Abstract
The nonreceptor tyrosine kinase Src is expressed at a high level in cells that are specialized for regulated secretion, such as the neuron, and is concentrated on secretory vesicles or at the site of exocytosis. To investigate the possibility that Src may play a role in regulating membrane traffic, we searched for neuronal proteins that will interact with Src. The SH3 domain of Src, but not that of the splice variant N-Src, bound to three proteins from mouse synaptosomes or PC12 cells: dynamin, synapsin Ia, and synapsin Ib. Dynamin and the synapsins coprecipitated with Src from PC12 cell extracts, and they colocalized with a subset of Src in the PC12 cell by immunofluorescence. Neither dynamin nor the synapsins were phosphorylated by Src, suggesting that the interaction of these proteins serves to direct the kinase activity of Src toward other proteins in the vesicle population. In immunoprecipitates containing Src and dynamin, the clathrin adaptor protein α-adaptin was also found. The association of Src and synapsin suggests a role for Src in the life cycle of the synaptic vesicle. The identification of a complex containing Src, dynamin, and α-adaptin indicates that Src may play a more general role in membrane traffic as well.
Keywords: synaptic vesicles, Src-homology domain 3, membrane traffic
The Src protein is the archetype for a family of nonreceptor protein tyrosine kinases (1). Src is expressed at a low level in many cell types, but it is expressed at a high level in certain cells that are specialized for regulated secretion, including neurons, endocrine cells, platelets, and osteoclasts (2, 3). The subcellular localization of Src corresponds to organelles involved in membrane traffic, such as the endosome (4, 5), the synaptic vesicle (6), and the secretory granule (7). Neurons express both Src itself and a neuron-specific splice variant, N-Src (8, 9). Therefore, Src may play a regulatory role in membrane traffic events that are particularly critical in the neuron.
Src interacts with proline-rich domains of other proteins by means of a domain known as SH3 (1). Using an in vitro binding assay, we found that the SH3 domain of Src, but not of N-Src, interacts with three known proline-rich proteins: dynamin, synapsin Ia, and synapsin Ib. Moreover, both immunofluorescence and immunoprecipitation demonstrated that these proteins interact in vivo. In addition, the clathrin assembly protein α-adaptin, which had previously been shown to bind to dynamin, coprecipitated with Src. These protein associations suggest a role for Src in membrane traffic events mediated by dynamin and the synapsins in the neuron. While this manuscript was in preparation, Onofri et al. (10) reported that the SH3 domain of Src interacts with the synapsins and that the association of Src and synapsin results in stimulation of the kinase activity of Src (see Discussion).
MATERIALS AND METHODS
Materials.
Cell lines used in this work include PC12 cells, mouse embryo fibroblasts derived from src−/− mice (kindly provided by H. Varmus, National Institutes of Health), and 3T3 cells overexpressing Src (generously provided by D. Shalloway, Cornell Univ.). Wild-type FVB/N female mice, or age-matched src−/− mice (provided by C. Lowell, Univ. of California, San Francisco) were used for mouse synaptosome preparations. The following antibodies and antisera were used: monoclonal antibodies to Src, 2-17 and 327 (provided by S. Courtneidge, Sugen, Redwood City, CA, and J. Brugge, Arrad, Cambridge, MA, respectively); Fyn3 polyclonal antiserum (Santa Cruz Biotechnology); monoclonal antibody 2-7 to Yes (J. Brugge); anti-dynamin antibodies (Transduction Laboratories); anti-synapsin I (Biogenesis, Bournemouth, U.K.); various antibodies to synaptic vesicle proteins (provided by R. Kelly, Univ. of California, San Francisco); monoclonal antibody to transferrin receptor (I. Trowbridge, Arrad, Cambridge, MA); antiserum against pyruvate kinase (made by standard procedures); antibodies to α-adaptin and the clathrin heavy chain (provided by F. Brodsky, Univ. of California, San Francisco); an antibody to γ-adaptin (provided by E. Ungewickell, Washington Univ., St. Louis); monoclonal antibody to Rab3a (R. Jahn, Yale Univ.); and polyclonal antiserum against glutathione S-transferase (GST), Grb2, and phospholipase C-γ (Transduction Laboratories, Lexington, KY).
