Abstract
KNOTTED1-like homeobox (KNOX) genes promote stem cell activity and must be repressed to form determinate lateral organs. Stable KNOX gene silencing during organogenesis is known to involve the predicted DNA binding proteins ASYMMETRIC LEAVES1 (AS1) and AS2 as well as the chromatin-remodeling factor HIRA. However, the mechanism of silencing is unknown. Here, we show that AS1 and AS2 form a repressor complex that binds directly to the regulatory motifs CWGTTD and KMKTTGAHW present at two sites in the promoters of the KNOX genes BREVIPEDICELLUS (BP) and KNAT2. The two binding sites act nonredundantly, and interaction between AS1-AS2 complexes at these sites is required to repress BP. Promoter deletion analysis further indicates that enhancer elements required for BP expression in the leaf are located between the AS1-AS2 complex binding sites. We propose that AS1-AS2 complexes interact to create a loop in the KNOX promoter and, likely through recruitment of HIRA, form a repressive chromatin state that blocks enhancer activity during organogenesis. Our model for AS1-AS2–mediated KNOX gene silencing is conceptually similar to the action of an insulator. This regulatory mechanism may be conserved in simple leafed species of monocot and dicot lineages and constitutes a potential key determinant in the evolution of compound leaves.
INTRODUCTION
The reiterative process of organogenesis characteristic of plants depends on the activity of a population of self-renewing, pluripotent stem cells present in meristems at the growing tips. Meristem activity in the shoot apex is specified in part by the class I KNOTTED1-like homeobox (KNOX) genes (Long et al., 1996; Vollbrecht et al., 2000; Scofield and Murray, 2006). Lateral organs, such as leaves, initiate on the flank of the shoot apical meristem (SAM), and downregulation of KNOX gene expression is essential to facilitate this process (Jackson et al., 1994; Long et al., 1996). Moreover, acquisition of determinacy in developing organs requires the continued silencing of KNOX genes, as ectopic KNOX expression during organogenesis results in patterning defects and overproliferation of cells (Sinha et al., 1993; Chuck et al., 1996; Kidner et al., 2002). Thus, in plants, the precise balance between stem cell proliferation and differentiation that is critical for development is attained, in part, through the proper regulation of KNOX gene expression.
KNOX repression during organogenesis is mediated by the orthologous MYB domain proteins ROUGH SHEATH2 (RS2) and ASYMMETRIC LEAVES1 (AS1) from maize (Zea mays) and Arabidopsis thaliana, respectively (Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000; Ori et al., 2000). These proteins are expressed in a pattern complementary to the KNOX genes in organ founder cells and developing primordia. Loss-of-function mutations in RS2 and AS1 lead to perturbations in cell determination typical of ectopic KNOX accumulation; however, the initial downregulation in KNOX expression associated with organ initiation is unaffected in these mutants. RS2 and AS1 are therefore thought to act after organ founder cell specification to maintain KNOX gene silencing during subsequent leaf development.
Despite numerous studies addressing the role of RS2/AS1 in leaf development, the mechanism with which these proteins maintain KNOX gene silencing and determinacy during organogenesis is not currently understood. In rs2, KNOX genes become reactivated randomly in a variegated clonal pattern, such that rs2 null leaves are mosaics of KNOX+ and KNOX− sectors (Timmermans et al., 1999). This pattern of KNOX reactivation is reminiscent of several classic epigenetic phenomena associated with a failure to stably maintain a repressive chromatin state in all cells of a lineage. Consistent with an epigenetic mode of KNOX gene repression, RS2 and AS1 interact with the chromatin-remodeling factor HIRA, and reduced HIRA function in Arabidopsis results in ectopic KNOX expression in developing leaves (Phelps-Durr et al., 2005).
In addition to HIRA, RS2 and AS1 interact with the LOB domain protein AS2 (Xu et al., 2003; Phelps-Durr et al., 2005). Both RS2/AS1and AS2 are predicted DNA binding proteins and may serve as specificity factors to recruit HIRA to target loci, similar to the scenario of target recognition by the Polycomb repressor complex (Ringrose and Paro, 2007). HIRA proteins are known to modulate chromatin structure during both heterochromatic and euchromatic gene silencing in yeast (Saccharomyces cerevisiae) and mammalian cells (Spector et al., 1997; Magnaghi et al., 1998; Sharp et al., 2001; Roberts et al., 2002; Zhang et al., 2005). A similar role for HIRA in plants presents the possibility that RS2/AS1 complexes act directly at the KNOX loci to establish a repressive chromatin state that is stably inherited throughout organ development. However, efforts to demonstrate binding of RS2/AS1 or AS2 to promoters of KNOX genes have thus far been unsuccessful (Theodoris et al., 2003). Therefore, the action of the RS2/AS1 complexes may be indirect. Indeed, recent studies indicate a unique role for HIRA in the deposition of the histone variant H3.3 at target loci, which is associated with transcriptionally active states (Ahmad and Henikoff, 2002; Tagami et al., 2004; Loppin et al., 2005; Nakayama et al., 2007). Such an activity for HIRA in plants suggests an alternative hypothesis, namely, that RS2/AS1 complexes regulate KNOX expression indirectly through the activation of a repressor.
Here, we investigate the mechanism of AS1 complex–mediated KNOX gene silencing in Arabidopsis. We show that AS1 functions as a transcriptional repressor and binds directly to its KNOX targets when in a complex with AS2. We also define the DNA motifs that mediate AS1and AS2 binding and demonstrate that silencing of the KNOX gene BREVIPEDICELLUS (BP) in developing leaves requires binding of AS1-AS2 complexes at two sites in its promoter. Our observations suggest that AS1 and AS2 establish a loop in the KNOX promoter that represses KNOX expression during leaf development. We propose that AS1, AS2, and HIRA are part of a novel cellular memory system required for determinacy in plants that silences KNOX genes via a mechanism that is conceptually similar to the action of a genetic insulator (Gaszner and Felsenfeld, 2006).
RESULTS
AS1 Functions as a Transcriptional Repressor
Genetic analyses indicate that AS1 acts together with AS2 and HIRA in the stable silencing of KNOX targets during organogenesis (Ori et al., 2000; Semiarti et al., 2001; Byrne et al., 2002; Lin et al., 2003; Phelps-Durr et al., 2005). Because HIRA mediates epigenetic transitions associated with the activation and repression of target loci (Spector et al., 1997; Magnaghi et al., 1998; Sharp et al., 2001; Loppin et al., 2005; Zhang et al., 2005; Nakayama et al., 2007), AS1 could conceivably assemble into a transcriptional activator or repressor complex. To distinguish between these possibilities, we generated transgenic lines that express a chimeric protein, LFYDB:AS1CTD (Figure 1A), in which the C-terminal, non-MYB domain of AS1 (AS1CTD) that mediates the interactions with AS2 and HIRA is fused to the LEAFY DNA binding domain (LFYDB) (Maizel et al., 2005; Phelps-Durr et al., 2005). LFY specifies floral meristem fate and controls the activation of homeotic genes in the flower (Weigel et al., 1992; Parcy et al., 1998; Lamb et al., 2002). We reasoned that if AS1 functions as a transcriptional activator, placing this chimeric protein under control of the LFY regulatory sequences might lead to floral defects reminiscent of those observed upon expression of a constitutively activated form of LFY, such as LFY-VP16 (Parcy et al., 1998). On the other hand, if AS1 functions to repress its targets, expression of the LFYDB:AS1CTD transgene may lead to lfy loss-of-function phenotypes (Weigel et al., 1992).
