a semi-autonomous computer program (or a human for that matter), using only the words or phrases, to determine if this is recording the name of a drug, a spy, or a realtor. The solution to this problem is to formally bind the words and phrases used for human readability to a concept in a controlled terminology. At the implementation level, this means using a series of ordered, alphanumeric concept codes that are invariant identifiers for a concept in a controlled terminology. This terminology is supplied by the NCI EVS to indicate the meaning of the components of a CDE. The caDSR binds ISO 11179 components to controlled terminology, as indicated in Figure 4 by the NCI Thesaurus Concept Codes or Concept Unique Identifiers (CUIs).
Mapping concepts to metadata is not a simple one-to-one process. Consider the example of Patient Birth Date or Patient Death Date. The Object Class is ‘Patient’, and the property has to represent the date of birth or death. There are two possible ways to represent this property in controlled terminology. One option is to create specific concepts for date of birth or death (pre-coordination). The alternative is to use two concepts, a primary concept (date) and a qualifier (birth or death). This latter case is referred to as post-coordination and is commonly used in the caDSR for Object Classes and Properties. The caDSR currently supports the use of multiple, ordered qualifiers with Boolean operators to fully capture complex semantics through post-coordination of atomic concepts. The decision to choose a pre-coordinated versus a post-coordinated mapping is made when the model is annotated, informed by the controlled terminology but is still somewhat arbitrary. Within caCORE, the decision is currently based on a determination of the utility to the controlled terminology (generally NCI Thesaurus) of the pre-coordinated concept. If the new concept is considered valuable to the terminology where it would reside, the pre-coordinated concept is used; if not, post-coordination is used. This has the additional benefit of preventing unnecessary growth in the size (and hence complexity) of the terminology.
Mapping object oriented software systems to components of the ISO 11179 metamodel
The process of creating a set of formal descriptions of domain objects in an ISO 11179 metadata repository involves co-relating, or mapping, the semantics of an information model described in UML to the components of the ISO 11179 metamodel. In a UML domain model, a class represents something in the real world; the equivalent in the ISO 11179 metamodel is an Object Class. UML class attributes represent characteristics that serve to help distinguish instances of the classes; the ISO11179 equivalent is a Property. Thus, by mapping the concepts bound to the UML classes to the ISO 11179 Object Class and the UML attributes to the ISO 11179 property, it is possible to create a set of ISO 11179 Data Element Concepts to describe each UML class plus attribute pair. When combined with an appropriate Value Domain, each class plus attribute in the software system is mapped to one Common Data Element. This mapping is shown schematically in Figure 5.
Figure 5.

Binding of controlled terminology to the classes and attributes of a class derived from a domain model in UML. This binding is then used to derive the components of an ISO 11179 Common Data Element in the caDSR (bottom).
The mapping in Figure 5 fully describes the semantics of an attribute. Systems built using MDA, however, also provide associations, and for full interoperability, it is necessary to supply semantics for these as well. The ISO 11179 model does not specify relationships between Object Classes, but it does specify relationships between other types of Administered Items such as Data Elements, Value Domains and Data Element Concepts. Administered Items are items within an ISO 11179 registry for which there is administrative information, including naming, identification and definitional attributes. Consequently, the caDSR extended 11179 to include Object Class relationships in which the UML Class associations are recorded in caDSR.
At a practical level, this binding occurs not by creating a series of data elements and mapping them to the object oriented data system, but rather by mapping concepts to the classes and attributes of the object oriented data system and creating the metadata of the data elements from this mapping. Using the caCORE-enabling technology called the Semantic Integration Workbench (SIW), a series of concepts is mapped to the components of the information model of the software system, and a caCORE program called the UML Loader creates or reuses existing data elements and all of their subcomponents based on a comparison of the bound concepts to preexisting CDEs. Readers interested in the details of the UML loader’s design are directed toward [2] and those interested in the practical aspects of its operation are directed to the caCORE Software Development Kit Developers Guide [3].
Computational Determination of Equivalence and Semantic Interoperability
One characteristic of data elements that are defined by a series of concept codes is that it is possible to determine equivalence by comparing their semantic signatures, the series of ordered concept codes (plus the Value Domain) that are bound to the data element. As a result, it is simple for the UML loader to determine if a particular class plus attribute is semantically equivalent as one in another system that has already been loaded, regardless of the name of that class plus attribute. The only relevant question is whether the semantic signature is the same: if it is, the original CDE is reused; if not, a new CDE is created. This makes CDEs created via the UML loader ‘self-harmonizing’ without human intervention.
This combination of computable equivalence and runtime access (via caDSR API) to a human- understandable definition for a data element and its instance values is what makes caCORE semantically interoperable. A user or computer system that accesses a component of caCORE can retrieve information, determine which data elements it has in common with other systems, and retrieve an unambiguous definition of the meaning of each object, attribute and instance value for those attributes. With that information, a user or computer system with no a priori knowledge of the specific circumstances under which the data was collected can still make an appropriate decision regarding data reuse.
Section 3 Provenance
Data provenance describes the origin and chain of ownership of information in a system. The caBIO provenance model is hierarchical; that is, a site is responsible for storing provenance information that describes the source from which it obtained the data. In a world that utilizes such a model, a user would follow the provenance trail to the previous source and then retrieve the provenance information from that site to extend the search.
Ideally, three levels of provenance should be provided for data:
Supplying Source: The group/system that is supplying the information to the end user (i.e. the current system). Useful for distinguishing data from multiple instances of the same information sources.
Immediate Source: The group/system that supplied the information to the supplying source. If the supplying source generated the information than they are the immediate and supplying source.