Subcellular Fractionation and Cell Lysis.
PC12 cells were lysed and fractionated by differential centrifugation as previously described (6). Mouse brain synaptosomes were prepared by a modification of a protocol described for preparing rat brain synaptosomes (11). Membrane populations were lysed in a Nonidet P-40-based lysis buffer previously described. The lysate was then subjected to centrifugation at 100,000 × g for 1 hr to remove any nonsolubilized membranes.
Overlay Assays.
Overlay assays were performed with GST-fusion proteins and polyclonal antiserum to GST as previously described (17).
Western Blotting.
Western blotting was performed as previously described (12). Primary antibodies were detected with either a horseradish peroxidase-conjugated secondary antibody and chemiluminescent detection system (Amersham), or an 125I-conjugated secondary antibody (ICN), which was then quantified with a PhosphorImager (Molecular Dynamics).
Immunoprecipitation.
Immunoprecipitation was performed as previously described (12), except that all lysates were first centrifuged at 100,000 × g for 1 hr to remove any nonsolubilized membranes.
Immunofluorescence.
PC12 cells were plated on glass coverslips coated with poly-d-lysine and collagen IV, then differentiated by stimulation with nerve growth factor. Cells were washed in PBS, then fixed for 10 min in 3% paraformaldehyde. The samples were blocked for 15 min in 10% normal goat serum (PBS/NGS), then incubated with primary antibody for 1.5 hr. Washing with PBS/NGS was followed by a 30-min incubation with a secondary antibody conjugated to the red fluorophore Cy3 (Molecular Probes). The slides were mounted in 70% glycerol with the antifade agent n-propyl gallate.
RESULTS
Dynamin and Synapsins Ia and Ib Bind to the SH3 Domain of Src.
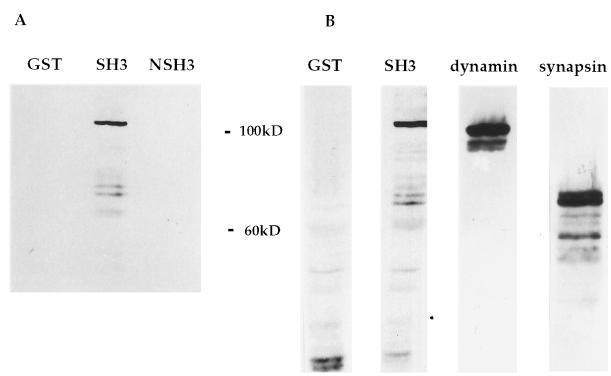
To identify proteins from the neuron that would interact with Src, SH3 domains of Src and N-Src (SrcSH3 and NSrcSH3, respectively) were expressed in bacteria as GST fusion proteins and used as probes in an overlay assay. Equal amounts of solubilized protein from a membrane preparation of PC12 cells or from a synaptosome preparation of mouse brain were probed with the GST fusion proteins. The SrcSH3 fusion protein, but not the NSrcSH3 fusion protein, bound to three proteins, approximately 100, 80, and 75 kDa, in preparations of mouse brain synaptosomes (Fig. 1A). The same three bands were detected in a small vesicle fraction from PC12 cells. These three proteins comigrated with three known proteins identified by specific antibodies: dynamin and synapsins Ia and Ib (Fig. 1B).
Figure 1.
Mouse synaptosome proteins that interact with the SH3 domain of Src. (A) Mouse brain synaptosomes were lysed, resolved by electrophoresis in a polyacrylamide gel, and used in an overlay assay with the indicated GST fusion proteins (GST, GST alone; SH3, GSTSrcSH3; NSH3, GSTNSrcSH3). (B) The same preparation was Western blotted with antibodies to dynamin and synapsins Ia and Ib.
Src, the Synapsins, and Dynamin Colocalize in PC12 Cells.
