Abstract
The centromere in eukaryotes is defined by the presence of a special histone H3 variant, CENH3. Centromeric chromatin consists of blocks of CENH3-containing nucleosomes interspersed with blocks of canonical H3-containing nucleosomes. However, it is not known how CENH3 is precisely deposited in the centromeres. It has been suggested that epigenetic modifications of the centromeric chromatin may play a role in centromere identity. The centromeres of Arabidopsis thaliana are composed of megabase-sized arrays of a 178-bp satellite repeat. Here, we report that the 178-bp repeats associated with the CENH3-containing chromatin (CEN chromatin) are hypomethylated compared with the same repeats located in the flanking pericentromeric regions. A similar hypomethylation of DNA in CEN chromatin was also revealed in maize (Zea mays). Hypomethylation of the DNA in CEN chromatin is correlated with a significantly reduced level of H3K9me2 in Arabidopsis. We demonstrate that the 178-bp repeats from CEN chromatin display a distinct distribution pattern of the CG and CNG sites, which may provide a foundation for the differential methylation of these repeats. Our results suggest that DNA methylation plays an important role in epigenetic demarcation of the CEN chromatin.
INTRODUCTION
The centromere is the chromosomal domain that directs the formation of the kinetochore, a proteinaceous structure that interacts with the spindle microtubules to ensure proper chromosomal segregation. Centromeric chromatin is unique due to the presence of a centromere-specific histone variant, CENH3 (CENP-A in mammals and centromere identifier [CID] in Drosophila melanogaster), in place of the canonical H3. Centromeres in multicellular eukaryotes are composed of alternating blocks of nucleosomes that contain either H3 or CENH3 (Blower et al., 2002; Sullivan and Karpen, 2004; Chueh et al., 2005; Alonso et al., 2007). CENP-A nucleosomes directly recruit a CENP-A nucleosome-associated complex that in turn recruits a number of centromeric proteins (Foltz et al., 2006; Okada et al., 2006). Despite the extensive recent research on CENH3 function and CENH3 nucleosome assembly, it remains unclear how CENH3 is precisely deposited in the centromeres, which has become the most intriguing question in centromere biology.
It has been well demonstrated in several model eukaryotes that the specification and propagation of the centromeres are not defined by the underlying DNA sequences but rather are determined by epigenetic mechanisms (Cleveland et al., 2003; Amor et al., 2004). The CENP-A/CID-associated centromeric chromatin (CEN chromatin) in human and D. melanogaster displays histone modification patterns that are distinct from those of both euchromatin and heterochromatin (Sullivan and Karpen, 2004). Specifically, CEN chromatin is enriched with H3 dimethylated at K4 (H3K4me2), a euchromatic mark associated with permissive transcription. Interestingly, the CEN chromatin is not associated with heterochromatic marks H3K9me2 and H3K9me3, nor with H3K4me3, a mark associated with active transcription. By contrast, the pericentromeric chromatin that flanks the CEN chromatin is enriched with H3K9me2 (Sullivan and Karpen, 2004). A similar association of CEN chromatin with H3K4me but not H3K9me was also reported in Schizosaccharomyces pombe (Cam et al., 2005).
DNA methylation is another potential epigenetic mark for centromere identity. However, methylation of centromeric DNA can hardly be assessed since the centromeres of most multicellular eukaryotes contain only highly repetitive DNA sequences (Henikoff et al., 2001; Jiang et al., 2003). Wong et al. (2006) recently analyzed the methylation status of 2041 CpG dinucleotides distributed across a 6.76-Mb chromosomal region that spans a human neocentromere. An overall hypermethylation was found to be associated with these CpG dinucleotides after neocentromere formation (Wong et al., 2006). However, it is unclear if the DNA sequences in the CENP-A binding domain are more or less methylated compared with the DNA sequences in the perineocentromeric region. In addition, human neocentromeres do not contain the 171-bp α satellite repeat that is the dominant DNA component of native human centromeres. It is not known if the overall hypermethylation in human neocentromeres is also associated with native human centromeres.
The centromeres of the model plant Arabidopsis thaliana contain megabase-sized arrays of a 178-bp satellite repeat (Maluszynsak and Heslop-Harrison, 1991; Murata et al., 1994; Round et al., 1997; Heslop-Harrison et al., 1999). Here, we report that the 178-bp repeats associated with CEN chromatin are surprisingly hypomethylated compared with the same repeats located in the flanking pericentromeric heterochromatin. We confirmed a similar hypomethylation of DNA sequences in CEN chromatin in maize (Zea mays). The hypomethylation of DNA in CEN chromatin in Arabidopsis is correlated with a significantly reduced level of H3K9me2. We demonstrate that the differential DNA methylation of the 178-bp repeats is possibly associated with a distinct distribution pattern of CG and CNG sites within these repeats. We propose that DNA methylation plays an important role in demarcating the CEN chromatin from its flanking pericentromeric heterochromatin.
RESULTS
DNA Sequences in the CEN Chromatin and Its Flanking Pericentromeric Heterochromatin Are Differentially Methylated in Arabidopsis
The centromeric regions of all five Arabidopsis chromosomes are composed of cytologically distinct heterochromatin that stains brightly with 4′,6-diamidino-2-phenylindole (DAPI) (Fransz et al., 1998, 2003). The centromeric heterochromatin forms the chromocenters in interphase nuclei and is highly enriched with H3K9me2 (Soppe et al., 2002; Jasencakova et al., 2003; Probst et al., 2003; Fransz et al., 2006). The DNA sequences in the centromeric regions are extensively methylated based on both cytological data (Soppe et al., 2002; Jasencakova et al., 2003; Probst et al., 2003) and DNA chip-based genomics data (Zhang et al., 2006; Zilberman et al., 2007).