Figure 1.
AS1 Is a Transcriptional Repressor.
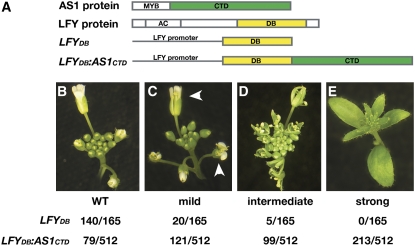
(A) Schematic representations of the AS1 and LFY proteins and the LFYDB and LFYDB:AS1CTD transgenes. The relative positions of the AS1 MYB domain, AS1 C-terminal domain (CTD), LFY activation domain (AC), and LFY DNA binding domain (DB) are indicated. The C-terminal domain of AS1, comprising amino acids 107 to 367, and the LFY DB domain, comprising amino acids 228 to 420, are highlighted in green and yellow, respectively.
(B) to (E) Inflorescence phenotypes observed among transgenic plants harboring the LFYDB or LFYDB:AS1CTD transgene. Examples of a wild-type inflorescence (B) and inflorescences with a mild (C), intermediate (D), or strong (E) lfy phenotype are shown. The frequencies with which each phenotypic class was observed are indicated below. Arrowheads in (C) mark flowers with missing petals.
Nearly 85% (433/512) of primary transformants carrying the LFYDB:AS1CTD transgene exhibited lfy-like floral defects. The phenotypes ranged in severity from weak, with minor defects in petal and stamen number (Figure 1C), to intermediate in which flowers formed fewer and homeotically transformed floral organs (Figure 1D), to severe in which floral meristems were completely transformed into inflorescence shoots with leaf-like lateral organs arranged in a spiral phyllotaxis (Figure 1E) (Weigel et al., 1992). No gain-of-function phenotypes were observed. Consistent with prior reports, nearly all plants transformed with the control transgene LFYDB, in which the LFY promoter drives expression of just the LFY DNA binding domain, were phenotypically normal (Figures 1A and 1B) (Parcy et al., 1998). This suggests that expression of LFYDB alone does not interfere with LFY function, whether through dominant-negative competition with LFY-mediated activation of its targets or through the induction of posttranscriptional gene silencing. Moreover, the lfy loss-of-function defects induced by the LFYDB:AS1CTD transgene are unlikely to result from transcriptional squelching as such defects are not observed upon overexpression of LFY or LFY-VP16 (Weigel and Nilsson, 1995; Parcy et al., 1998). Thus, replacement of the LFY activation domain with the C-terminal domain of AS1 blocks the activation of LFY targets, consistent with the hypothesis that AS1 functions as a transcriptional repressor.
AS1 Complexes Bind to Two Sites in the Promoter of the KNOX Target BP
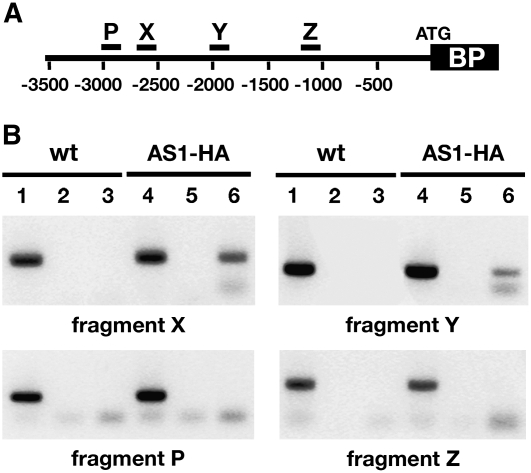
A repressive function for AS1 suggests that the AS1 complex may act directly at the KNOX target loci to maintain their silencing during organogenesis. To test this possibility, we used chromatin immunoprecipitation (ChIP) to identify elements at the Arabidopsis KNOX target BP that can mediate AS1 complex binding. We generated transgenic lines in which the AS1regulatory sequences drive expression of an HA epitope–tagged version of AS1 that is specifically recognized by HA antibodies (see Supplemental Figure 1 online). A line in which this AS1pro>AS1-HA transgene fully complements the as1-1 null allele was used for ChIP experiments. Previous studies have shown that a 5-kb region upstream of the BP start codon is sufficient for normal BP expression in the SAM and contains cis-acting sequences sufficient for AS1-, AS2-, and HIRA-mediated repression of BP in leaves (Ori et al., 2000; Phelps-Durr et al., 2005). ChIP samples of wild-type and AS1pro>AS1-HA seedlings were tested with primer pairs that allow amplification of ∼200- to 300-bp fragments spanning most of the 5-kb BP promoter. Only promoter regions that are unusually AT rich were omitted, as these could not be amplified efficiently or specifically. Out of 16 regions tested, we identified two fragments in the BP promoter that reproducibly amplified from AS1pro >AS1-HA chromatin samples immunoprecipitated using HA antibodies but not from mock-treated chromatin samples or samples prepared from wild-type seedlings (Figure 2). These promoter fragments, referred to below as X and Y, are located between nucleotides 2707 to 2522 and 2038 to 1788 upstream of the BP translation start site, respectively (Figure 2A). These results indicate that an AS1 complex binds to target sequences in the BP promoter and, together with the results of Figure 1, supports the notion that AS1 is part of a repressor complex that acts directly at the KNOX targets to maintain their silencing during leaf development.
Figure 2.
An AS1 Complex Binds in Vivo to Two Sites in the BP Promoter.
(A) Diagram of the BP promoter region showing the relative positions of four of the 16 promoter fragments analyzed by ChIP: X, −2707 to −2522; Y, −2038 to −1788; P, −3021 to −2720; and Z, −1299 to −1071. Numbers indicate distance in base pairs from the translation initiation site.
(B) Immunoprecipitation of wild-type and AS1pro>AS1-HA chromatin samples with HA monoclonal antibodies shows a specific association of the AS1 complex with fragments X and Y of the BP promoter. ChIP results for promoter fragments P and Z, which do not interact with the AS1 complex, are shown for comparison. Lanes 1 to 3, ChIP performed on wild-type chromatin samples; lanes 4 to 6, ChIP performed on chromatin from AS1pro >AS1-HA transgenic seedlings; lanes 1 and 4, total DNA; lanes 2 and 5, mock ChIP; lanes 3 and 6, ChIP with HA antibodies.