Src, dynamin, and the synapsins have been previously reported to be enriched in nerve terminals from brain (6, 13, 14). We compared the immunofluorescent staining patterns of these proteins in PC12 cells.
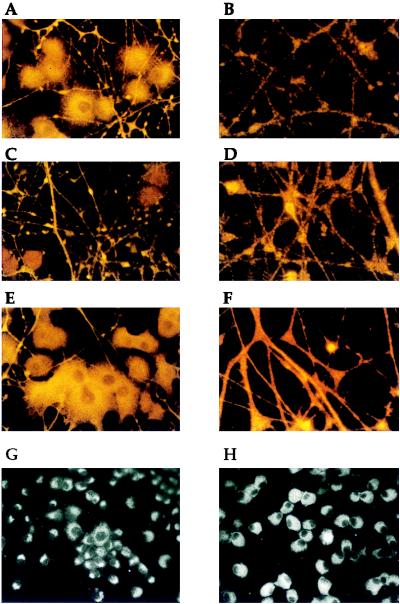
The immunofluorescent staining pattern of Src in PC12 cells included a bright perinuclear staining (Fig. 2A) as well as a punctate staining pattern throughout the neurites (Fig. 2B). The cell body staining patterns of dynamin (Fig. 2C) and the synapsins (Fig. 2E) in PC12 cells were less intense than that of Src. Instead, in these cells, the staining patterns in the neurite predominated. The neurite staining of dynamin in PC12 cells (Fig. 2D) was very similar to that of Src. The neurites were intensely stained in a punctate pattern. The neurite staining of the synapsins in PC12 cells (Fig. 2F) was also quite intense, but it exhibited less of a punctate character than Src or dynamin.
Figure 2.
Immunofluorescence patterns of Src, dynamin, and the synapsins in PC12 cells. PC12 cells were induced to differentiate with nerve growth factor and were prepared for immunofluorescence with antibodies to the following proteins: Src (A and B), dynamin (C and D), synapsin I (E and F), the transferrin receptor (G), and pyruvate kinase (H). (Final magnification for A, C, and E is ×100; for B, D, and F it is ×250; and for G and H it is ×50.)
The staining patterns of a predominantly endosomal protein, transferrin (Fig. 2G), and a cytoplasmic protein, pyruvate kinase (Fig. 2H), differed dramatically from those of Src, dynamin, and the synapsins. Antibodies to transferrin and pyruvate kinase did not stain the neurites of differentiated PC12 cells at all (Fig. 2 G and H). In contrast, antibodies to Src, dynamin, and the synapsins stained the neurites intensely. Therefore, a subset of the total Src in the PC12 cell colocalizes with dynamin and the synapsins. We were unable to perform double-label immunofluorescence on PC12 cells with the available antibodies.
Dynamin and Synapsins Ia and b Coprecipitate with Src from PC12 Cell Lysates.
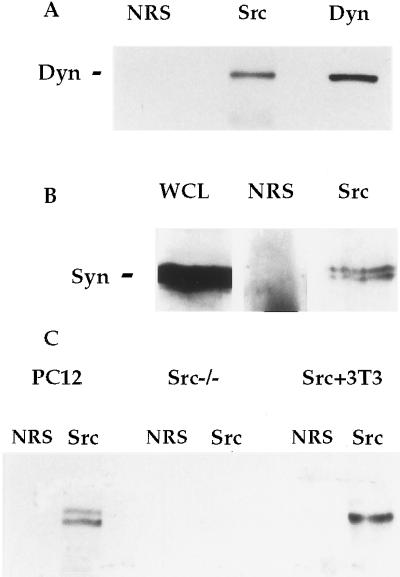
To determine whether the interactions detected between Src, dynamin, and the synapsins in vitro were physiologically relevant, we assessed the association of the proteins in PC12 cells. Lysates of PC12 cells were prepared and analyzed by immunoprecipitation with normal rabbit serum or an antibody to Src, followed by Western blotting for dynamin and the synapsins (Fig. 3 A and B). Both dynamin and the synapsins were precipitated reproducibly by a number of different Src-specific antibodies, but not by normal rabbit serum. These results were the same in untreated and fully differentiated PC12 cells (not shown).