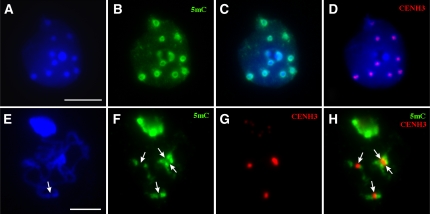
We conducted immunofluorescence assays using an antibody against 5-methylcytosine (5mC) and observed bright and uniform signals associated with the chromocenters in most cells prepared from young anthers. However, we frequently observed nonuniform and hollow-heart signals in some large nuclei that may be from endoreduplicated cells (Figures 1A to 1C). We suspected that the hollow centers of such signals are occupied by the CENH3-associated centromeric chromatin. This prediction was confirmed by immunofluorescence assays using an anti-CENH3 antibody (Figure 1D).
Figure 1.
Cytological Mapping of 5mC in the Interphase Nucleus and Meiotic Pachytene Chromosomes in Arabidopsis.
(A) DAPI staining of an interphase nucleus. Bar = 10 μm.
(B) Detection of 5mC (green).
(C) Merge of (A) and (B).
(D) Detection of CENH3 (red) in the same nucleus. The hollow centers of the 5mC signals in (B) are occupied by the CENH3 signals.
(E) DAPI staining of chromosomes from a meiotic cell at early pachytene stage. The arrow points to a centromeric region that shows reduced DAPI staining than its flanking regions. Bar = 5 μm.
(F) Detection of 5mC (green). Arrows point to the significantly less stained domains that divide each of the bright centromeric signals into two sections.
(G) Detection of CENH3 (red).
(H) Merge of (F) and (G). Note that the less-stained domains in (F) are occupied by the CENH3 signals (arrows).
We then conducted immunofluorescence assays on highly extended early pachytene chromosomes. The bright signals associated with each centromere were clearly separated by a domain that is significantly less stained by the anti-5mC antibody (Figure 1F). These less-stained domains were occupied by the CENH3 signals (Figures 1G and 1H). These results confirmed that the DNA sequences in the CENH3-associated regions are differentially methylated from the flanking pericentromeric regions.
The 178-bp Satellite Repeats in the Centromeric and Pericentromeric Regions Are Differentially Methylated
The centromeres of all five Arabidopsis chromosomes contain megabase-sized arrays of the 178-bp centromere-specific satellite repeat (Kumekawa et al., 2000, 2001; Hosouchi et al., 2002). Chromatin immunoprecipitation (ChIP) analysis showed that this 178-bp repeat is the main DNA component of the CEN chromatin (Nagaki et al., 2003). Cytological mapping of CENH3 on extended chromatin fibers suggested that only the middle portion of the megabase-sized repeat array in each centromere appeared to be associated with CENH3 (Shibata and Murata, 2004).
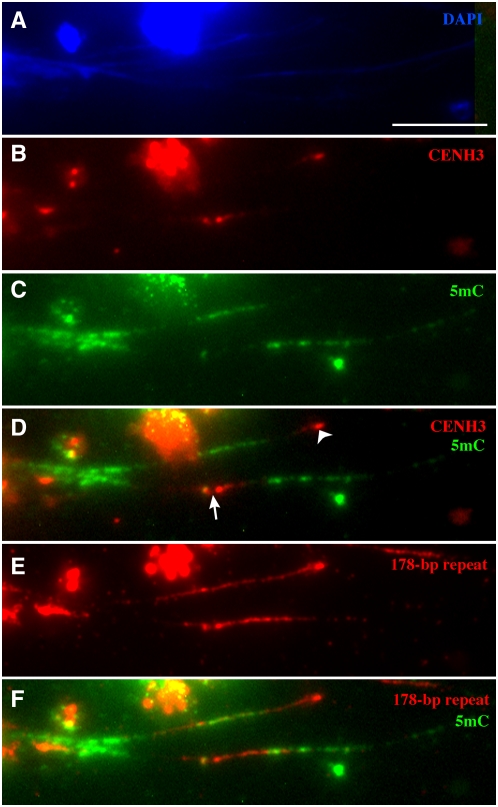
To inspect whether the 178-bp repeat arrays are uniformly methylated, we performed immunofluorescence assays on extended chromatin fibers (Figure 2). The CENH3 binding domains and the regions immediately adjacent showed significantly reduced signals of 5mC (Figure 2D). These results corroborated the immunofluorescence results derived from both interphase nuclei and pachytene chromosomes. Fluorescence in situ hybridization (FISH) mapping on the same chromatin fibers showed that the 178-bp repeats corresponding to the CENH3 binding domains and their immediate flanking regions are either unmethylated or significantly less methylated, while the rest of the 178-bp repeat arrays are highly methylated (Figure 2F). The highly methylated regions extended beyond the 178-bp repeat arrays on the chromatin fibers.
Figure 2.
Mapping of CENH3, 5mC, and the 178-bp Repeats on Extended Chromatin Fibers in Arabidopsis.
(A) DAPI staining of extended chromatin fibers. Bar = 10 μm.
(B) Detection of CENH3 (red) on the extended chromatin fibers.
(C) Detection of 5mC (green) on the extended chromatin fibers.
(D) Merge of (B) and (C). The CENH3 signal (arrowhead) on one fiber is flanked by 5mC signals on one side only. The second CENH3 signal (arrow) is flanked by 5mC signals on both sides.
(E) FISH mapping of the 178-bp repeat on the same chromatin fibers.
(F) Digital merge of (C) and (E). Note that only some of the 178-bp repeat sections are associated with the 5mC signals.
On pachytene chromosomes, bright 5mC signals covered the entire DAPI-bright heterochromatin domains. The intensity of the 5mC signals was gradually reduced beyond the heterochromatin-euchromatin boundaries (see Supplemental Figure 1 online). This 5mC distribution pattern based on immunoassaying on chromosomes is well correlated with data from DNA chip-based genome-wide DNA methylation mapping (Zhang et al., 2006; Zilberman et al., 2007), except that the DNA chip-based data did not reveal the hypomethylation of the repeats associated with CEN chromatin.