Original Source: The farthest source from the supplying source that is known by the supplying source; ideally the group that collected the initial data. If not known, the immediate source is considered the original source. Note that in the case of information supplied by the group that created it, that group would be the Supplying Source, Immediate Source and Original Source.
The caCORE Provenance model is shown in Figure 6. The central class is the Provenance class. Provenance information is uniquely identified by the combination of the fully qualified class name and the object identifier for the information that one wishes to track. The three sources are indicated by the associations to the Source class. The Source class is abstract. All actual data will be encapsulated in one of the three implementing subclasses: Publication Source, ResearchInstitutionSource and InternetSource. In addition there is a SourceReference class that is used to obtain a mechanism to retrieve the data from the immediate source. Since this model is hierarchical, this is the only reference that is maintained. This reference is either in the form of a URL or a Web services call. To access provenance information, a program would create a Provenance object and set the class name and object ID of the specific instance of the data that one is interested in. Currently, only the caBIO Protein class has this information available.
Figure 6.

Hierarchical Provenance Model implemented in caCORE version 3. The central class is the Provenance class that has associations to a SourceReference (a means to retrieve the information that was used to create a particular instance of a class) and three Source objects that describe the supplying source (the entity supplying the data to the user of the API), the immediate source (the source that was used by the supplying source) and the original source (the original creators or publishers of the data).
Section 4 Using caCORE
All caCORE APIs are implemented as Query By Example (QBE) and are all accessed in a consistent fashion. Figure 7 demonstrates the use of of the QBE mechanism by querying the caBIO service for information about the Breast Cancer Susceptibility Gene BRCA1. In the example, a reference to the service endpoint where the caBIO middleware resides is created and then an empty instance of the object type that is to be used as the query (in this case a Gene object). In order to search, the query parameters on the object are set (wildcards can be used), and the object is passed to the application service. Query objects in caCORE can search using the attributes that are part of the classes (in the example, a gene symbol) or attributes of classes with associations to the original class. In the example shown in Figure 7, the Taxon class is associated with the Gene class, and is used as a search attribute for a Gene class. The search request has two parameters, a string and the object that is used as a query. The former lists the type of object that is being requested, in this case a Gene object, the same type as the query. There is, however, no requirement that the return object type be the same as the requesting object type. As long as a traversable path exists between the query object and the return object, the search service will find all of the instances of that object that meet the search criteria, and then use the association methods to retrieve the associate objects of the return type. More details about querying a caCORE version 3 data system (including the use of non-Java APIs and the Provenance API) can be found in the caCORE Technical Guide [3].
Figure 7.

A Query By Example (QBE) query into the caBIO service utilizing the Java Bean API. A QBE service, wherein a prototype of the query object is created and appropriate attributes have values set to incorporate the boundaries of the query. This code sample is not complete, certain setup components have been removed for clarity. See the caCORE Technical Guide [3] for complete details.
Section 5 Enterprise Vocabulary Services (EVS)
Introduction
NCI Enterprise Vocabulary Services encompass terminology development, terminology licensing, software development and licensing and operations support activities. The EVS is a joint project of the NCI Office of Communications and the NCICB. From its inception, EVS has strived to address the broad spectrum of terminology needs at NCI. (See for example,[4]) This section describes the architecture of the EVS and its relationship to the larger caCORE architecture, the terminology content that EVS makes available to caCORE, and those aspects of EVS operations, such as data and software updates and quality assurance, that have direct bearing on the quality of the semantic support EVS provides to caCORE.
Architectural Considerations
The NCI EVS produces two terminology products: the NCI Thesaurus (NCIt), a cancer focused terminology, and the NCI Metathesaurus (Meta) a mapping between many terminologies with the goal of providing a comprehensive source of terminology needed by the other caCORE components and users of the caCORE, served respectively by the NCI Terminology Server and the NCI Metathesaurus Server. The NCI Terminology Server provides access to individual terminologies while the NCI Metathesaurus Server provides access to numerous terminologies integrated in a single data structure via n-way mapping to concepts [5]. Both environments are currently based on proprietary terminology service software: the former uses the Apelon Distributed Terminology Server (DTS), and the Metathesaurus Server utilizes the Apelon Metaphrase server. The caCORE client side API provides a unified open source interface to both EVS server environments. Applications that rely on EVS for terminology support make use of the caCORE API rather than the native Apelon APIs.
At the time EVS was originally constituted, there were few open source tools available for either terminology development or terminology server application, and none that appeared adequate for operational use in a large enterprise. The EVS was therefore constructed using proprietary commercial software. Consistent with NCICB’s commitment to open software and content, EVS is moving to architecture that employs only open components. Currently, the EVS has migrated development of NCI Metathesaurus to open software (MEME/Jekyll, described below), and is engaged in migration of NCI Thesaurus development to open software (Protégé/OWL, also described below). Migration of the EVS servers to open software awaits the availability of lexBIG server (see http://cabigcvs.nci.nih.gov/viewcvs/viewcvs.cgi/lexgrid/) being built by the Mayo Clinic. The EVS will be the launch customer for this product.
NCI Thesaurus
The NCI Thesaurus (NCIt) is a product of the NCI, described previously [4, 6, 7]. NCIt is one of the individual terminologies that are served on the NCI Terminology Server. It is also one of the approximately 50 source terminologies that comprise the NCI Metathesaurus.
The NCI Thesaurus (NCIt) is a concept-based terminology with broad coverage of the cancer research and clinical domain. It is a Federal Standard Terminology that is a reference terminology for the NCI and its partners. NCIt contains more than 48,000 distinct concepts with definitions and description-logic based associations (‘roles’) that relate the different concepts. The NCIt is available under a non-viral, open content license in a variety of formats, including OWL, XML and flat files.