Both AS1 Complex Binding Sites Contribute to KNOX Silencing in the Leaf
Next, we tested in vivo the requirement of the AS1 complex binding sites for BP silencing in leaves by analyzing the expression pattern resulting from various BP promoter fragments. As a starting point, we used a 3.5-kb region upstream of the BP translation initiation site to drive expression of the β-glucuronidase (GUS) reporter. This promoter fragment includes both AS1 complex binding sites identified by ChIP and, in a wild-type background, drives GUS expression in the root and SAM but not in developing leaves (Figure 3A). In the as1 and as2 mutants, GUS expression was observed also in the major vascular bundles and petioles of leaves (Figures 3B and 3C). Therefore, this 3.5-kb regulatory region recapitulates the described BP mRNA expression patterns in wild-type and as1 and as2 backgrounds (Lincoln et al., 1994; Ori et al., 2000). This indicates that this promoter fragment contains the regulatory elements sufficient not only for BP expression in the SAM but also for the stable silencing of BP in developing leaves mediated by AS1 and AS2.
Figure 3.
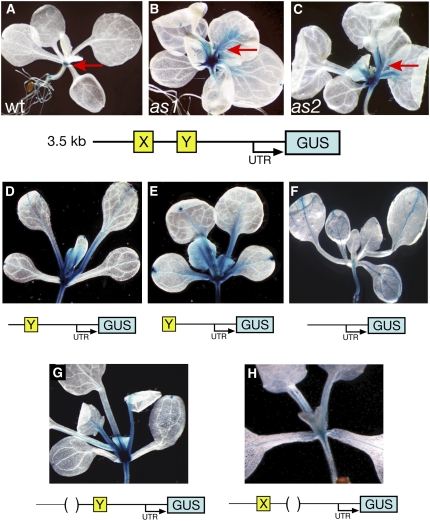
Both AS1 Complex Binding Sites Are Required to Repress BP Expression in Leaves.
Representative expression patterns of transgenic plants carrying distinct BPpro>GUS reporter constructs as diagramed below each panel. The expression pattern conditioned by a 3.5-kb region upstream of the BP translation initiation site resembles that of the endogenous BP gene. In the wild type (A), expression is restricted to the SAM (arrow), but in as1 (B) and as2 (C), expression extends into the leaves (arrows). Progressive 5′ end truncations of the BP promoter that delete one ([D] and [E]) or both (F) of the AS1 complex binding sites show GUS activity in leaves. Internal deletions of AS1 complex binding site X (G) or Y (H) reveals a requirement for both sites in the silencing of BP during leaf development.
Progressive 5′ end truncations of the BP promoter revealed that deletion of the sequences immediately upstream of site X had no effect on the GUS expression pattern (see Supplemental Figure 2A online). However, deletion of AS1 complex binding site X leads to GUS expression not just in the SAM but also in developing leaf primordia (Figure 3D). In older leaves, expression from this promoter occurs predominantly in the major veins and petioles, similar to that of the 3.5-kb BP reporter construct in as1 and as2 leaves. This ectopic expression pattern is consistent with the ChIP data and suggests that sequences involved in AS1 complex–mediated repression of BP in leaves are present in fragment X. Additional deletion of the region between the AS1 complex binding sites does not alter the GUS expression pattern further (Figure 3E). However, in plants transformed with a BP reporter construct in which both sites X and Y are deleted, ectopic GUS activity was reduced and became restricted to the vasculature of developing leaves (Figure 3F). This suggests that site Y contributes to the ectopic expression of BP in the leaf petioles and young leaf primordia. Upon further deletion of nucleotides 1788 to 1080 upstream of the BP start codon, GUS expression was lost in all aerial parts of the plant but persisted in the root (see Supplemental Figure 2B online). This ∼700-bp promoter region thus includes regulatory elements required for expression in the SAM and, along with sequence motifs in site Y, for misexpression in leaves.
To assess specifically the contributions of the AS1 complex binding sites to BP repression in leaves, we analyzed the effects of individual internal deletions of site X and Y on the expression domain of the 3.5-kb BPpro>GUS reporter. Deletion of site X alone was sufficient to induce ectopic GUS expression in a pattern that resembles the BP expression pattern in as1 and as2, throughout young leaf primordia and in the petioles and large vascular bundles of older leaves (Figure 3G). Deletion of site Y also leads to ectopic GUS expression in the leaf, indicating that site X is not sufficient to restrict BP expression to the SAM (Figure 3G). However, upon deletion of site Y, ectopic GUS expression is limited to the vasculature and occurs in a more restrictive pattern than that of BP in as1 and as2. Thus, although sites X and Y act nonredundantly in AS1 complex–mediated repression of BP during organogenesis, fragment Y includes additional regulatory motifs that direct BP misexpression outside the vasculature, in petioles and young leaf primordia.
Interaction between AS1 and AS2 Facilitates Binding to the BP Promoter
The ChIP experiments indicate that AS1 complexes bind directly to the X and Y sites in the BP promoter. AS1 is a MYB domain protein and could conceivably mediate the recruitment of HIRA and other potential complex components to the KNOX targets. However, several of the amino acids in the third helix of the R3 MYB motif that are critical for MYB–DNA interaction are not conserved in AS1, and attempts to demonstrate binding of this protein to DNA in vitro have thus far been unsuccessful (Romero et al., 1998; Waites et al., 1998; Rabinowicz et al., 1999; Theodoris et al., 2003). Moreover, the synergistic interaction between as1 and hira indicates that AS1 requires cofactors to recruit HIRA to the KNOX loci (Phelps-Durr et al., 2005). AS2 would be an obvious candidate. AS2 contains a Zn finger and leucine zipper–like motif that could mediate protein–protein and/or protein–DNA interactions (Iwakawa et al., 2002; Shuai et al., 2002). Also, the epistatic interaction between as1 and as2 indicates that AS1 function depends on AS2 (Serrano-Cartagena et al., 1999; Byrne et al., 2002), which presents the possibility that AS2 aids the targeting of AS1 repressor complexes to the KNOX loci.
To define the cis-elements and DNA binding factors required for binding of the AS1 complex to the KNOX loci, we performed electrophoretic mobility shift assays (EMSAs). AS1 and AS2 proteins were expressed in an in vitro wheat germ system. When translated separately, neither AS1 nor AS2 was able to bind fragment X in vitro (Figure 4A). Even when individually translated AS1 and AS2 proteins were mixed immediately prior to the binding assay, these proteins were unable to bind to fragment X. However, when AS1 and AS2 were cotranslated, these proteins were able to bind as a complex to site X of the BP promoter (Figure 4A). Similarly, cotranslated AS1 and AS2 proteins bound to fragment Y (Figure 4A). The specificity of these interactions was tested using competition assays. Increasing concentrations of unlabeled fragment X was able to compete for AS1-AS2 binding to site X and site Y, whereas regions of the BP promoter that, based on ChIP, do not interact with the AS1 complex in vivo were unable to compete for AS1-AS2 binding to these sites (see Supplemental Figure 3A online). Together, these data indicate that interaction between AS1 and AS2 facilitates their direct binding to two sites in the BP promoter required for stable KNOX repression and acquisition of determinacy in leaves.