Figure 3.
Coprecipitation of dynamin and synapsin with Src from PC12 cells. (A and B) PC12 cells were lysed and prepared for immunoprecipitation with normal rabbit serum (NRS) and antibodies to Src and dynamin. Whole cell lysate (WCL) and the immunoprecipitates were Western blotted for dynamin or the synapsins as indicated. (C) Lysates from PC12 cells, fibroblasts from src−/− mice, and 3T3 cells overexpressing Src were immunoprecipitated with NRS and a Src antibody, and Western blotted for dynamin.
Dynamin coprecipitated with Src from a fibroblast cell line, indicating that this interaction was not limited to neuronal cell types (Fig. 3C). The synapsins are not expressed in nonneuronal cell types. When an antibody to Src was used in a precipitation from a src−/− cell lysate, dynamin was not precipitated (Fig. 3C). Thus, the coprecipitation of dynamin with Src was not an artifact of antibody cross-reactivity, but rather required the presence of the Src protein.
We were unable to detect tyrosine phosphorylation on dynamin or the synapsins in PC12 cell lysate by using several appropriate techniques (not shown).
The Interaction Between Src and Dynamin and the Synapsins Is Specific and Direct.
To exclude the possibility that the coprecipitation of dynamin and the synapsins with Src was the result of an artifactual precipitation of whole synaptic vesicles or other membranes, two approaches were taken. First, all lysates were subjected to a high speed centrifugation that was sufficient to sediment synaptic vesicles, and should remove any poorly solubilized membranes prior to immunoprecipitation. Second, the Src immunoprecipitate was tested for the presence of other synaptic vesicle, endosome, or coated vesicle antigens. Lysates were immunoprecipitated with normal rabbit serum or the Src antibody. Whole cell lysates and the immunoprecipitates were then Western blotted with antibodies to a number of abundant synaptic vesicle proteins and proteins found on endosomes and coated vesicles. None of the antigens tested was detectable in the Src immunoprecipitate from at least 10-fold more lysate than was required for detection of the antigen by Western blotting (data summarized in Table 1).
Table 1.
Summary of coimmunoprecipitations
| Protein | IP with Src Ab |
|---|---|
| Dynamin | + |
| Snapsins Ia and Ib | + |
| Synaptophysin | − |
| Synaptotagmin | − |
| Syntaxin | − |
| Rab3a | − |
| Transferrin receptor | − |
| Grb2 | − |
| Phospholipase C-γ | − |
| α-Adaptin | + |
| γ-Adaptin | − |
| Clathrin | − |
PC12 cells were lysed and prepared for immunoprecipitation (IP) with normal rabbit serum or Src antibody. Whole cell lysate and the immunoprecipitates were Western blotted with the indicated antibodies. A + indicates that the antigen was detected in the immunoprecipitate of Src but not normal rabbit serum when 10-fold more protein than required to detect the antigen by Western blotting was used. A − indicates that the antigen was not detected.
These results indicate that the membranes in the PC12 lysates were likely to be fully solubilized, and that dynamin and the synapsins were not precipitating nonspecifically. The associations of dynamin and the synapsins were therefore the result of specific and direct interactions of these proteins with Src.
α-Adaptin Coprecipitates with Src.
Dynamin binds to Grb2, phospholipase C-γ, and α-adaptin (15–20). We tested for the presence of these proteins in the Src immunoprecipitate from PC12 cells (Table 1). Neither Grb2 nor phospholipase C-γ was detected in the Src immunoprecipitates. Although complexes containing Grb2 and dynamin may exist in PC12 cells, the complex containing Src and dynamin is apparently different.