NCIt is widely used within NCI for a variety of purposes including indexing the contents of PDQ [8] and the NCI’s Web portal (see http://cancer.gov), for support of enterprise-wide science management initiatives as the National Institutes of Health Knowledge Management Disease Coding Project http://era.nih.gov/eranews/news_article1.cfm?lobjectid=F12ABEF5-101C-4874-B9A87DE7882300B0), and for providing the concepts upon which semantic consistency in the caCORE is based. Increasingly NCIt is being used by organizations external to NCI including the caBIG™ program and the Food and Drug Administration (FDA), which has standardized on the NCIt as the source of terminology used in creation of Structured Product Label (SPL). The NCIt’s rapid turnaround time for new concepts (often less than 24 hours when the requests come from collaborators) helps prevent terminology development from causing major delays in the software development or information deployment process.
The NCIt provides four features which, taken together, have proven sufficient to underpin the semantic integration of the caCORE: 1) concept codes that exhibit concept orientation, permanence and non-semantic construction [9]; 2) preferred terms for each concept that are the NCI’s canonical lexical token for that concept; 3) English language definitions; and 4) synonyms. Of these, the concept code is the most important, as it provides unambiguous linkage to a specific meaning and is machine interpretable. The description logic roles that are used to assert relationships have the potential to become a resource for data discovery, data management, and making inferences about data. CaCORE may, in future releases, leverage NCIt for these purposes. NCIt exhibits a number of semantic shortcomings common among contemporary terminologies [10]. Changes planned to address these shortcomings may ultimately enable NCIt to be used to reason about instances, or be employed in decision support systems.
Data sources, updates and business practices
The NCI EVS employs a group of full time terminology curators who maintain and expand the NCIt. The focus of the NCIt editors is to monitor the literature, other terminology products, and professional meetings for new concepts and changes in understanding of existing concepts as they apply to cancer science. This background research, combined with the direct requirements of users, collaborators and the results of critiques and analyses [10–12] are the primary drivers of the NCIt content. In general, EVS resists adding content when that content is available in another terminology, especially one that is already widely used and is maintained by another organization. Nonetheless, the needs of community collaborations such as the joint FDA/CDISC/NCI efforts and caCORE-like data systems dominate content development decisions.
Maintainability is the major driver of business practices related to NCIt production. For example, cancer researchers from Iceland, Japan, and elsewhere wishing to use the caDSR and other caCORE tools have expressed the desire to add terms in their native languages to NCIt concepts (currently as synonyms, but once our migration to the LexBIG terminology server is complete as first rank attributes). While NCI EVS is willing to add such content, we require that the researchers commit to a long term collaboration aimed at maintenance of such extensions to the terminology.
Information Model and Application Programmer Interfaces
The description logic used to create the NCIt has been described elsewhere[13]. The major semantic constructs used in the NCIt information model are Concept, Kind, Role, Association and Property. The NCIt is a self defining3 namespace in that each semantic construct is present in the NCIt as a concept. There are about 90 specific instances of roles defined in Thesaurus, 21 Kinds, about 40 Properties and Associations. Interested readers may find a more complete description of the semantics of the NCIt is available online ftp://ftp1.nci.nih.gov/pub/cacore/EVS/ThesaurusSemantics/).
The caCORE client API (model available at http://gforge.nci.nih.gov/frs/download.php/214/EVS_v3-1_rv1.zip) provides methods to enable calling applications to search the NCIt by concept, role, property, and association, as well as by the domain and range values. The API also provides the ability to perform transitive closures and other operations across NCIt’s subsumption hierarchies. Certain caCORE-enabled applications leverage the subsumption hierarchies that descend from the kinds. But with respect to caCORE, the most important NCIt semantic constructions are the concept code, which is found in the concept entity, and among the properties. Definition, synonym and preferred term are all types of property. Especially for annotation of UML model entities, it is helpful if concepts in the NCIt are explicitly linked to corresponding nodes in other terminologies. Accordingly, properties have been created to link NCIt concepts to other information sources (GO, Swissprot, drug formularies, trial protocols, and so on).
NCI Metathesaurus
The NCI Metathesaurus [14] is a specialized version of the Unified Medical Language System [15] Metathesaurus [5]. NCI uses a recent version of NLM’s MEME [16] to produce the NCI Metathesaurus (NCI Meta). Terminology coverage by the UMLS Metathesaurus is expanding, and covers much of basic research and clinical research and practice. The broad scope of the UMLS Metathesaurus, however, means that it includes terminology sources not relevant to NCI’s needs, and also that it excludes certain narrowly focused, cancer-specific sources. By retaining only cancer-relevant sources in NCI Meta, semantic drift [17] is reduced, and by incorporating cancer-specific sources, NCI Meta provides support of terminology needs of the cancer community.
NCI Metathesaurus data sources, updates and business practices
The source vocabularies included in NCI Meta include those UMLS Metathesaurus sources (see http://www.nlm.nih.gov/research/umls/about_umls.html) known to be used by NCI, its grantees and business partners, or by other components of the greater cancer community. Additional sources, called local sources, are added to NCI Meta by NCI upon request. NCI obtains local sources from its publisher, or it develops the source itself, depending on the availability of suitable terminology.