Figure 4.
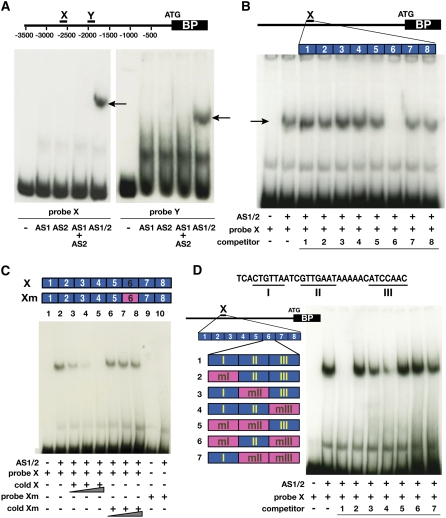
AS1-AS2 Heterodimers Bind to Specific Sequence Motifs in the BP Promoter.
(A) Interaction between AS1 and AS2 is required for binding to DNA. EMSA using in vitro–translated AS1 or AS2 proteins individually, as a mix (AS1+AS2) or as cotranslated proteins (AS1/2), shows that AS1 and AS2 can bind to BP promoter fragments X (left panel) and Y (right panel) but only when translated together (arrows). The diagram shows the relative positions of AS1 complex binding sites X and Y in the BP promoter. EMSA with in vitro–translated Luciferase protein was used as a nonspecific binding control (lanes marked “−”).
(B) Competition assays delineate a 32-bp sequence involved in AS1-AS2 complex binding. X was divided into eight duplexes as indicated in the diagram, and the ability of each duplex to block binding of the AS1-AS2 complex to site X is shown. Only duplex number 6 competes with fragment X for binding to AS1-AS2. Duplexes were added to the binding assay at 500-fold molar excess as indicated below each lane.
(C) Sequences in duplex 6 are essential and sufficient for AS1-AS2 binding. As illustrated in the diagram, fragment X of the BP promoter was mutagenized to change the sequence encompassing duplex 6 (Xm). Increasing amounts (50×, 100×, and 250×) of unlabeled wild-type fragment X compete effectively with AS1-AS2 binding to probe X (lanes 3 to 5). By contrast, addition of unlabeled fragment Xm to the binding assay whether at 50-, 100-, or 250-fold molar excess has no effect on AS1-AS2 binding to X (lanes 6 to 8). Cotranslated AS1-AS2 proteins were also unable to bind to probe Xm (lane 10).
(D) Two sequence motifs in duplex 6 contribute to AS1-AS2 binding. Duplex 6 was divided into regions I to III. Region 6-I includes a consensus c-Myb binding site (underlined), and regions 6-II and 6-III are partially palindromic (underlined). Regions were mutagenized individually or in combinations, as indicated in pink in the diagram (see also Supplemental Figure 3 online). The ability of each duplex 6 derivative to block AS1-AS2 binding to fragment X is shown in the gel on the right. Mutagenesis of region I renders duplex 6 an ineffective competitor, and mutations in regions II and III reduce the effectiveness of duplex 6 as competitor for AS1-AS2 binding to probe X. Duplexes were added to the binding assay at 500-fold molar excess as indicated below each lane.
Binding of AS1-AS2 to BP Is Mediated by Two Specific cis-Regulatory Motifs
The observation that fragment X can compete for binding of AS1-AS2 to fragment Y further suggests that sites X and Y contain conserved DNA sequence motifs that mediate AS1-AS2 binding to BP. To define such cis-regulatory elements, eight short duplexes corresponding to overlapping regions of the 185-bp X fragment were used as cold competitors in EMSA. Only duplex 6 was able to compete for binding of the AS1-AS2 complex to site X (Figure 4B), indicating that this 32-bp fragment contains cis-elements involved in AS1-AS2–mediated gene regulation. Consistent with the notion that sites X and Y include related AS1-AS2 DNA binding motifs, duplex 6 could also successfully compete with binding of these proteins to fragment Y. However, a higher molar excess of duplex 6 is required to obtain full competition of AS1-AS2 binding to fragment X than to fragment Y, suggesting that the protein complex binds with higher affinity to site X (see Supplemental Figures 3B and 3C online). To verify that duplex 6 mediates AS1-AS2 binding to site X in BP, we introduced mutations in fragment X at the position of duplex 6 (see Supplemental Figure 4 online). In contrast with the wild-type X fragment, this mutated version (Xm) was not bound by the cotranslated AS1-AS2 protein complex and failed to compete with its binding to site X (Figure 4C). This confirms that the 32-bp region corresponding to duplex 6 is critical for AS1-AS2 binding to site X in the BP promoter. Moreover, these data show that sequences outside of duplex 6 negligibly contribute to binding of AS1-AS2, suggesting that the region encompassing duplex 6 is also sufficient for recruitment of the AS1 complex to site X in BP.
Next, we used several mutant versions of this 32-bp fragment as unlabeled competitors in EMSA to precisely identify the regulatory sequences that facilitate AS1-AS2 binding (see Supplemental Figure 4 online). Duplex 6 was divided into three regions. Sequence analysis showed that region 6-I includes the consensus animal c-Myb binding site CNGTTR. Plant R2R3-MYB proteins typically recognize DNA sequence motifs closely related to this canonical MYB binding site that share a consensus sequence BNGTWR (e.g., Grotewold et al., 1994; Abe et al., 2003; Ryu et al., 2005; E. Grotewold, personal communication). Mutations that disrupt the presumptive MYB binding site abolished the ability of duplex 6 to compete for binding of AS1-AS2 to fragment X, indicating that this sequence is essential for AS1 complex binding (Figure 4D). Interestingly, regions 6-II and 6-III are partially palindromic. Mutations in region 6-II or both regions 6-II and 6-III that disrupt this palindrome significantly diminished the effectiveness of duplex 6 as a competitor of AS1-AS2 binding to X, whereas mutations in site 6-III had a relatively minor effect on complex binding (Figure 4D). Thus, the consensus MYB binding site in 6-I alone is not sufficient to completely disrupt AS1-AS2 binding to X, which indicates that sequences in regions 6-II and 6-III increase the binding affinity of these proteins to site X in the BP promoter. In addition, because region 6-II acts as a more effective competitor for AS1-AS2 binding to X than region 6-III, sequences in 6-II likely contribute more to binding of the AS1 complex to BP.