Hypomethylation of DNA Associated with CEN Chromatin in Maize
On some Arabidopsis early pachytene chromosomes, the middle portion of the centromeric heterochromatin showed a reduced intensity of DAPI staining compared with the flanking regions (Figure 1E). This region is correlated with the hypomethylation domain (Figure 1F). Thus, we wonder if the relatively faint 5mC signals in these domains are caused by less condensation of the chromatin and, thus, less DNA associated with these domains. To address this possibility, we conducted a similar immunofluorescence assay on the pachytene chromosomes of maize.
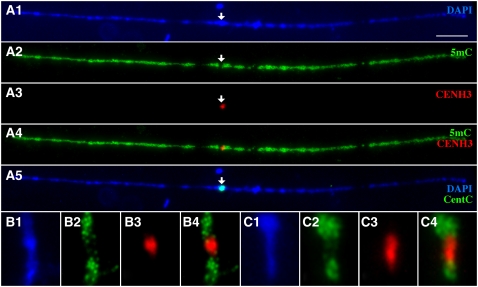
The heterochromatin in Arabidopsis is almost exclusively located in the centromeric regions (Fransz et al., 1998). By contrast, the heterochromatin in maize is distributed throughout the genome. DAPI staining of maize pachytene chromosomes revealed only subtle differences between the distal and the proximal halves of the chromosomes (Shi and Dawe, 2006; Wang et al., 2006). To increase the mapping resolution, we performed immunofluorescence assays on mechanically stretched pachytene chromosomes. Signals derived from the anti-5mC antibody were almost uniformly distributed along the entire length of the pachytene chromosomes (Figure 3). However, on the pachytene chromosomes where the centromeric regions were well stretched, significantly reduced 5mC signal was observed in the domains occupied by maize CENH3. The DAPI staining intensity from the same domains was not reduced compared with the flanking domains (Figure 3). These results suggest that the reduced 5mC signals were not caused by the reduced amount of DNA associated with these domains. Thus, the DNA sequences in the CEN chromatin in maize are also hypomethylated compared with the DNA sequences associated with the flanking pericentromeric chromatin.
Figure 3.
Cytological Mapping of 5mC and the Centromeric Satellite Repeat CentC (Ananiev et al., 1998) in the Centromeric and Pericentromeric Regions on Stretched Pachytene Chromosomes of Maize.
(A1) DAPI staining of a mechanically stretched pachytene chromosome. Bar = 10 μm.
(A2) Detection of 5mC (green).
(A3) Detection of CENH3 (red).
(A4) Merge of (A2) and (A3). The arrows in (A1) to (A3) point to the same position on the stretched pachytene chromosome.
(A5) FISH mapping of the centromeric satellite repeat CentC (green) on the same pachytene chromosome after immunofluorescence assay. The arrows in (A1) to (A3) and (A5) point to the same position on the pachytene chromosome.
(B1) and (C1) DAPI staining of the centromeric segments from two different stretched maize pachytene chromosomes.
(B2) and (C2) Detection of 5mC (green).
(B3) and (C3) Detection of CENH3 (red).
(B4) and (C4) Merges of 5mC and CENH3 signals from (B2) and (B3) and from (C2) and (C3).
Differential H3K9 Methylation Associated with CEN Chromatin and the Flanking Pericentromeric Heterochromatin in Arabidopsis
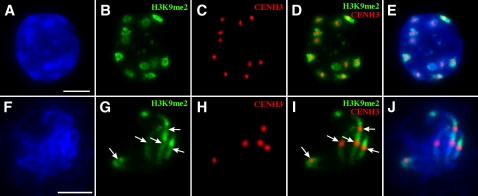
The chromocenters on the interphase nuclei of Arabidopsis were previously demonstrated to be highly enriched with H3K9me2 (Soppe et al., 2002; Jasencakova et al., 2003; Probst et al., 2003; Lindroth et al., 2004). We found that the immunofluorescence signal pattern derived from an anti-H3K9me2 antibody was similar to that from the anti-5mC antibody (Figures 4B and 4G). The H3K9me2 signals associated with the chromocenters were clearly not uniform in some nuclei and often appeared in a hollow-heart pattern (Figure 4B). The hollow centers of such signals overlapped with the locations of the CENH3 signals (Figures 4C to 4E). Immunofluorescence assays on early pachytene chromosomes revealed that the CENH3 binding domains displayed significantly reduced H3K9me2 signals compared with the flanking domains (Figures 4G to 4I). These results showed that the locations of enriched/reduced signals from H3K9me2 and those from 5mC were correlated with each other.
Figure 4.
Mapping of H3K9me2 Associated with Centromeric and Pericentromeric Chromatin in Arabidopsis.
(A) DAPI staining of an interphase nucleus. Bar = 10 μm.
(B) Detection of H3K9me2 (green).
(C) Detection of CENH3 (red).
(D) Merge of (B) and (C). The hollow centers of the signals in (B) are occupied by the CENH3 signals.
(E) Merge of (A) to (C).
(F) DAPI staining of chromosomes from a meiotic cell at the early pachytene stage. Bar = 5 μm.
(G) Detection of H3K9me2 (green). Arrows point to the domains with reduced immunofluorescence signals.
(H) Detection of CENH3 (red).
(I) Merge of (G) and (H). The domains with reduced signals in (G) are corresponding to the regions with CENH3 signals.
(J) Merge of (F) to (H).
Distribution of CG and CNG Sites within the 178-bp Repeats in Arabidopsis
Immunofluorescence assays showed that different sections of the 178-bp repeat arrays are differentially methylated. We next investigated whether the 178-bp repeats located within and outside of the CENH3 binding domains are associated with different sequence motifs that would allow differential methylation. We first isolated the 178-bp repeats from the CENH3-associated nucleosomes using a ChIP-based cloning method (Lee et al., 2005). We extracted 108 complete monomers from a total of 96 sequenced plasmid clones derived from ChIP cloning. Pairwise alignment of the sequences by the Clustal method revealed 70.5 to 100% sequence similarity among the monomers.