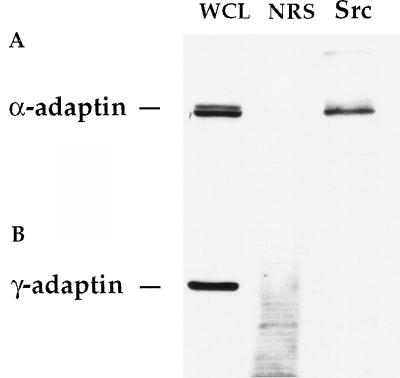
We were able to detect an association of Src and α-adaptin in PC12 cell lysates (Fig. 4A). As a control, we checked for the association of a related adaptor protein, γ-adaptin, that is not expected to be enriched in the nerve terminal. We were unable to detect γ-adaptin in our immunoprecipitates, indicating that the association with α-adaptin is specific (Fig. 4B). We found no evidence for a direct interaction between the SrcSH3 fusion protein and α-adaptin (not shown). It is likely that the presence of α-adaptin in the immunoprecipitate is a result of binding to dynamin (20). Despite the presence of the adaptor protein, we were unable to detect clathrin in the immunoprecipitates.
Figure 4.
α-Adaptin coprecipitates with Src from PC12 cells. PC12 cells were lysed and prepared by immunoprecipitation with NRS or Src antibody. Whole cell lysate (WCL) and the immunoprecipitates were Western blotted with antibodies to α-adaptin (A) and γ-adaptin (B).
Other Src Family Members Do Not Bind to Dynamin in Neuronal Cells.
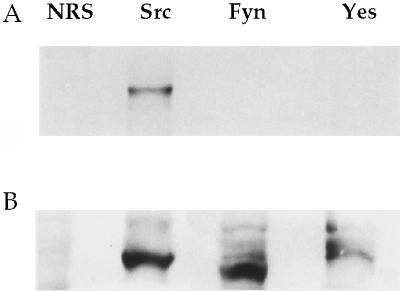
Because of the similarity in the sequences and the subcellular localization of Src family members in PC12 cells, we looked for the association of other endogenously expressed Src family members with dynamin. Dynamin failed to coprecipitate with Fyn and Yes from PC12 cell lysate (Fig. 5A) or mouse synaptosomes (not shown). Western blotting with an antibody to the carboxyl-terminal tail of Src that cross-reacts with Fyn and Yes confirmed that the Src family member proteins were precipitated by their specific antibodies (Fig. 5B). Similar results were obtained when the immunoprecipitates were Western blotted for synapsin (not shown), suggesting that this interaction was also unique to Src in the neuron.
Figure 5.
Association of dynamin with other Src family members. PC12 cells were lysed and prepared for immunoprecipitation with NRS and antibodies to Src, Fyn, and Yes. The immunoprecipitates were Western blotted with an antibody to dynamin (A) and with a pan-Src-family antiserum (B).
To determine the effect of loss of Src expression on the other Src family members in neurons, we analyzed the expression level of other Src family members and their ability to associate with dynamin in preparations of brain from src−/− mice. We found no change in the expression level of other Src family members in src−/− brain preparations. In addition, we found no evidence for a compensatory change in the ability of other Src family members to associate with dynamin in the absence of Src (not shown).
DISCUSSION
A Role for Src in Neuronal Membrane Traffic.
There is mounting evidence to suggest a role for Src in membrane traffic. Src is expressed at a high level in a variety of cell types that are specialized for regulated secretion, including the neuron (2). In these cells, Src is concentrated on the organelles involved in exocytosis or endocytosis, such as the synaptic vesicle in neurons and the endosome in fibroblasts (4–6). Overexpression of Src in bovine adrenal chromaffin cells leads to an increased secretory response to acetylcholine receptor stimulation (21). Stenberg et al. (22) have recently shown that some Src family members colocalize with clathrin in coated pits and vesicles in the platelet. We and Onofri et al. (10) have now shown that Src interacts with dynamin and the synapsins, proteins known to be involved in membrane regulation in the nerve terminal. In addition we find that Src interacts indirectly with the clathrin adaptor protein, α-adaptin.