Considering that duplex 6 competes effectively for binding of AS1-AS2 to fragment Y, we performed matrix analysis to search for potential DNA sequence motifs that are conserved between duplex 6 and fragment Y and that may mediate AS1 complex binding to both sites X and Y in the BP promoter. Fragment Y lacks the palindromic sequence of region 6-III, but this AS1-AS2 binding fragment does contain the c-Myb–related sequence CTGTTt and the sequence motif TCtTTGAAT, which is closely related to region 6-II in fragment X (Figure 5E). These two sequence elements, referred to below as motifs I and II, respectively, are present in the same order in both fragments X and Y. While the c-Myb–related binding sequence is positioned upstream of motif II in both fragments, it is located directly adjacent to motif II in fragment X and 55 bp upstream of motif II in fragment Y. A similar arrangement of these two sequence motifs is not found elsewhere in the BP promoter, supporting the notion that recruitment of the AS1 complex to BP is mediated by binding of AS1-AS2 to the MYB and TCg/tTTGAAT cis-elements in sites X and Y in the promoter.
Figure 5.
Related Sequence Motifs Mediate AS1-AS2 Binding to BP and KNAT2.
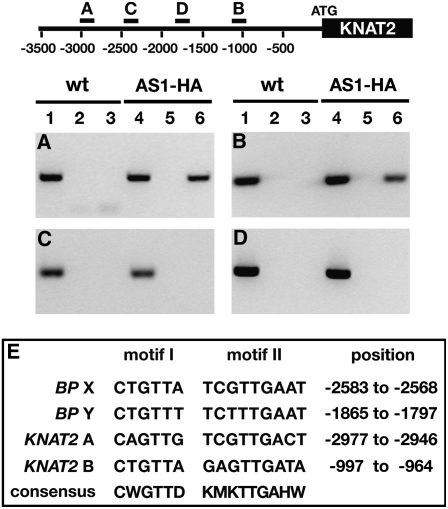
(A) to (D) The AS1 complex binds to two sites in the KNAT2 promoter. The diagram of the KNAT2 promoter indicates the relative positions of four of the 14 promoter fragments analyzed by ChIP. Numbers indicate distance in base pairs from the translation initiation site. Lanes 1 to 3, ChIP on wild-type seedlings; lanes 4 to 6, ChIP on AS1pro >AS1-HA transgenic seedlings; lanes 1 and 4, total DNA; lanes 2 and 5, mock ChIP; lanes 3 and 6, ChIP with HA antibodies.
(A) and (B) ChIP on wild-type and AS1pro >AS1-HA seedlings shows association of the AS1 complex with KNAT2 promoter fragments A and B.
(C) and (D) ChIP results for two promoter fragments that do not interact with the AS1 complex.
(E) Sequence motifs related to the AS1-AS2 cis-regulatory elements in BP are present in the AS1 complex binding sites A and B of the KNAT2 promoter. Inferred consensus sequences for the AS1 and AS2 binding motifs and their positions relative to the start codon of BP or KNAT2 are also shown.
Conserved Regulatory Elements Mediate AS1-AS2 Binding to KNAT2
To strengthen the hypothesis that AS1 complex binding to the KNOX targets is mediated by the AS1-AS2 binding motifs I and II, we examined whether similar regulatory sequences are present in AS1 complex binding sites at KNAT2, the other class I KNOX gene whose expression in developing leaves is repressed by AS1, AS2, and HIRA (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001; Phelps-Durr et al., 2005). We used ChIP to scan a 3.5-kb KNAT2 upstream region for AS1 complex binding sites. As for BP, we identified two fragments in the KNAT2 promoter that are enriched specifically in HA-ChIP samples from AS1pro >AS1-HA seedlings (Figures 5A to 5D). These AS1 complex binding sites are located between nucleotides 3048 to 2761 (designated fragment A) and 1260 to 914 (designated fragment B) upstream of the KNAT2 translation initiation site. Sequence analysis revealed c-Myb and motif II–related elements in each binding site that are separated by 22 to 24 bp but otherwise have the same arrangements as in the AS1 complex binding sites of BP (Figure 5E). Importantly, these motifs are not present in this arrangement elsewhere in the KNAT2 promoter. Taken together with the observation that duplex 6 is necessary and sufficient for binding of AS1-AS2 to BP, these data suggest that recruitment of the AS1 complex to its KNOX targets is mediated through a specific configuration of two regulatory elements with consensus sequences CWGTTD and KMKTTGAHW. The MYB binding site is positioned upstream of motif II in each of the four AS1 complex binding sites, but the spacing between these motifs is variable. However, as AS1 and AS2 must interact to bind DNA, the variability in the spacing of these sites is likely constrained.
DISCUSSION
AS1 and AS2 Form a Repressor Complex That Acts Directly at KNOX Targets
Stem cell homeostasis in plants is attained in part through the controlled expression of the class I KNOX homeodomain transcription factors. These proteins promote stem cell proliferation and indeterminacy, whereas acquisition of determinacy during organogenesis requires the continued silencing of KNOX gene activity (see Kidner et al., 2002; Scofield and Murray, 2006). Despite a recognized role for AS1, AS2, and HIRA in this process (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001; Lin et al., 2003; Phelps-Durr et al., 2005), insights into the molecular mechanism bringing about this repression were lacking. Here, we show that AS1 functions as a transcriptional repressor. Fusion to the C-terminal domain of AS1 can convert the LFY DNA binding protein into a dominant repressor. Similar to the use of the EAR domain (Hiratsu et al., 2003), the AS1 C-terminal domain provides a powerful tool to modulate transcription factor activity or characterize the biological functions of DNA binding proteins.
We further show that an AS1 repressor complex binds directly to two sites in the promoters of the KNOX targets BP and KNAT2. Complex binding at each site is mediated by the regulatory motif arrangement CWGTTD-KMKTTGAHW and requires interaction between AS1 and AS2. Although plant R2R3 MYB domain proteins may require heterodimerization with other transcription factors, such as bHLH proteins, to activate gene expression, they typically do not require auxiliary factors to bind DNA (see Stracke et al., 2001; Ramsay and Glover, 2005). Several of the DNA-contacting amino acid residues in the AS1 MYB domain have diverged from other plant R2R3 MYB domain proteins, which could affect the binding affinity of AS1 to DNA (Romero et al., 1998; Waites et al., 1998; Rabinowicz et al., 1999; Timmermans et al., 1999). In this regard, it is interesting to note that the MYB binding site in motif I is essential but not sufficient for AS1-AS2 binding. Our data indicate that motif II increases the binding affinity of AS1-AS2 to site X in the BP promoter. These data present the likely possibility that AS2, through interaction with motif II, stabilizes AS1 complex binding to the KNOX promoters. This scenario is consistent with the genetic interactions between as1 and as2 as well as the requirement for AS1 function to induce AS2 misexpression phenotypes (Serrano-Cartagena et al., 1999; Byrne et al., 2002; Lin et al., 2003; Xu et al., 2003; Phelps-Durr et al., 2005).