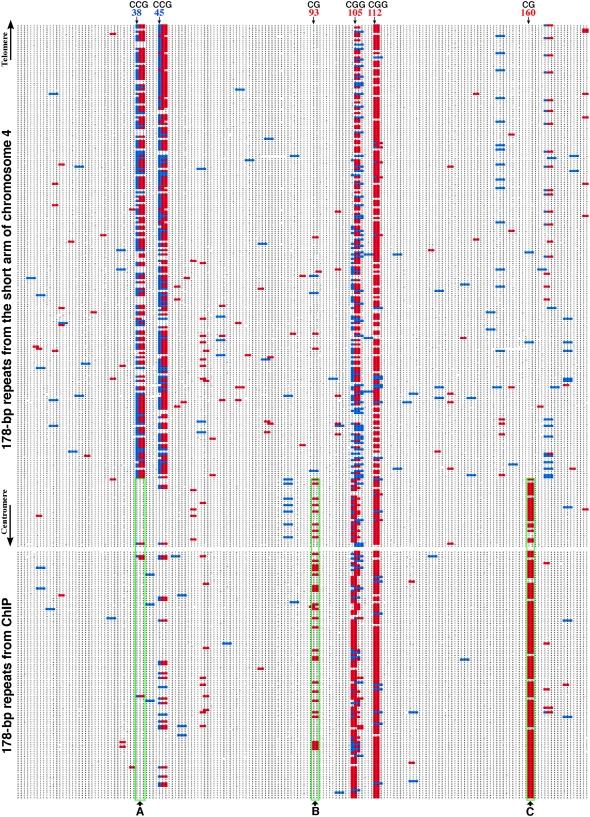
In plants, cytosine can be methylated in the CG, CNG (where N is any nucleotide), and CHH (where H is A, C, or T) sequence contexts (Gruenbaum et al., 1981; Chan et al., 2005). We mapped all the CG and CNG sites within the 108 monomers (Figure 5). We observed two CG sites at 93 to 94th bp and 160 to 161th bp, respectively. These two CG sites are present in 23.2 and 92.6% of the 108 monomers. One CCG (45 to 47th bp) and two CGG sites (105 to 108th bp; 112 to 114th bp) were observed and their frequencies were 18.5, 75, and 93.5%, respectively (Figure 5).
Figure 5.
Distribution of CG and CNG Sites within the 178-bp Repeats of Arabidopsis.
Top panel: CG and CNG sites located in the 178-bp repeats from the short arm of Arabidopsis chromosome 4. Only the proximal (centromeric side) 244 monomers are included in the figure; the distal (telomeric side) 194 monomers are not included.
Bottom panel: CG and CNG sites located in the 108 monomers derived from ChIP. These repeats are derived from different centromeres and are randomly arranged in the alignment.
All CG sites are marked in red. CAG and CTG sites are marked in blue. The CG from each CGG is marked in red and the second G is not marked. For CCG sites, the first C is marked in blue and the CG is marked in red. The green square A covers a CCG site (38 to 40 bp) that is missing from the ChIPed repeats. The green squares B and C cover two CG sites (93 to 94 bp; 160 to 161 bp) that are highly enriched in the ChIPed repeats.
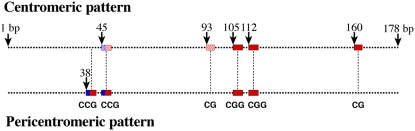
We then analyzed the CG and CNG sites within all available 178-bp repeats from the Arabidopsis sequence map (Arabidopsis Genome Initiative, 2000). The current sequence map contains only portions of the 178-bp repeat array in each of the five centromeres, ranging from 28 monomers from the short arm of chromosome 1 to 787 monomers (∼140 kb) from the long arm of chromosome 5. Mapping of the CG and CNG sites within these available sequences revealed two strikingly distinct distribution patterns. The first pattern, named centromeric pattern, is similar to the pattern of the 178-bp repeats isolated by ChIP from the CENH3 binding domain (Figure 6). The second pattern, named pericentromeric pattern, lacks the two CG sites in the first pattern and contains an additional CCG site at the 38 to 40th bp (Figures 5 and 6). Furthermore, the CCG site at the 45 to 47th bp is more frequently present in the pericentromeric pattern than in the centromeric pattern (Figure 5). Several other CG and CNG sites were observed in the pericentromeric pattern (see Supplemental Figure 2 online). However, these sites are not universal and were only detected in the 178-bp repeats from a specific chromosomal arm. Interestingly, we observed a drastic transition between these two patterns in the 178-bp repeats located in several chromosomal arms, including the short arm of chromosome 4 (Figure 5), the long arm of chromosome 2, the long arm of chromosome 3, and both the short and long arms of chromosome 5 (see Supplemental Figure 2 online). We also mapped all CHH sites in the 178-bp repeats but were not able to find any distinct distribution patterns associated with the centromeric or the pericentromeric repeats.
Figure 6.
Diagrammatic Illustration of the Two Distinct CG and CNG Distribution Patterns in the 178-bp Centromeric Satellite Repeats from Arabidopsis.
The two CG sites, 93 to 94th bp and 160 to 161th, are unique to the centromeric pattern. The CCG site at 38 to 40th bp is missing in the centromeric pattern. The shaded squares indicated two less frequently occurring CCG (at 45 to 47th bp) and CG (93 to 94th bp) sites.
DISCUSSION
Centromeric satellite DNAs, including the 178-bp repeats in Arabidopsis, were often reported to be heavily methylated (Martinez-Zapater et al., 1986; Miniou et al., 1997; Dong et al., 1998; Okano et al., 1999). However, gel blot-based or DNA chip-based genome-wide DNA methylation analyses do not reveal whether a centromeric satellite repeat is uniformly methylated across an entire centromere or an entire genome. A megabase-sized centromeric satellite repeat array could potentially span the entire CENH3 binding domain and its flanking pericentromeric regions. Thus, the previous reports on methylation of centromeric satellite repeats could not reveal whether the repeats associated with CEN chromatin are methylated. Using immunofluorescence assays on early pachytene chromosomes and extended chromatin fibers, we demonstrate that DNA sequences associated with CEN chromatin in two different plant species are hypomethylated compared with the DNA sequences located in the adjacent heterochromatin.