NCI Metathesaurus Information Model and Application Programmer Interfaces
The major semantic constructions used in the NCI Meta are identical with those in the UMLS Metathesaurus [15]. The caCORE client API provides methods to enable calling applications to search the NCI Meta by source, concept, semantic type, various kinds of lexical units, inter- and intra-source relationships, attributes and identifiers. The major functional benefit that the NCI Meta provides to caCORE users is sophisticated lexical searching. Its structure enables the NCI Meta server API to find all the concepts in which any given lexical token occurs. For example, searching NCI Meta for the lexical unit “cold” returns six non-overlapping concepts ranging from Cold (cold temperature) to Cold (acute nasopharyngitis).
Other Vocabulary Content
NCI EVS provides runtime access to other non-NCI terminologies needed by the NCI and the larger cancer community using the Apelon DTS system. Currently the list includes GO[18], MGED Ontology [19], LOINC [20], SNOMED/CT [21], and VA NDF, among others. In addition to providing runtime access to these terminologies, the EVS provides support for intellectual property issues associated with the terminology (for example, obtaining licenses, redistribution permission, or permission to reformat the terminology in a standard form).
Section 6 cancer Data Standards Repository
Architectural considerations
The caDSR is designed to provide a complete infrastructure for creating and managing metadata, so it consists of a repository, accessible through APIs and a series of web based GUI applications for managing the content of the repository. The caDSR supports several options for inputting or creating content; both ETL processes (bulk loading using formatted Excel spreadsheets and UML model extraction and transformation) and transactional creation and editing of content.
At the system level, caDSR (like all caCORE systems) follows an n-tier architecture model. The metadata repository is persisted via an Oracle database server. At the next level, an Oracle application server provides database management and core content management such as creating lists of values (LOV) for populating various fields within the repository. The primary API is comprised of Java Entity beans. As with most caCORE systems, the caDSR supplies a Web Services API that is generated using the Axis toolkit.
As indicated above, the caDSR maintains a suite of web based tools for end users to view, create, edit and maintain content. These tools satisfy a number of use cases that were identified by users both within the internal NCI and wider research community. The tools supplied by caDSR 3 are:
CDE Browser: This tool (http://cdebrowser.nci.nih.gov/CDEBrowser/) provides a graphical interface to search and view metadata content. The tool provides tabs that describe the data element, and all of its component parts. In addition, the CDE Browser provides access to Clinical Case Report Forms (CRFs) and Data Collection Instruments (DCIs) that use Data Elements contained in the caDSR.
UML Model Browser: This tool (http://umlmodelbrowser.nci.nih.gov/umlmodelbrowser/) is similar to the CDE browser in that it provides a means to search and view metadata content. Unlike the CDE Browser, the UML Model Browser presents caDSR content in the context of a UML model.
Curation Tool: This is the primary tool used by curators of metadata content to manually create metadata. In addition to data entry and editing screens, it contains tools for accessing the content of the EVS for the purposes of identifying appropriate concepts to annotate metadata. The curation tool programmatically implements many caDSR business rules.
Administration Tool: This is a fully featured content editing tool that is not bound by the caDSR business rules. The Administration Tool is used by system administrators to correct errors where corrections are not allowed in the Curation Tool or where an undesirable versioning event would occur.
Sentinel Tool: The Sentinel Tool allows users to register their interest in a particular data element or other Administered Component. If the owner of that content makes a change, an alert is sent to interested users.
Data sources and business rules
The metadata content in the caDSR is derived from a variety of sources, including existing or forthcoming data systems and electronic or paper based clinical case report forms or data collection instruments (CRFs and DCIs). The current metadata initiatives support a wide range of clinical and research domains within the cancer community. In addition, the National Institute of Dental and Craniofacial Research, the National Heart, Lung and Blood Institute and the Clinical Data Interchange Standards Consortium (CDISC) are conducting pilot programs to store their metadata in the caDSR, expanding its content beyond the area of cancer research and treatment.
The caDSR maintains two distinct sets of business rules: those that apply to all content, and those that apply only within a specific context or content ownership group. The former set includes such rules as the prohibition against creating duplicate metadata content within a context and the versioning rules that apply to metadata changes. Context-specific business rules include processes for determining the need for new content and the conditions under which certain changes are allowed. Some of the business rules are implemented in software (particularly with regard to duplications in content and versioning rules). The remainder of the business rules are encapsulated as procedural rules and are promulgated through Standard Operating Procedures and a formal training program for metadata curators.
Since data collection needs change over time, the caDSR has mechanisms to support metadata lifecycle. There are two lifecycle management components: a business status and a registration status. The business status covers lifecycle and runs from ‘Draft New’ through a series of statuses ending with ‘Retired’ when the CDE is eventually phased out. The registration status indicates whether the CDE is a data standard, or other special status on a registry-wide basis. The particular series of business statuses are not a characteristic of the caDSR, but rather driven by the needs of a particular caDSR context. Thus, different contexts can adopt different sets of lifecycle rules that meet their specific requirements rather than being tied to a specific caDSR wide implementation.
Versioning is part of the lifecycle of a CDE as changes are made to it or its parts. The rules that determine whether a change warrants a versioning event are encapsulated in the caDSR and its tools, but were determined by the user community, guided by the ISO 11179 Parts 5 and 6, and the 20943 Part 1, Procedures for achieving metadata registry content consistency Part 1: Data Elements. When a versioning or other lifecycle event occurs, the older version of the data element is not destroyed. It is retained for support of systems that used the older version, but its business status is such that it is indicated that the CDE has been superseded by a newer version.
Information Model
The metadata registry information model is based on the ISO 11179 metamodel that defines the semantics of data elements. As previously noted, these semantics are comprised of a component describing a thing in the real world (Object Class), a characteristic that can distinguish one instance of the thing from another (Property), and a representation of the instance of the characteristic (Value Domain). This information model provides the structure around which the data descriptors (metadata) can be interpreted by computers without human intervention.