AS1 and AS2 Are Part of a Cellular Memory System
Deletion of the AS1-AS2 binding sites or loss of AS1 or AS2 function results in ectopic expression of BP and KNAT2 throughout young leaf primordia and in the petiole region and vasculature of older leaves (Figure 3; Ori et al., 2000). However, the expression domains of AS1 and AS2 overlap only in the very young leaf primordia (Iwakawa et al., 2007). The observed KNOX misexpression in older leaves is thus unlikely a direct reflection of lost AS1-AS2 complex activity. Considering that AS1 interacts with the chromatin-remodeling factor HIRA and its involvement in KNOX gene repression during organogenesis (Phelps-Durr et al., 2005), the AS1-AS2 complex may act early in leaf development to recruit HIRA and establish a somatically stable silenced state at the KNOX targets that is maintained throughout leaf development, even though AS1-AS2 activity does not persist. Similar to the variegated pattern of KNOX reactivation in rs2 (Timmermans et al., 1999; Phelps-Durr et al., 2005), the pattern of KNOX misexpression in older Arabidopsis leaves may thus reflect a predisposition of certain cells to reactivate KNOX genes in the absence of a somatically heritable silenced state.
In addition to this repressive system, promoter deletion analysis showed that regulatory elements within fragment Y and a 708-bp fragment located ∼1 kb upstream of the BP start codon are required for BP misexpression in leaves upon loss of AS1-AS2 regulation. Accordingly, KNOX misexpression caused by loss of AS1-AS2 regulation also reflects the spatiotemporal activation resulting from specific enhancer elements. Such AS1-AS2–independent regulatory mechanisms may explain why in the C24 ecotype, expression resulting from BP promoter fragments lacking both AS1 complex binding sites remains restricted to the SAM (Heyer et al., 2004; Truernit et al., 2006). Perhaps factors required for BP activation in the leaf are missing in C24. Similarly, expression of the KNOX family member SHOOTMERISTEMLESS (STM) remains restricted to the SAM even in an as1 or as2 background (Byrne et al., 2000; Ori et al., 2000; Semiarti et al., 2001). In fact, STM lacks AS1 complex binding motifs.
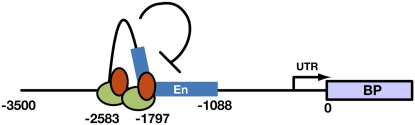
Deletion of either AS1 complex binding site X or Y results in ectopic expression of BP in developing leaves, indicating that the two sites act nonredundantly despite their analogous AS1-AS2 binding properties. Accordingly, interaction between AS1 complexes at each site appears to be required, suggesting that a repressive loop may be formed in the KNOX promoters that mediates stable KNOX gene silencing during organogenesis. The observation that AS1 can form homodimers (Theodoris et al., 2003; Phelps-Durr et al., 2005) presents a possible mechanism via which AS1 complexes can interact. Based on the position of the AS1-AS2 binding motifs (Figure 5E), such a loop in the promoter of BP would include enhancer elements required for expression in the leaf, as deletion of site Y prevents BP misexpression in the petioles and young leaf primordia. We propose that by binding to two sites, AS1 repressor complexes establish a loop in the promoter of its KNOX targets and through recruitment of the chromatin-remodeling factor HIRA establish a repressive chromatin state that blocks enhancer activity in the leaf and that is stably inherited throughout the many rounds of cell division associated with leaf development (Figure 6).
Figure 6.
Model for AS1-AS2–Mediated KNOX Gene Silencing in the Leaf.
The AS1-AS2 complex binds to the regulatory motifs CWGTTD and KMKTTGAHW, which are present at two sites in the promoters of KNOX targets immediately upstream and surrounding an enhancer region required for expression in developing leaves. Interaction between the AS1 complexes is required for stable KNOX gene silencing, suggesting formation of a loop in the KNOX promoter that, likely through recruitment of HIRA, leads to formation of a stable repressive chromatin state that blocks enhancer activity throughout leaf development. Green ovals, AS1; red ovals, AS2; blue box, leaf enhancers.
This model suggests that, within the context of KNOX gene silencing, AS1 and AS2 are part of a cellular memory system that is conceptually similar to the action of genetic insulators, which form chromatin loop domains that sequester enhancer elements and block their action on promoters (Gaszner and Felsenfeld, 2006). Identifying the proposed epigenetic modifications associated with AS1 complex–mediated KNOX repression will be the next challenge in understanding how cells progress from indeterminate stem cells to their final differentiated state. Several recent studies imply a role for AS1 and AS2 in adaxial-abaxial patterning of the leaf by spatially restricting the expression domain of specific abaxial determinants (Lin et al., 2003; Xu et al., 2003; Li et al., 2005; Garcia et al., 2006; Iwakawa et al., 2007; Ueno et al., 2007). Whether AS1 and AS2 control the spatiotemporal expression of polarity genes directly and through a similar silencing mechanism remains to be determined.
Evolutionary and Developmental Considerations of AS1-AS2–Mediated KNOX Gene Silencing
Recruitment of AS1-AS2 to BP and KNAT2 is essential to repress KNOX activity and establish determinacy during organogenesis. In other simple leafed species, such as maize and snapdragon (Antirrhinum majus), AS1 orthologs similarly confine KNOX activity to the SAM (Timmermans et al., 1999; Tsiantis et al., 1999). In maize, expression of the KNOX family members rs1 and liguleless3 in the leaf is suppressed by RS2 and the AS2 homolog INDETERMINATE GAMETOPHYTE1 (Schneeberger et al., 1998; Scanlon et al., 2002; Evans, 2007). Through preliminary sequence analysis, we identified correctly arranged motif I and motif II consensus binding sites in the promoters and/or large third introns of these KNOX genes. Along with the fact that RS2 interacts with HIRA (Phelps-Durr et al., 2005), this suggests that conservation in the mechanism of KNOX gene silencing during organogenesis exists between these monocot and dicot species.
Unlike Arabidopsis, its close relative Cardamine hirsuta develops compound leaves. This difference in leaf shape is at least partially attributable to divergent regulation of KNOX genes, including BP, which in C. hirsuta are expressed in the leaf (Hay and Tsiantis, 2006). The AS1 ortholog of C. hirsuta can complement the as1-1 mutation in Arabidopsis, indicating functional conservation of AS1 between the two species (Hay and Tsiantis, 2006). Furthermore, AS2 function may be conserved, as expression resulting from the Arabidopsis BP promoter in C. hirsuta remains confined to the SAM. Consensus motif I and motif II sequences are present in the C. hirsuta BP promoter, but these cis-regulatory elements occur only once in the specific arrangement known to mediate AS1-AS2 binding (CTGTTT and TATTTGATA at 1653 to 1589 bp upstream of the translation start site). Mutation of one of the AS1-AS2 cis-regulatory sequences may thus have contributed to the divergent patterns of KNOX expression between Arabidopsis and C. hirsuta. Considering that compound leafed species that exhibit KNOX expression in the leaf arose multiple times independently during evolution (Bharathan et al., 2002), such cis-regulatory polymorphisms that abrogate AS1-AS2 binding may constitute a key determinant in the evolution of leaf morphologies.