Wong et al. (2006) investigated the DNA methylation associated with a neocentromere formed at q25 of human chromosome 10 using sodium bisulfite PCR and sequencing. The authors analyzed the methylation status of 129 GC-rich sequences, which included 2041 CpG dinucleotides, within a 6.76-Mb DNA region spanning the neocentromere and compared them to the methylation pattern of the same DNA sequences in a normal chromosome 10. The majority of these GC-rich sequences showed an increase in methylation after neocentromere formation (Wong et al., 2006). Some interspersed repeats, including long interspersed nucleotide elements, short interspersed nucleotide elements, and mammalian interspersed repeats, as well as a selected number of CpG-orphan sites (CpG dinucleotides in GC-poor regions) also showed an overall increase in their methylation status after neocentromere formation. However, the data did not reveal whether the DNA in the CENP-A region of this neocentromere, which spans ∼330 kb (Lo et al., 2001), is equally methylated compared with the DNA in the flanking regions. In addition, the majority of the CpG dinucleotides studied were associated with single or low copy sequences and with GC-rich regions, whereas the methylation status is unknown for the majority of the repetitive DNA sequences, which are often AT rich. Thus, it is still possible that the DNA sequences in the CENP-A binding region of this neocentromere are less methylated than the DNA in the flanking regions. The potential hypomethylation of human centromeric DNA is indicated by a recent report that methylation of the two CpG dinucleotides within the 17-bp CENP-B box of the human α satellite repeat reduced the binding of human centromere protein B (CENP-B) (Tanaka et al., 2005).
Mapping of CG and CNG sites within the 178-bp centromeric repeats revealed two distinct distribution patterns (Figures 5 and 6). The 108 monomer repeats derived from ChIP are likely to be randomly derived from all five centromeres, yet these monomers all share a strikingly similar CG/CNG distribution pattern (Figure 5). We also analyzed the distribution of these distinct CG and CNG sites in the previously reported 178-bp repeats isolated from the Columbia ecotype of Arabidopsis by PCR (Hall et al., 2003). We expect that the PCR-amplified 178-bp repeats would be randomly derived from both centromeric and pericentromeric regions. We aligned the monomeric repeats extracted from 13 clones (AF494926 to AF494941). We found that six of the 13 monomers show the centromeric CG/CNG distribution pattern, and the other seven monomers show the pericentromeric pattern (see Supplemental Figure 3 online).
In Arabidopsis, cytosine methylation at CGG sites is the same as CG sites and is dependent on the DNA methyltransferase MET1. For each CCG site, the first C is a CNG and is dependent on DNA methyltransferases CMT3/DRM2; the second C is a CG and is dependent on MET1 (Chan et al., 2005; Simon Chan, personal communication). Although the centromeric CG/CNG distribution pattern includes two extra CG sites (Figure 6), the pericentromeric pattern contains more cytosine residues that could potentially be methylated due to the presence of an extra CCG site (38 to 40th bp) and to the relatively low frequency of the CGG (45 to 47th bp) and CG (93 to 94th bp) sites in the centromeric pattern (Figure 5).
Another possibility is that the two CG sites associated with the centromeric pattern may not be methylated. Most noteworthy is the periodicity of the two pairs of CG/CCG sites within the pericentromeric pattern (Figure 5). Each pair is separated by four to five base pairs. A recent study showed that the mammalian DNA methyltransferase Dnmt3a preferentially methylases pairs of CGs that are eight to 10 base pairs apart (Jia et al., 2007). The two CG sites in the centromeric pattern may not be methylated if plant DNA methyltransferases employ a similar CG periodicity-based sequence specificity. Therefore, the number of cytosine residues associated with CG/CNG and/or the different distribution patterns of these cytosine residues can provide a foundation for differential methylation of the 178-bp repeats located in the centromeric and pericentromeric regions. Woo et al. (2007) recently showed that VIM1, a methylcytosine binding protein, can specifically impact the methylation of centromeric DNA in Arabidopsis. Thus, differential methylation of centromeric and pericentromeric DNA sequences could also be controlled by an unknown mechanism.
We observed a drastic transition between the centromeric and the pericentromeric distribution pattern in the middle of the 178-bp repeat arrays from several chromosomal arms (Figure 5; see Supplemental Figure 2 online). The repeats in the section showing the centromeric pattern are all located toward the centromeric direction in these chromosomal arms (see Supplemental Figure 2 online). Only the 178-bp repeat array from the long arm of chromosome 4 showed a slightly modified pattern, which contains only one of the two CG sites, one of the two CCG sites, and one of the two CGG sites (see Supplemental Figure 2 online). Since this pattern includes fewer cytosine residues compared with both the centromeric and the pericentromeric pattern, we suspect that this entire array is associated with CEN chromatin. The binding of CENP-A/CENH3 toward one end of a long centromeric satellite array was observed in the centromere of human X chromosome (Spence et al., 2002) as well as in maize centromeres (Jin et al., 2004).
The CEN chromatin is flanked by heterochromatin in humans, Drosophila, and S. pombe (Sullivan and Karpen, 2004; Cam et al., 2005). We demonstrated that the CEN chromatin in Arabidopsis is also flanked by heterochromatin enriched with H3K9me2. The flanking heterochromatin may provide a boundary to prevent the spread of CENH3 deposition. In contrast with the flanking heterochromatin, CEN chromatin often shows a euchromatic appearance cytologically (Dawe 2003), which is also indicated by the presence of H3K4me and absence of H3K9me in animal centromeres (Sullivan and Karpen, 2004; Cam et al., 2005). In S. pombe, Swi6 (a homolog of heterochromatin protein 1) is enriched in pericentromeric heterochromatin but not detected in CEN chromatin (central core regions) (Cam et al., 2005). In addition, marker genes inserted in the pericentromeric heterochromatin are transcriptionally silenced. However, insertion of the same marker genes in the CEN chromatin showed a much weaker silencing effect (Allshire et al., 1994, 1995). In Arabidopsis, the CEN chromatin, although composed almost exclusively of 178-bp satellite repeats, shows a drastically reduced amount of H3K9me2. It appears that CENH3 deposition cannot spread into heterochromatin. Interestingly, overexpressed CID in Drosophila localized only to euchromatic regions and was excluded from heterochromatin (Ahmad and Henikoff, 2002; Heun et al., 2006).