As noted earlier in this manuscript, although the caDSR is consistent with the ISO 11179 Edition 2 Part 3, there are extensions that are specific to the NCI’s implementation, in particular, Object Class relationships, the binding of controlled terminology to ISO 11179 Administered Items. Thus, there is some degree of variance between the caDSR UML model and the ISO 11179 standard. The complete caDSR UML model is available at http://ncicb.nci.nih.gov/NCICB/content/ncicblfs/EA/caCORE3-1Model/index.htm.
Section 7: cancer Bioinformatics Infrastructure Objects (caBIO)
Information models and Data Types
The caBIO system provides a wide range of basic bioinformatics information. It is not the purpose of this paper to exhaustively list all of the classes, attributes and associations in the information model contained in the caBIO system, as this information is available in the caCORE Technical Guide [3], online documentation (http://ncicb.nci.nih.gov/NCICB/content/ncicblfs/caCORE3-1_JavaDocs), and UML models, but rather to describe the main functional areas covered by the caBIO system.
The caBIO system is at its core a repository of molecular biology data, and so it is not surprising that the entities that concern the Central Dogma of Molecular Biology are well represented in its model. Data for these classes come from a variety of sources. Gene, chromosome, nucleic acid sequence, taxon and cytogenetic location information for Genes comes from the NCBI UniGene data resource [22] via the Cancer Gene Anatomy Project (CGAP), an NCI activity that maintains information about expression of genes in tumors based on EST sequencing frequency [23, 24]. Protein data (including data about sequences) is retrieved from the Protein Information Resource (PIR) at Georgetown University[25]. Gene Ontology [18] data is retrieved from the EVS which supplies the Gene Ontology at runtime.
The caCORE pathway data is primarily supplied from the BioCarta and its Proteomics Pathways Project via the NCI Cancer Molecular Analysis Project or CMAP [26]. This pathway data is integrated into four primary classes: ‘Gene’, ‘Pathway’, ‘Taxon’ and ‘Histopathology’. The presence of the latter class is designed to allow a relationship between a pathway and a disease to be annotated. The Pathway object contains an attribute that contains an image of the pathway as a Scalar Vector Graphic (SVG). Following the caCORE design philosophy, the methods for manipulating these SVG images are encapsulated into an SVGManipulator class that is kept distinct from the Pathway class in a service layer for easier code maintenance.
Single Nucleotide Polymorphism data are primarily supplied by dbSNP [27] and by Affymetrix (http://www.affymetrix.com). The core classes are SNP, PopulationFrequency and GeneRelativeLocation. Certain types of microarray information are maintained in the GenericReporter and GenericArray classes. The current primary source of this information is Affymetrix, although the structure of the classes are such that they should be capable of storing information about virtually any type of reporter.
The caBIO system provides information about libraries used as part of EST sequencing efforts. This set of classes (which includes Library, Tissue and Protocol) are populated with data from the NCI CGAP initiative. This group of classes provides an instructive example about the necessity of grounding metadata in concept-based controlled terminology. The Protocol object here represents the method used to create a library from a tissue (NCI Thesaurus concept code C41111) rather than the concept of a clinical trial protocol (C25320).
Clinical trial data in the caBIO system are obtained from the Cancer Therapeutics Evaluation Program (CTEP) of the NCI. This part of the model was originally driven by use cases from the NCI CMAP project, and as such, the classes contain only very basic information about ongoing clinical trials. Rather than attempt to expand the information for the caBIO model in the clinical trial space. Future releases of caBIO will be harmonized to clinically oriented object models to allow access to data via the interoperability built into the caCORE infrastructure. In addition, the caBIO system provides some very basic information on the mechanism of action of drug compounds, variations found in disease and the genetic targets of drug compounds.
Data update strategy, and business practices
The data that is supplied by the caBIO system comes from a wide range of both internal and external sources and is loaded into the system using a series of Extract, Transform, Load (ETL) processes; the specifics of which are dictated by the particular data source involved. Generally, the data is updated from the original sources during the first and third weeks each month and is visible to the user community at the end of the second and fourth week of each month. This gap (combined with a load process that utilizes two copies of the caBIO database) allows for time to perform quality assurance checks on the data and to perform the update in such a way that users do not experience downtime.
Section 8: Common Security Model
Introduction
Recognizing the need for a pluggable security framework within the caCORE, the NCICB developed the Common Security Model (CSM). The CSM provides three primary services that allow developers to integrate security in their applications with minimal effort:
Authentication: Validating a users identity for purposes of access to the system and its data.
Authorization: Providing a rule framework for access to data, methods and objects within the information system based on the users’ identity.
User Provisioning: Managing authentication and access rights in CSM enabled applications.
Architecture
The general architectural framework of the CSM is described in Figure 8, with the two primary security components (authentication and authorization). The CSM uses the Java Authentication and Authorization Service (JAAS) to authenticate and authorize within an application.
Figure 8.

Architecture of the Common Security Module (CSM). The CSM is designed to work with pluggable credential providers so that it can be incorporated into applications that will operate in diverse environments.
Authentication
The authentication service is designed to respond to external credential providers, and can be configured to respond to more than one credential service for a particular application. By default, CSM has provided implementation for authentications using Lightweight Directory Access Protocol (LDAP) and Relational Database Management Systems (RDBMS). Applications can configure authentication by customizing the ApplicationSecurityConfig.xml file which provides information used for instantiating the JAAS implementations used for performing authentication.