METHODS
Molecular Cloning
A transformation vector containing the LFY promoter plus LFY DNA binding domain (amino acids 228 to 420), referred to as pLFYDB, was kindly provided by Detlef Weigel (Max Planck Institute for Developmental Biology, Tübingen, Germany). The AS1 C-terminal domain (amino acids 107 to 367) was amplified to insert SalI restriction sites and cloned in frame into pLFYDB to make pLFYDB:AS1CTD. To generate an HA epitope–tagged version of AS1, a 5-kb fragment including the AS1 promoter and coding region was amplified and engineered to insert in frame upstream of a 3x HA-tag containing 5′ EcoRI and 3′ XhoI sites. Subsequently, a 1-kb AS1 fragment comprising the 3′ untranslated region and terminator regions was inserted at the C terminus of the 3x HA-tag, and the resulting fusion gene was cloned into pCambia2300 to give pAS1pro>AS1-HA. BP promoter deletion derivatives were generated as in-frame translational fusions of the GUS reporter to the ATG of BP. The various upstream regions of BP were amplified from Arabidopsis thaliana Columbia (Col) genomic DNA, cloned into Gateway TOPO pCR8 (Invitrogen), and subsequently recombined into pKGWFS7 (Plant Systems Biology) according to the manufacturer's protocol.
Plant Materials
All plants were grown at 21°C under long-day conditions. The pLFYDB, pLFYDB:AS1CTD, and pBPpro >GUS plasmids were transformed in Arabidopsis ecotype Col-0 using standard procedures. Selected BPpro>GUS transgenes were crossed into as1-1 and as2-4 previously introgressed into Col-0. GUS staining was performed as described (Sundaresan et al., 1995), and at least 20 independent T2 lines were analyzed for each construct. The pAS1pro>AS1-HA vector was transformed into as1-1/+ plants in the Landsberg erecta (Ler) background. T2 lines that were homozygous for as1-1 and the transgene and that were phenotypically normal were propagated for use in ChIP assays.
Protein Gel Blot Analysis
Protein extracts were prepared from Ler and AS1pro>AS1-HA plants. Approximately 0.25 g of inflorescence tissue or 1 g of seedling tissue were ground in 500 μL of extraction buffer (10% sucrose, 100 mM Tris HCl, pH 8.5, 5 mM EDTA, 5 mM EGTA, 40 mM β-mercaptoethanol, and 2 mM PMFS), centrifuged for 10 min, and 250 μL supernatant mixed with equal volume 2× SDS-PAGE loading dye. Ten microliters out of 300-μL chromatin samples prepared for ChIP assays was similarly mixed with 10 μL of 2× SDS-PAGE loading dye. After boiling, 20-μL aliquots were separated on a 10% SDS-PAGE gel, transferred to Trans-Blot membrane (Bio-Rad), and incubated with the primary monoclonal HA antibody 12CA5 (Abgent) at a 1:5000 dilution followed by horseradish peroxidase anti-mouse IgG secondary antibody (GE Healthcare) at 1:2000 dilution, using standard protocols. ECL Plus reagents (Amersham) were used for immunodetection according to the manufacturer's recommended protocol.
ChIP
ChIP was performed as described (Gendrel et al., 2002). Approximately 3 g of normal and AS1pro>AS1-HA seedlings at the four-leaf stage were used as starting materials. ChIP reactions were mock treated or incubated with 5 μL of 12CA5 monoclonal antibody (Abgent), and immunoprecipitates were collected using Dynabead protein G magnetic beads (Invitrogen). Final eluted DNA was resuspended in 50 μL of water, and 1 μL was amplified by PCR using standard protocols with an annealing temperature of 54°C and typically 35 cycles. Each promoter region was tested on five to six independent biological replicates. Primers for sequences are as follows: for BP, Xfor, 5′-TACACGAACACAGATGATGAT-3′; Xrev, 5′-CAGTGGAAGTGAGAGTAGG-3′; Yfor, 5′-TAGATCCATATGGTTATGGGT-3′; Yrev, 5′-CCTCTTATTTTCTGTTTCAGTA-3′; for KNAT2, Afor, 5′-CCTGAGCTAATTAAGTAGA-3′; Arev, 5′-GGTGCTAATTTTGCTTATG-3′; Bfor, 5′-CTGTCGTTTTTATAAGGTTTG-3′; Brev, 5′-CACTTATCGCACTTCTTGTT-3′.
EMSA
The AS1 and AS2 coding regions were amplified via RT-PCR from total RNA and cloned into the Luciferase-T7 control DNA vector from the TNT-coupled wheat germ extract systems kit (Promega) through engineered BamHI-SacI and BamHI-EcoRV restriction sites, respectively. Proteins were produced by in vitro transcription and translation according to the manufacturer's suggested protocol. DNA probes were amplified from Arabidopsis genomic DNA, cloned into the TOPOII plasmid (Invitrogen), and excised through SpeI-EcoRV digestion, and 100 ng (1.5 to 2 nM) were end-labeled with 32P-dCTP using standard Klenow fill-in reactions. Labeled probes were purified from nondenaturing PAGE gels and diluted to a final concentration of ∼80 pM. Binding reactions were in 16 μL and included 2 μL in vitro–translated protein, 1 to 2 fmol of radiolabeled probe, 10 mM Tris-HCl, pH 8.0, 100 mM KCl, 1 mM EDTA, 1 mM DTT, 8% glycerol, 500 ng single-stranded DNA, and 25 ng polydeoxyinosinic-deoxycytidylic acid. Reactions were preincubated at 4°C for 20 min and subsequently incubated for an additional 20 min with radiolabeled probe. Cold competitors were added at the preincubation step. Duplexes used in competition assays were annealed from complementary oligos by boiling them for 5 min followed by slow cooling to room temperature. Reactions were separated on 5% nondenaturing PAGE gels in 0.5× TBE buffer.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: BP, At4g08150; KNAT2, At1g70510; AS1, At2g37630; AS2, At1g65620.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Monoclonal HA Antibodies Specifically Recognize the AS1-HA Fusion Protein.
Supplemental Figure 2. Expression Analysis of Additional BP Promoter Deletion Constructs.
Supplemental Figure 3. Specific Binding of AS1-AS2 Heterodimers to BP Promoter Fragments X and Y.
Supplemental Figure 4. Sequences of Duplex 6 Variants Used in Competition Assays.