In S. pombe, the tRNA genes act as boundary elements to prevent the spreading of pericentromeric heterochromatin into the CEN domain (Noma et al., 2006; Scott et al., 2006). Such boundary elements may not be present in most multicellular eukaryotes because both the CEN chromatin and the pericentromeric heterochromatin often contain the same satellite repeat families. In rice (Oryza sativa), the DNA sequences associated with CEN chromatin span beyond the centromeric satellite repeat arrays, and the boundaries between the CEN and the pericentromeric domains do not contain specific repeats or gene families (Nagaki et al., 2004; Yan et al., 2006; Yan and Jiang, 2007). In the absence of such boundary elements, DNA methylation can serve as an epigenetic mark to demarcate the CEN chromatin and its flanking domains. It has been well demonstrated in Arabidopsis that DNA methylation controls H3K9 methylation because removal of CG methylation results in a loss of H3K9me2 in pericentromeric heterochromatin (Soppe et al., 2002; Tariq et al., 2003). Thus, H3K9me2 or other histone modifications associated with DNA methylation may further enforce the role of DNA methylation as the boundary mark to isolate the CEN chromatin domain.
METHODS
Cytological Preparations
The Columbia ecotype of Arabidopsis thaliana and maize (Zea mays) inbred line B73 were used in all experiments. Arabidopsis seeds for in vitro culture were sterilized for 10 min in 10% sodium-hypochlorite containing 0.1% Tween 20, followed by three washes with sterile deionized water. The seeds were dried and plated on half-strength Murashige and Skoog medium for germination under normal culture conditions, and the seedlings were then transferred to soil. Maize plants were maintained in greenhouses under typical greenhouse conditions. Maize anthers from young panicles were collected and fixed in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, and 0.3 mM sorbitol, pH 6.8) containing 2% paraformaldehyde. The fixed anthers were rinsed in 1× PBS for ∼20 min before chromosome preparation. Young Arabidopsis flower buds were fixed in 4% paraformaldehyde in 1× PBS buffer, pH 7.0, for immunofluorescence assays. The flower buds were thoroughly washed in 1× PBS buffer and digested with 4% cellulase (Yakult Honsha Co.) and 1% pectinase (Sigma-Aldrich) for 20 min at 37°C. Extended chromatin fibers were prepared according to the published protocol (Jin et al., 2004). Mechanically stretched maize pachytene chromosomes were prepared by gently and slightly dislocating the cover slips from the glass slides in squashes. The prepared slides were store at −80°C until use.
Immunofluorescence Assay and FISH
After removing the cover glasses, the slides were incubated with the primary antisera overnight at 4°C in a wet chamber. After washes in 1× PBS, the slides were incubated with the appropriate secondary antibodies at 37°C for 30 min. Mitotic interphase nuclei and pachytene chromosomes were counterstained with DAPI (2.0 g/mL). Antibodies included anti-CENH3 antibodies for Arabidopsis (Talbert et al., 2002) and maize (Zhong et al., 2002) and a mouse monoclonal antibody against H3K9me2 (Abcam ab1220).
To detect 5mC, the slides were denatured in 70% formamide in 2× SSC, at 80°C for 3 min, washed in ice-cold 70% ethanol for 5 min, and then incubated in 1% BSA in 1× PBS for 30 min at 37°C and subsequently incubated with mouse antiserum raised against 5mC (1:250) (Aviva Systems Biology) in 1× TNB (100 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.5% blocking reagent). The mouse antibodies were detected using fluorescein isothiocyanate–conjugated goat anti-mouse antibody (1:1000) (Jackson ImmunoResearch Labs).
For dual detections of modified histones and 5mC, the slides were post-fixed after histone detection, denatured and incubated with mouse anti-5mC, and detected with goat anti-mouse IgG conjugated with fluorescein isothiocyanate. To combine the immunofluorescence assay with FISH, the slides were first processed for detection of histones and/or 5mC and then were dehydrated in an ethanol series (5 min in 70%, 5 min in 90% and 5min in 100%) and air-dried. The FISH procedure followed published protocols (Jiang et al., 1995).
ChIP Cloning and Sequence Analyses
The ChIP cloning procedure using an Arabidopsis anti-CENH3 antibody followed published protocols (Lee et al., 2005). Immunoprecipitated DNA fragments were cloned into the pCR 2.1-TOPO vector (Invitrogen). Recombinant clones were transferred to 384-well microtiter plates containing 30 μL of Luria-Bertani freezing buffer. A plasmid library consisting of 1920 clones was developed and screened using ChIPed DNA as a probe. A total of 96 positive plasmid clones were randomly selected for sequencing. All clones, except three, contained exclusively 178-bp repeats. DNA sequencing was performed by the DNA Sequencing Facility at the Biotechnology Center at the University of Wisconsin–Madison. The 178-bp repeats from the five Arabidopsis centromeres were extracted from the Arabidopsis Genome Initiative sequence (ftp://ftp.Arabidopsis.org/%2Fhome/tair/home/tair/home/tair/Sequences/whole_chromosomes/) by in silico restriction digestion using MAPDRAW (DNASTAR) and EMBOSS programs (Rice et al., 2000) (http://emboss.sourceforge.net/). The 178-bp repeats were aligned by the Clustal method with a default parameter using the Megalign program (DNASTAR). The CG and CNG sites were searched and marked manually on the aligned sequences.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers EU359480 to EU359566.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mapping of 5-Methylcytosine and the 178-bp Repeats on Pachytene Chromosomes.
Supplemental Figure 2. Distribution of CG and CNG Sites within All Available 178-bp Repeats in the Current Sequence Map of Arabidopsis.