Authorization
The CSM authorization model is based on the concept of Role Based Access Control (RBAC) that operates on ‘Protection Elements’. A Protection Element is segment of an object oriented data system that requires authorization to access. Such elements could be a particular class or a particular attribute of a class. Protection Elements are aggregated into Protection Groups, and individual users are assigned roles that give them access to particular Protection Groups.
The CSM has developed a set of common privileges that are available to applications by default. The standard privileges are access, execute, read, write, create, update and delete, but applications have the ability to add or modify privileges if needed. By providing standard privileges, applications can integrate with CSM quickly and can benefit from shared meaning and common understanding of access control design among various applications.
User Provisioning Tool
The User Provisioning Tool (UPT) is a web application used to configure an application’s authentication and authorization data. The UPT provides functionality to create authorization data elements such as Roles, Protection Elements, Users, etc., and also provides functionality to associate them with each other. The runtime API can then use this authorization data to authorize user actions.
The UPT includes two modes – Super Admin and Admin. The Super Admin operations are typically performed first, as they register the application and application administrators. The primary mode operations, including authorization user provisioning, occur next.
Section 9 Discussion/Future Directions
Open Development
Open Software Development
Although caCORE is open source, to date the bulk of its development has been conducted by teams funded by and working under the supervision of the NCICB. Recognizing that this is not a scalable solution to the rapidly evolving needs of the cancer research community, the NCICB has created an Open Development Initiative (ODI) to move caCORE into becoming a true open source project. To participate in the caCORE ODI, a developer or group should contact the NCICB manager for the component that they were interested in and describe the enhancements that they are interested in providing. Qualified developers will be given access to the NCICB GForge site, and their contributions will become part of the caCORE code base.
Open Terminology Development
The EVS is planning to migrate to more open development of the NCIt, which is expected to have several positive effects on the Thesaurus. This move is expected to both enhance the adoption of NCIt by a wider set of specialized communities within the cancer community and increase the scope and depth of the knowledge modeling that has begun to implement in certain areas of the NCIt namespace [4]
Open, collaborative development is desirable because maintenance of modern, sophisticated terminological or ontological structures is beyond the capabilities of any single group. In addition, experience with biological ontologies (such as GO and MGED Ontology) has shown that adoption of terminologies is facilitated if the product is produced by prominent members of the community it is intended to support. Such collaborative development may require changes to the structure of NCIt, for example, so that it conforms to conventions such as the OBO edge semantics [10]. n addition, the NCI EVS is migrating from proprietary to open source software for both the development and deployment of terminology. Production and maintenance of the NCI Meta has migrated to MEME/Jekyll; NCIt production is currently being migrated to Protégé/OWL (see http://protege.stanford.edu/). During the next year, the existing terminology servers will be replaced with the open source LexBIG terminology server developed by the Mayo Clinic under the auspices of the caBIG program. (see http://informatics.mayo.edu/LexGrid/).
Terminology as data
An increasing number of terminologies served by EVS contain domain knowledge that is represented in the form of named semantic associations (roles) between concepts. Although this domain knowledge has been surfaced in a very basic way in caCORE (given a concept in NCIt, it is possible, for example, to obtain the list of roles and associated concepts), it has not been utilized to the fullest extent possible. With the introduction of LexGRID, EVS will have an open source terminology server that exposes the full range of data on terminology at runtime. The discovery services available in caGrid will be able to not only find all terminology services that are data servers but, more importantly, to determine the terminology services that can supply any given terminology, terminology version, concept, role, or other concept restriction available. Availability of terminology data through caGrid should enable use of domain knowledge in searches and in processes such as annotation of data being stored in repositories.
Terminology models are intended to represent something close to the totality of the useful relationships among the concepts in the namespace. In the future caCORE may leverage this rich web of semantic relationships. Doing so would be facilitated if instance data were directly tagged with concept codes, and in the future, caCORE may implement that tagging. Even currently however, caCORE-compliant class attributes can provide values that can be used to locate concepts in the terminology space. For example, a class that includes an attribute Locus_ID bound to the NCIt concept C43693 can be expected to provide an actual LocusLink identifier for each instance contained in data objects obtained thru the class. One can use the GeneID value or Locus identifier to retrieve the concept(s) possessing the identifier. Once one has identified a concept, its roles and property values can be leveraged. The roles could be used to obtain the set of concepts known to be related to a starting concept. These related concepts could be used to guide a search across multiple data models.
cancer Biomedical Informatics Grid (caBIG)
The caCORE infrastructure provides a strong degree of interoperability, but there are still barriers to the use of the resources that it provides. The caCORE 3 Java APIs require that a client JAR file be incorporated at compile time in any application that uses the system. In addition, there is the broader problem of discovering that the various components of the caCORE system exist and that they are available for use. To resolve these problems, the NCICB has launched the cancer Biomedical Informatics Grid or caBIG™, a voluntary network or grid of researchers that are building the infrastructure to support an interoperable network of data and analytical services for cancer research. caBIG™ builds upon many of the technologies that are part of caCORE 3 (including the EVS and caDSR) to create its interoperable network.
The caBIG™ infrastructure is based on Grid computing technology. A Grid has been described as “a system that coordinates resources that are not subject to centralized control using standard, open, general-purpose protocols and interfaces to deliver nontrivial qualities of service.” [28]; that is, it allows users to access a federated network of resources through standardized interfaces to solve informatics problems that are not solvable with conventional computing technology. The caGrid [29] is built upon the Open Source Globus Toolkit (http://www.globus.org) and OGSA-DAI extensions (http://www.ogsadai.org.uk) with additional components from Ohio State University and the caCORE.