Dynamin is a GTPase involved in the final steps of fission of a clathrin-coated vesicle from the membrane, and it may be involved in other, clathrin-independent, types of endocytosis. Dynamin plays an important role in receptor-mediated endocytosis in a variety of cells and in synaptic vesicle recycling in neurons (14, 23–25). SH3 domain-containing proteins, such as Grb2 and amphiphysin, are thought to recruit dynamin to the plasma membrane (26, 27). Shupliakov et al. (28) have blocked synaptic vesicle recycling in neurons with the injection of a bacterially expressed SH3 domain from amphiphysin or a peptide consisting of the proline-rich sequence of dynamin. Since we found that Src and dynamin interact in neurons and nonneuronal cells, Src may play a role similar to that hypothesized for amphiphysin and Grb2 in neurons and possibly in other cell types.
α-Adaptin is an assembly protein that is involved in recruiting clathrin to the plasma membrane (29). Like dynamin, α-adaptin plays a role in both receptor-mediated endocytosis and synaptic vesicle recycling (30). The interaction of Src and α-adaptin in vivo suggests a role for Src in the regulation of the endocytic process in neurons.
The synapsins are neuron-specific proteins that bind both to the synaptic vesicle and to actin. The binding is thought to cross-link a subpopulation of synaptic vesicles, known as the reserve pool, to an actin cytomatrix in the nerve terminal (31–33). Activation of a signal transduction pathway leading to the phosphorylation of synapsin on serine residues by CamKinase II is required for release of synaptic vesicles from this reserve pool (13, 34–37). The interaction of Src with the synapsins suggests that Src might play a role in regulating the formation, stability, or dissolution of this actin-bound population of synaptic vesicles.
Despite the fact that dynamin and the synapsins coprecipitated with Src, we have no evidence that the proteins were tyrosine-phosphorylated in PC12 cells. Neither protein was recognized by an anti-phosphotyrosine antibody when coprecipitated with Src or when immunoprecipitated with a specific antibody to dynamin or the synapsins from whole cell lysate. Neither protein became phosphorylated in an in vitro kinase assay using Src immunoprecipitates. There is no published evidence for the tyrosine phosphorylation of the synapsins. Dynamin is tyrosine-phosphorylated and associated with the Lyn tyrosine kinase in resting mast cells, but becomes dephosphorylated when mast cells are stimulated to degranulate (38). Although dynamin and the synapsins appear not to be substrates for Src, the interaction with synapsin stimulates the kinase activity of Src (10). We sought but failed to find such stimulation, possibly because of idiosyncrasies of our serological reagents.
Other Src Family Members in Neurons Do Not Bind to Dynamin or the Synapsins.
Whereas antibodies to Src coprecipitated dynamin and the synapsins, antibodies to other Src family members expressed in neurons failed to coprecipitate these proteins. The fact that the protein interactions observed here seem to be unique to Src suggests that, at least in this case, the Src family members are not redundant. In addition, we have found that there is no direct compensatory activity of the other Src family members present in a src−/− mouse.
To date, there are no reports of defects in the function of differentiated neurons from the brain of src−/− mice. Previously, this had been explained by the possible redundant or compensatory functions of other Src family members. Our results require alternative explanations. It is possible that other, unrelated, proteins play the same role as Src in the neuron. Alternatively, Src may play a role in the neuron that is not essential for normal function.
N-Src May Play a Different Role Than Src in Neurons.
The NSrcSH3 domain contains an insert of six amino acids in the region responsible for binding to the polyproline sequence. This insert has been previously shown to change the binding specificity of the SH3 domain, preventing N-Src from binding to many of the proteins to which Src binds (39). Accordingly, the SH3 domain of N-Src failed to bind the proline-rich domains of dynamin and the synapsins. This result sustains the conclusion that the association of dynamin and the synapsins with the SH3 domain of Src is most likely mediated by an interaction with polyproline sequences. Onofri et al. (10) have substantiated this view by mapping the interaction to a carboxyl-terminal domain of synapsin that contains two canonical binding sites for the SH3 domain of Src.
Most neurons of the central nervous system express both the Src and N-Src proteins (8, 9). If the neuronal variant of Src is not involved in interactions with dynamin and the synapsins, then perhaps it plays a different role in the cell. The insert within the SH3 domain of N-Src might serve to free N-Src from some protein associations, allowing it to bind to other proteins specifically expressed in the neuron.