Supplemental Figure 3. Distribution of CG and CNG Sites within the 178-bp Repeats Isolated by PCR from the Columbia Ecotype of Arabidopsis by Hall et al. (2003).
Supplementary Material
Acknowledgments
This research was supported by Grants DBI-0553417 and DBI-0421671 from the National Science Foundation. We are grateful to the valuable comments on our manuscript from an anonymous reviewer.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jiming Jiang (jjiang1@wisc.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ahmad, K., and Henikoff, S. (2002). Histone H3 variants specify modes of chromatin assembly. Proc. Natl. Acad. Sci. USA 99 16477–16484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire, R.C., Javerzat, J.P., Redhead, N.J., and Cranston, G. (1994). Position effect variegation at fission yeast centromeres. Cell 76 157–169. [DOI] [PubMed] [Google Scholar]
- Allshire, R.C., Nimmo, E.R., Ekwall, K., Javerzat, J.P., and Cranston, G. (1995). Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9 218–233. [DOI] [PubMed] [Google Scholar]
- Alonso, A., Fritz, B., Hasson, D., Abrusan, G., Cheung, F., Yoda, K., Radlwimmer, B., Ladurner, A.G., and Warburton, P.E. (2007). Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol. 8 R148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor, D.J., Kalitsis, P., Sumer, H., and Choo, K.H.A. (2004). Building the centromere: From foundation proteins to 3D organization. Trends Cell Biol. 14 359–368. [DOI] [PubMed] [Google Scholar]
- Ananiev, E.V., Phillips, R.L., and Rines, H.W. (1998). Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Blower, M.D., Sullivan, B.A., and Karpen, G.H. (2002). Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam, H.P., Sugiyama, T., Chen, E.S., Chen, X., FitzGerald, P.C., and Grewal, S.I.S. (2005). Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37 809–819. [DOI] [PubMed] [Google Scholar]
- Chan, S.W.L., Henderson, I.R., and Jacobsen, S.E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6 351–360. [DOI] [PubMed] [Google Scholar]
- Chueh, A.C., Wong, L.H., Wong, N., and Choo, K.H.A. (2005). Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere. Hum. Mol. Genet. 14 85–93. [DOI] [PubMed] [Google Scholar]
- Cleveland, D.W., Mao, Y.H., and Sullivan, K.F. (2003). Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell 112 407–421. [DOI] [PubMed] [Google Scholar]
- Dawe, R.K. (2003). RNA interference, transposons, and the centromere. Plant Cell 15 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, F., Miller, J.T., Jackson, S.A., Wang, G.L., Ronald, P.C., and Jiang, J. (1998). Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc. Natl. Acad. Sci. USA 95 8135–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz, D.R., Jansen, L.E.T., Black, B.E., Bailey, A.O., Yates, J.R., and Cleveland, D.W. (2006). The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 8 458–469. [DOI] [PubMed] [Google Scholar]
- Fransz, P., Armstrong, S., Alonso-Blanco, C., Fischer, T.C., Torres-Ruiz, R.A., and Jones, G.H. (1998). Cytogenetics for the model system Arabidopsis thaliana. Plant J. 13 867–876. [DOI] [PubMed] [Google Scholar]
- Fransz, P., Soppe, W., and Schubert, I. (2003). Heterochromatin in interphase nuclei of Arabidopsis thaliana. Chromosome Res. 11 227–240. [DOI] [PubMed] [Google Scholar]
- Fransz, P., ten Hoopen, R., and Tessadori, F. (2006). Composition and formation of heterochromatin in Arabidopsis thaliana. Chromosome Res. 14 71–82. [DOI] [PubMed] [Google Scholar]
- Gruenbaum, Y., Naveh-Many, T., Cedar, H., and Razin, A. (1981). Sequence specificity of methylation in higher plant DNA. Nature 292 860–862. [DOI] [PubMed] [Google Scholar]
- Hall, S.E., Kettler, G., and Preuss, D. (2003). Centromere satellites from Arabidopsis populations: Maintenance of conserved and variable domains. Genome Res. 13 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H.S. (2001). The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293 1098–1102. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison, J.S., Murata, M., Ogura, Y., Schwarzacher, T., and Motoyoshi, F. (1999). Polymorphisms and genomic organization of repetitive DNA from centromeric regions of Arabidopsis chromosomes. Plant Cell 11 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun, P., Erhardt, S., Blower, M.D., Weiss, S., Skora, A.D., and Karpen, G.H. (2006). Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell 10 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosouchi, T., Kumekawa, N., Tsuruoka, H., and Kotani, H. (2002). Physical map-based sizes of the centromeric regions of Arabidopsis thaliana chromosomes 1, 2, and 3. DNA Res. 9 117–121. [DOI] [PubMed] [Google Scholar]
- Jasencakova, Z., Soppe, W.J.J., Meister, A., Gernand, D., Turner, B.M., and Schubert, I. (2003). Histone modifications in Arabidopsis - High methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J. 33 471–480. [DOI] [PubMed] [Google Scholar]
- Jia, D., Jurkowska, R.Z., Zhang, X., Jeltsch, A., and Cheng, X.D. (2007). Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449 248–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J., Birchler, J.A., Parrott, W.A., and Dawe, R.K. (2003). A molecular view of plant centromeres. Trends Plant Sci. 8 570–575. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Gill, B.S., Wang, G.L., Ronald, P.C., and Ward, D.C. (1995). Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 92 4487–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, W.W., Melo, J.R., Nagaki, K., Talbert, P.B., Henikoff, S., Dawe, R.K., and Jiang, J. (2004). Maize centromeres: Organization and functional adaptation in the genetic background of oat. Plant Cell 16 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumekawa, N., Hosouchi, T., Tsuruoka, H., and Kotani, H. (2000). The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 5. DNA Res. 7 315–321. [DOI] [PubMed] [Google Scholar]
- Kumekawa, N., Hosouchi, T., Tsuruoka, H., and Kotani, H. (2001). The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 4. DNA Res. 8 285–290. [DOI] [PubMed] [Google Scholar]