The use of Grid technology solves the two major remaining interoperability barriers. First, the caGrid provides for advertising and discovery of services based on the metadata that they register in the caDSR. Second, the caGrid provides access to all services (i.e. software systems) that connect to the caGrid through a single API that does not need to be updated when a new service is added.
Software systems that wish to connect to the caGrid must meet a set of compatibility guidelines (available from the caBIG web site at https://cabig.nci.nih.gov/guidelines_documentation) that describe requirements (but not the specifics of the implementation) in the areas of interface integration (APIs), information models, controlled terminology and data elements. Because interoperability is not Boolean, there is a series of levels of compatibility, starting with ‘Legacy’ (not interoperable) through ‘Bronze’, ‘Silver’ and ‘Gold’ (fully interoperable on the caGrid). The compatibility guidelines were informed by the NCICB experience with the caCORE infrastructure. Semantically annotated systems like caCORE 3 that are built using the caCORE SDK, are typically compatible at the ‘Silver’ level. When they are connected to the caGrid, and have met additional harmonization requirements they would achieve ‘Gold’ and be fully interoperable within the limits of current Grid technology.
The coupling of the caCORE semantic methodology with the Grid technology that will be deployed in caBIG provides an opportunity for true semantic interoperability, from discovery to the creation of federated data sets maintained in distinct resources created by disparate research groups. This technology provides a semantic Service Oriented Architecture, supporting both syntactic and semantic interoperation, and the infrastructure to implement several interesting use cases:
Simplified Workflows: As has been indicated, all caCORE-like data and analytical services maintain full semantics describing their input and output objects, and the caGrid provides a uniform transport mechanism for these objects, regardless of the implementation details. This should greatly improve the ease of creating complex workflows between disparate data and analytical resources maintained by different entities. Figure 9 (top) describes a sample workflow on the caGrid. In this example two data services are queried for objects that are used as the inputs in an analytical service.
-
Aggregation of federated query results: The caCORE extension of the model driven paradigm is well suited to allow the aggregation of results from disparate data systems. Consider the two hypothetical data systems described in Figure 9 (bottom). On the left is a model that closely resembles the caBIO Agent Class (the id attribute has been removed for clarity). On the right is a class from a hypothetical data system that provides information about Drugs. Using the caCORE infrastructure, both classes have been mapped to the concept Agent, which has NCI Thesaurus Concept Code C1708. From this we can conclude that despite the fact that the classes have distinct names that they are supplying information about the same semantic entity (i.e. they are describing instances of the same concept, but using a different term to describe that concept).
By inspection, none of the attributes have the same name. Under normal conditions, the creators of the data services would need to be consulted to determine if any of the attributes actually represented the same data. Since the caCORE infrastructure provides a mapping to controlled terminology and then uses that controlled terminology to map class/attribute pairs to CDEs, it is possible to computationally determine equivalence merely by examining the data elements that are used to describe the systems. In this case, Agent.NSCNumber and Drug.fdaCode are both mapped to the same CDE with public ID 2223866 and version 3.0. The implication of this identity and our semantic model are profound; it implies that when the value of Agent.NSCNumber equals Drug.fdaCode that the two instances of this class are carrying information about the same instance of the same entity (i.e. they have information about the same drug). This would allow the creation of a data set that is a Cartesian product of the two data systems, akin to the data structures that can be created using a relational database management system. We refer to these common CDEs as ‘Grid keys’ in reference to the foreign and primary keys in relational database management systems.
It is important to note that while this method provides a relatively automated mechanism to identify touch points between object oriented data systems it does not eliminate the need for scientific judgment when creating these ‘Grid joins’. For instance, if both classes contained an FDA Approval Date attribute, it would be possible to perform a join on the two systems. However, it is quite likely that equivalence on this attribute will not indicate that the two objects contain data about the same instance of the class (although such a join might be useful for other purposes). Recognizing this limitation, the registered metadata allows a user of the caGrid to be able to determine if the particular attribute is sufficient to allow for aggregation based on their specific use case.
Distributed Queries: An obvious extension of the two above scenarios above is the distributed query. In this instance, a result (or portion of a result) from a query into one system is used as the input into a query in a second system. Since caCORE-like systems have registered metadata for each attribute, it is straightforward to determine which values/objects to use as inputs from each system.
Figure 9.

Two caGrid use cases: The top panel describes a set of workflows in which objects retrieved from one system are used as inputs into another system. The bottom panel describes aggregation of data from two distinct resources based on common CDEs in each system. The resulting aggregate using a Grid key is functionally equivalent to a Cartesian join using foreign keys in a relational database management system.
Acknowledgements
Tara Akhavan, Michael Connelly, Sasikumar Thangaraj and Doug Mason contributed to the development of caBIO. Kim Ong and Iris Guo contributed to the development of the EVS server and client software. Steve Alred, Edmond Mulaire, Prerna Aggarwal, Shaji Kakkodi, Larry Hebel, Sumana Hedge and Jennifer Brush contributed to the development of the caDSR. Vinay Kumar and Stephen Hunter contributed to the development of the Common Security Module. Kathleen Gundry, Tommie Curtis, Brenda Maeske, Mary Cooper and Nicole Thomas were instrumental in the semantic integration process. Ye Wu supervised the Quality Assurance Testing for caBIO and the APIs for caDSR and EVS, assisted by Scott Zhang. The caCORE documentation was edited by Wendy Erickson-Hirons. This work was supported by the direct operating expenditures of the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Any model driven, object-oriented data system could be at the top of the caCORE primary technology stack. In a sense, the caBIO system is a ‘reference implementation’ of ‘caCORE-like’ data system. We include it in the discussion of caCORE because it is the example implementation, and because the caBIO system is developed and released concurrently with the other components of caCORE.