Acknowledgments
We thank the many investigators who provided us with materials, members of the Kelly and Brodsky lab for invaluable advice and discussion, and members of the Bishop lab for support, advice, and encouragement. This work was supported by funds from the National Institutes of Health (CA 44338) and by the G. W. Hooper Research Foundation. A.F.-B. was supported by a Medical Scientist Training Grant from the National Institutes of Health.
ABBREVIATIONS
- SH3
Src-homology domain 3
- NSrcSH3
neuronal variant of SH3
- GST
glutathione S-transferase
- NRS
normal rabbit serum
References
- 1.Brown M T, Cooper J A. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 2.Brugge J S, Yonemoto W, Lustig A, Golden A. Proc Int Symp Princess Takamatsu Cancer Res Fund. 1986;17:241–249. [PubMed] [Google Scholar]
- 3.Horne W C, Neff L, Chaterjee D, Lomri A, Levy J B, Baron R. J Cell Biol. 1992;119:1003–1013. doi: 10.1083/jcb.119.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan K B, Swedlow J S, Varmus H E, Morgan D O. J Cell Biol. 1992;118:321–333. doi: 10.1083/jcb.118.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David P T, Nouvian D Y. J Cell Biol. 1990;111:3097–3116. doi: 10.1083/jcb.111.6.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linstedt A D, Vetter M L, Bishop J M, Kelly R B. J Cell Biol. 1992;117:1077–1084. doi: 10.1083/jcb.117.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grandori C, Hanafusa H. J Cell Biol. 1988;107:2125–2135. doi: 10.1083/jcb.107.6.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brugge J S, Cotton P C, Queral A E, Barrett J N, Nonner D, Keane R W. Nature (London) 1985;316:554–557. doi: 10.1038/316554a0. [DOI] [PubMed] [Google Scholar]
- 9.Brugge J, Cotton P, Lustig A, Yonemoto W, Lipsich L, Coussens P, Barrett J N, Nonner D, Keane R W. Genes Dev. 1987;1:287–296. doi: 10.1101/gad.1.3.287. [DOI] [PubMed] [Google Scholar]
- 10.Onofri F, Giovedi S, Vaccaro P, Czernik A J, Valtorta F, Greengard P, Benfenati F. Proc Natl Acad Sci USA. 1997;94:12168–12173. doi: 10.1073/pnas.94.22.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maycox P R, Link E, Reetz A, Morris S A, Jahn R. J Cell Biol. 1992;118:1379–1388. doi: 10.1083/jcb.118.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins S M, Quintrell N A, Bishop J M. Mol Cell Biol. 1995;15:3507–3515. doi: 10.1128/mcb.15.7.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahler M, Benfenati F, Valtorta F, Greengard P. BioEssays. 1990;12:259–263. doi: 10.1002/bies.950120603. [DOI] [PubMed] [Google Scholar]
- 14.Kelly R B. Nature (London) 1995;374:116–117. doi: 10.1038/374116a0. [DOI] [PubMed] [Google Scholar]
- 15.David C, McPherson P S, Mundigl O, De Camilli P. Proc Natl Acad Sci USA. 1996;93:331–335. doi: 10.1073/pnas.93.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gout I, Dhand R, Hiles I D, Fry M J, Panayotou G, Das P, Truong O, Totty N F, Hsuan J, Booker G W, et al. Cell. 1993;75:25–36. [PubMed] [Google Scholar]
- 17.McPherson P S, Czernik A J, Chilcote T J, Onofri F, Benfenati F, Greengard P, Schlessinger J, De Camilli P. Proc Natl Acad Sci USA. 1994;91:6486–6490. doi: 10.1073/pnas.91.14.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McPherson P S, Takei K, Schmid S L, De Camilli P. J Biol Chem. 1994;269:30132–30139. [PubMed] [Google Scholar]