- Lee, H.R., Zhang, W.L., Langdon, T., Jin, W.W., Yan, H.H., Cheng, Z.K., and Jiang, J. (2005). Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc. Natl. Acad. Sci. USA 102 11793–11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth, A.M., et al. (2004). Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 23 4146–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo, A.W., Craig, J.M., Saffery, R., Kalitsis, P., Irvine, D.V., Earle, E., Magliano, D.J., and Choo, K.H. (2001). A 330 kb CENP-A binding domain and altered replication timing at a human neocentromere. EMBO J. 20 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluszynsak, J., and Heslop-Harrison, J.S. (1991). Localization of tandemly repeated DNA sequences in Arabidopsis thaliana. Plant J. 1 159–166. [Google Scholar]
- Martinez-Zapater, J.M., Estelle, M.A., and Somerville, C.R. (1986). A high repeated DNA sequence in Arabidopsis thaliana. Mol. Gen. Genet. 204 417–423. [Google Scholar]
- Miniou, P., Jeanpierre, M., Bourc'his, D., Coutinho Barbosa, A.C., Blanquet, V., and Viegas-Pequignot, E. (1997). alpha-satellite DNA methylation in normal individuals and in ICF patients: Heterogeneous methylation of constitutive heterochromatin in adult and fetal tissues. Hum. Genet. 99 738–745. [DOI] [PubMed] [Google Scholar]
- Murata, M., Ogura, Y., and Motoyoshi, F. (1994). Centromeric repetitive sequences in Arabidopsis thaliana. Jpn. J. Genet. 69 361–370. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Cheng, Z.K., Ouyang, S., Talbert, P.B., Kim, M., Jones, K.M., Henikoff, S., Buell, C.R., and Jiang, J. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36 138–145. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Talbert, P.B., Zhong, C.X., Dawe, R.K., Henikoff, S., and Jiang, J.M. (2003). Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma, K.I., Cam, H.P., Maraia, R.J., and Grewal, S.I.S. (2006). A role for TFIIIC transcription factor complex in genome organization. Cell 125 859–872. [DOI] [PubMed] [Google Scholar]
- Okada, M., Cheeseman, I.M., Hori, T., Okawa, K., McLeod, I.X., Yates, J.R., Desai, A., and Fukagawa, T. (2006). The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 8 446–457. [DOI] [PubMed] [Google Scholar]
- Okano, M., Bell, D.W., Haber, D.A., and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99 247–257. [DOI] [PubMed] [Google Scholar]
- Probst, A.V., Fransz, P.F., Paszkowski, J., and Scheid, O.M. (2003). Two means of transcriptional reactivation within heterochromatin. Plant J. 33 743–749. [DOI] [PubMed] [Google Scholar]
- Rice, P., Longden, I., and Bleasby, A. (2000). EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16 276–277. [DOI] [PubMed] [Google Scholar]
- Round, E.K., Flowers, S.K., and Richards, E.J. (1997). Arabidopsis thaliana centromere regions: Genetic map positions and repetitive DNA structure. Genome Res. 7 1045–1053. [DOI] [PubMed] [Google Scholar]
- Scott, K.C., Merrett, S.L., and Willard, H.F. (2006). A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 16 119–129. [DOI] [PubMed] [Google Scholar]
- Shi, J., and Dawe, R.K. (2006). Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics 173 1571–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, F., and Murata, M. (2004). Differential localization of the centromere-specific proteins in the major centromeric satellite of Arabidopsis thaliana. J. Cell Sci. 117 2963–2970. [DOI] [PubMed] [Google Scholar]
- Soppe, W.J.J., Jasencakova, Z., Houben, A., Kakutani, T., Meister, A., Huang, M.S., Jacobsen, S.E., Schubert, I., and Fransz, P.F. (2002). DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 21 6549–6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence, J.M., Critcher, R., Ebersole, T.A., Valdivia, M.M., Earnshaw, W.C., Fukagawa, T., and Farr, C.J. (2002). Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X alpha satellite array. EMBO J. 21 5269–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, B.A., and Karpen, G.H. (2004). Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Biol. 11 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert, P.B., Masuelli, R., Tyagi, A.P., Comai, L., and Henikoff, S. (2002). Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, Y., Kurumizaka, H., and Yokoyama, S. (2005). CpG methylation of the CENP-B box reduces human CENP-B binding. FEBS J. 272 282–289. [DOI] [PubMed] [Google Scholar]
- Tariq, M., Saze, H., Probst, A.V., Lichota, J., Habu, Y., and Paszkowski, J. (2003). Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. USA 100 8823–8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.-J.R., Harper, L., and Cande, Z.W. (2006). High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell 18 529–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, N.C., Wong, L.H., Quach, J.M., Canham, P., Craig, J.M., Song, J.Z., Clark, S.J., and Choo, K.H.A. (2006). Permissive transcriptional activity at the centromere through pockets of DNA hypomethylation. PLoS Genet. 2 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, H.R., Pontes, O., Pikaard, C.S., and Richards, E.J. (2007). VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev. 21 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H.H., et al. (2006). Genomic and genetic characterization of rice Cen3 reveals extensive transcription and evolutionary implications of a complex centromere. Plant Cell 18 2123–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, H.H., and Jiang, J.M. (2007). Rice as a model for centromere and heterochromatin research. Chromosome Res. 15 77–84. [DOI] [PubMed] [Google Scholar]
- Zhang, X.Y., Yazaki, J., Sundaresan, A., Cokus, S., Chan, S.W.L., Chen, H.M., Henderson, I.R., Shinn, P., Pellegrini, M., Jacobsen, S.E., and Ecker, J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zhong, C.X., Marshall, J.B., Topp, C., Mroczek, R., Kato, A., Nagaki, K., Birchler, J.A., Jiang, J.M., and Dawe, R.K. (2002). Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14 2825–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman, D., Gehring, M., Tran, R.K., Ballinger, T., and Henikoff, S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39 61–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.