Users interested in the full ISO11179 specification are directed to the ISO web site.
This notion is commonly used by authors writing about RDF, XML and ontology, but is seldom defined. See "Reflection in logic, functional and object-oriented programming: a Short Comparative Study" http://citeseer.ist.psu.edu/106401.html for a formal, general definition
References
- 1.Staff, I.o.E.a.E.E. IEEE Computer Dictionary - Compilation of IEEE Standard Computer Glossaries. 1990. pp. 610–1990. [Google Scholar]
- 2.Phillips J, et al. The caCORE Software Development Kit: streamlining construction of interoperable biomedical information services. BMC Med Inform Decis Mak. 2006;6:2. doi: 10.1186/1472-6947-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bioinformatics. N.C.I.C.f., caCORE 3.1 Technical Guide. Rockville, MD: 2006. p. 202. [Google Scholar]
- 4.Sioutos N, et al. NCI thesaurus: A semantic model integrating cancer-related clinical and molecular information. J Biomed Inform. 2006 doi: 10.1016/j.jbi.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Tuttle MS, et al. The homogenization of the Metathesaurus schema and distribution format; Proc Annu Symp Comput Appl Med Care; 1992. pp. 299–303. [PMC free article] [PubMed] [Google Scholar]
- 6.Hartel FW, et al. Modeling a description logic vocabulary for cancer research. J Biomed Inform. 2005;38(2):114–129. doi: 10.1016/j.jbi.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Fragoso G, de Coronado S, Haber M, Hartel F, Wright L. Overview and Utilization of the NCI thesaurus. Comparative and Functional Genomics. 2004;5 doi: 10.1002/cfg.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Coronado S, et al. NCI Thesaurus: using science-based terminology to integrate cancer research results. Medinfo. 2004;11(Pt 1):33–37. [PubMed] [Google Scholar]
- 9.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998;37(4–5):394–403. [PMC free article] [PubMed] [Google Scholar]
- 10.Ceusters W, Smith B, Goldberg L. A terminological and ontological analysis of the NCI Thesaurus. Methods Inf Med. 2005;44(4):498–507. [PubMed] [Google Scholar]
- 11.Bodenreider O, et al. Of mice and men: aligning mouse and human anatomies; AMIA Annu Symp Proc; 2005. pp. 61–65. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S, Bodenreider O. Alignment of multiple ontologies of anatomy: deriving indirect mappings from direct mappings to a reference; AMIA Annu Symp Proc; 2005. pp. 864–868. [PMC free article] [PubMed] [Google Scholar]
- 13.Hartel FW, Denise B, Covitz P. W3C Workshop on Semantic Web for Life Sciences. Cambridge, Massachusetts USA: 2004. OWL/RDF/LSID Utilization in NCI Cancer Research Infrastructure. [Google Scholar]
- 14.Covitz PA, et al. caCORE: a common infrastructure for cancer informatics. Bioinformatics. 2003;19(18):2404–2412. doi: 10.1093/bioinformatics/btg335. [DOI] [PubMed] [Google Scholar]
- 15.Lindberg C. The Unified Medical Language System (UMLS) of the National Library of Medicine. J Am Med Rec Assoc. 1990;61(5):40–42. [PubMed] [Google Scholar]
- 16.Suarez-Munist ON, et al. MEME-II supports the cooperative management of terminology; Proc AMIA Annu Fall Symp; 1996. pp. 84–88. [PMC free article] [PubMed] [Google Scholar]
- 17.Solbrig HR, et al. A formal approach to integrating synonyms with a reference terminology; Proc AMIA Symp; 2000. pp. 814–818. [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whetzel PL, et al. The MGED Ontology: a resource for semantics-based description of microarray experiments. Bioinformatics. 2006;22(7):866–873. doi: 10.1093/bioinformatics/btl005. [DOI] [PubMed] [Google Scholar]
- 20.McDonald CJ, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem. 2003;49(4):624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 21.Kudla KM, Rallins MC. SNOMED: a controlled vocabulary for computer-based patient records. J Ahima. 1998;69(5):40–44. quiz 45-6. [PubMed] [Google Scholar]
- 22.Pontius JU, W L, Schuler GD N.C.f.B. Information, Editor. The NCBI Handbook National Center for Biotechnology. Bethesda (MD): 2003. UniGene: a unified view of the transcriptome. [Google Scholar]
- 23.Strausberg RL, et al. In silico analysis of cancer through the Cancer Genome Anatomy Project. Trends Cell Biol. 2001;11(11):S66–S71. doi: 10.1016/s0962-8924(01)02104-3. [DOI] [PubMed] [Google Scholar]
- 24.Strausberg RL. The Cancer Genome Anatomy Project: new resources for reading the molecular signatures of cancer. J Pathol. 2001;195(1):31–40. doi: 10.1002/1096-9896(200109)195:1<31::AID-PATH920>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 25.Wu CH, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34(Database issue):D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buetow KH, et al. Cancer Molecular Analysis Project: weaving a rich cancer research tapestry. Cancer Cell. 2002;1(4):315–318. doi: 10.1016/s1535-6108(02)00065-x. [DOI] [PubMed] [Google Scholar]
- 27.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foster I. What is the Grid? A three point checklist. Grid Today. Volume [Google Scholar]
- 29.Saltz J, et al. caGrid: design and implementation of the core architecture of the cancer biomedical informatics grid. Bioinformatics. 2006;22(15):1910–1916. doi: 10.1093/bioinformatics/btl272. [DOI] [PubMed] [Google Scholar]