- 19.Seedorf K, Kostka G, Lammers R, Bashkin P, Daly R, Burgess W H, van der Bliek A M, Schlessinger J, Ullrich A. J Biol Chem. 1994;269:16009–16014. [PubMed] [Google Scholar]
- 20.Wang L H, Sudhof T C, Anderson R G. J Biol Chem. 1995;270:10079–10083. doi: 10.1074/jbc.270.17.10079. [DOI] [PubMed] [Google Scholar]
- 21.Ely C M, Tomiak W M, Allen C M, Thomas L, Thomas G, Parsons S J. J Neurochem. 1994;62:923–933. doi: 10.1046/j.1471-4159.1994.62030923.x. [DOI] [PubMed] [Google Scholar]
- 22.Stenberg P E, Pestina T I, Barrie R J, Jackson C W. Blood. 1997;89:2384–2393. [PubMed] [Google Scholar]
- 23.Bauerfeind R, Galli T, De Camilli P. J Neurocytol. 1996;25:701–715. doi: 10.1007/BF02284836. [DOI] [PubMed] [Google Scholar]
- 24.Lamaze C, Schmid S L. Curr Opin Cell Biol. 1995;7:573–580. doi: 10.1016/0955-0674(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 25.Urrutia R, Henley J R, Cook T, McNiven M A. Proc Natl Acad Sci USA. 1997;94:377–384. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shpetner H S, Herskovits J S, Vallee R B. J Biol Chem. 1996;271:13–16. doi: 10.1074/jbc.271.1.13. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto P M, Herskovits J S, Vallee R B. J Biol Chem. 1997;272:11629–11635. doi: 10.1074/jbc.272.17.11629. [DOI] [PubMed] [Google Scholar]
- 28.Shupliakov O, Low P, Grabs D, Gad H, Chen H, David C, Takei K, De Camilli P, Brodin L. Science. 1997;276:259–263. doi: 10.1126/science.276.5310.259. [DOI] [PubMed] [Google Scholar]
- 29.Chang M P, Mallet W G, Mostov K E, Brodsky F M. EMBO J. 1993;12:2169–2180. doi: 10.1002/j.1460-2075.1993.tb05865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez-Gaitan M, Jackle H. Cell. 1997;88:767–776. doi: 10.1016/s0092-8674(00)81923-6. [DOI] [PubMed] [Google Scholar]
- 31.Valtorta F, Greengard P, Fesce R, Chieregatti E, Benfenati F. J Biol Chem. 1992;267:11281–8. [PubMed] [Google Scholar]
- 32.Benfenati F, Valtorta F, Chieregatti E, Greengard P. Neuron. 1992;8:377–386. doi: 10.1016/0896-6273(92)90303-u. [DOI] [PubMed] [Google Scholar]
- 33.Ceccaldi P E, Grohovaz F, Benfenati F, Chieregatti E, Greengard P, Valtorta F. J Cell Biol. 1995;128:905–912. doi: 10.1083/jcb.128.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyamoto S. Biochim Biophys Acta. 1995;1244:85–91. doi: 10.1016/0304-4165(94)00199-8. [DOI] [PubMed] [Google Scholar]
- 35.Greengard P, Benfenati F, Valtorta F. Adv Second Messenger Phosphoprotein Res. 1994;29:31–45. doi: 10.1016/s1040-7952(06)80005-4. [DOI] [PubMed] [Google Scholar]
- 36.Pieribone V A, Shupliakov O, Brodin L, Hilfiker-Rothenfluh S, Czernik A J, Greengard P. Nature (London) 1995;375:493–497. doi: 10.1038/375493a0. [DOI] [PubMed] [Google Scholar]
- 37.Rosahl T W, Spillane D, Missler M, Herz J, Selig D K, Wolff J R, Hammer R E, Malenka R C, Sudhof T C. Nature (London) 1995;375:488–493. doi: 10.1038/375488a0. [DOI] [PubMed] [Google Scholar]
- 38.Eiseman E, Bolen J B. Nature (London) 1992;355:78–80. doi: 10.1038/355078a0. [DOI] [PubMed] [Google Scholar]
- 39.Ren R, Mayer B J, Cicchetti P, Baltimore D. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]