Supplementary Material
Acknowledgments
We thank Shahinez Madi and Tara Phelps-Durr for technical assistance in characterizing the BPpro>GUS and LFYDB:AS1CTD transgenic lines and Tim Mulligan for plant care. We also thank Detlef Weigel for providing the LFYDB expression vector; Zach Lippman, Mikel Zaratiegui, and Alexis Maizel for technical advice; and Cris Kuhlemeier and Erich Grotewold for useful discussions. This work was supported by a grant from the National Science Foundation (MCB-0616114) to M.C.P.T.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Marja C.P. Timmermans (timmerma@cshl.edu).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad, K., and Henikoff, S. (2002). The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9 1191–1200. [DOI] [PubMed] [Google Scholar]
- Bharathan, G., Goliber, T.E., Moore, C., Kessler, S., Pham, T., and Sinha, N.R. (2002). Homologies in leaf form inferred from KNOXI gene expression during development. Science 296 1858–1860. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). ASYMMETRIC LEAVES1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Simorowski, J., and Martienssen, R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129 1957–1965. [DOI] [PubMed] [Google Scholar]
- Chuck, G., Lincoln, C., and Hake, S. (1996). KNAT1 induces lobed leaves with ectopic meristems when overexpressed in Arabidopsis. Plant Cell 8 1277–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, M. (2007). The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell 19 46–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D., Collier, S.A., Byrne, M.E., and Martienssen, R.A. (2006). Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr. Biol. 16 933–938. [DOI] [PubMed] [Google Scholar]
- Gaszner, M., and Felsenfeld, G. (2006). Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7 703–713. [DOI] [PubMed] [Google Scholar]
- Gendrel, A.V., Lippman, Z., Yordan, C., Colot, V., and Martienssen, R.A. (2002). Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 297 1871–1873. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., Drummond, B.J., Bowen, B., and Peterson, T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76 453–553. [DOI] [PubMed] [Google Scholar]
- Hay, A., and Tsiantis, M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 38 942–947. [DOI] [PubMed] [Google Scholar]
- Heyer, A.G., Raap, M., Schroeer, B., Marty, B., and Willmitzer, L. (2004). Cell wall invertase expression at the apical meristem alters floral, architectural, and reproductive traits in Arabidopsis thaliana. Plant J. 39 161–169. [DOI] [PubMed] [Google Scholar]
- Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H., Iwasaki, M., Kojima, S., Ueno, Y., Soma, T., Tanaka, H., Semiarti, E., Machida, Y., and Machida, C. (2007). Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 51 173–184. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H., Ueno, Y., Semiarti, E., Onouchi, H., Kojima, S., Tsukaya, H., Hasebe, M., Soma, T., Ikezaki, M., Machida, C., and Machida, Y. (2002). The ASYMMETRIC LEAVES1 gene of Arabidopsis thaliana, required for formation of symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 43 467–478. [DOI] [PubMed] [Google Scholar]
- Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120 405–413. [Google Scholar]
- Kidner, C.A., Timmermans, M.C.P., Byrne, M.E., and Martienssen, R.A. (2002). Developmental genetics of the angiosperm leaf. Adv. Bot. Res. 38 191–234. [Google Scholar]
- Lamb, R.S., Hill, T.A., Tan, Q.K.-G., and Irish, V.F. (2002). Regulation of APETALA3 floral homeotic gene expression by meristem identity genes. Development 129 2079–2086. [DOI] [PubMed] [Google Scholar]
- Li, H., Xu, L., Wang, H., Yuan, Z., Cao, X.F., Yang, Z.N., Zhang, D.B., Xu, Y.Q., and Huang, H. (2005). The putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and microRNA165/166 in Arabidopsis leaf development. Plant Cell 17 2157–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W.C., Shuai, B., and Springer, P.S. (2003). The Arabidopsis LATERAL ORGAN BOUNDARYS gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell 15 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 37 66–69. [DOI] [PubMed] [Google Scholar]
- Loppin, B., Bonnefoy, E., Anselme, C., Laurencon, A., Karr, T.L., and Couble, P. (2005). The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437 1386–1390. [DOI] [PubMed] [Google Scholar]
- Magnaghi, P., Roberts, C., Lorain, S., Lipinski, M., and Scambler, P.J. (1998). Hira, a mammalian homologue of Saccharomyces cerevisiae transcriptional co-repressors, interacts with Pax3. Nat. Genet. 20 74–77. [DOI] [PubMed] [Google Scholar]
- Maizel, A., Busch, M.A., Tanahashi, T., Perkovic, J., Kato, M., Hasebe, M., and Weigel, D. (2005). The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308 260–263. [DOI] [PubMed] [Google Scholar]
- Nakayama, T., Nishioka, K., Dong, Y.X., Shimojima, T., and Hirose, S. (2007). Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev. 21 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori, N., Eshed, Y., Chuck, G., Bowman, J.L., and Hake, S. (2000). Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127 5523–5532. [DOI] [PubMed] [Google Scholar]
- Parcy, F., Nilsson, O., Busch, M., Lee, I., and Weigel, D. (1998). A genetic framework for floral patterning. Nature 395 561–566. [DOI] [PubMed] [Google Scholar]
- Phelps-Durr, T.L., Thomas, J., Vahab, P., and Timmermans, M.C.P. (2005). Maize ROUGH SHEATH2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17 2886–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowicz, P.D., Braun, E.L., Wolfe, A.D., Bowen, B., and Grotewold, E. (1999). Maize R2R3 MYB genes: Sequence analysis reveals amplification in the higher plants. Genetics 153 427–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay, N., and Glover, B.J. (2005). The MYB/MYC/bHLH complex and the evolution of cellular diversity. Trends Plant Sci. 10 63–70. [DOI] [PubMed] [Google Scholar]
- Ringrose, L., and Paro, R. (2007). Polycomb/trithorax response elements and epigenetic memory of cell identity. Development 134 223–232. [DOI] [PubMed] [Google Scholar]
- Roberts, C., Sutherland, H.F., Farmer, H., Kimber, W., Halford, S., Carey, A., Brickman, J.M., Wynshaw-Boris, A., and Scambler, P.J. (2002). Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol. Cell. Biol. 22 2318–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, I., Fuertes, A., Benito, M.J., Malpica, J.M., Leyva, A., and Paz-Ares, J. (1998). More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 14 273–284. [DOI] [PubMed] [Google Scholar]
- Ryu, K.H., Kang, Y.H., Park, Y.H., Hwang, D., Schiefelbein, J., and Lee, M.M. (2005). The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development 132 4765–4775. [DOI] [PubMed] [Google Scholar]
- Scanlon, M.J., Henderson, D.C., and Bernstein, B. (2002). SEMAPHORE1 functions during the regulation of ancestrally duplicated knox genes and polar auxin transport in maize. Development 129 2663–2673. [DOI] [PubMed] [Google Scholar]
- Schneeberger, R., Tsiantis, M., Freeling, M., and Langdale, J.A. (1998). The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 125 2857–2865. [DOI] [PubMed] [Google Scholar]
- Scofield, S., and Murray, J.A.H. (2006). KNOX gene function in plant stem cell niches. Plant Mol. Biol. 60 929–946. [DOI] [PubMed] [Google Scholar]
- Semiarti, E., Ueno, Y., Tsukaya, H., Iwakawa, H., Machida, C., and Machida, Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128 1771–1783. [DOI] [PubMed] [Google Scholar]
- Serrano-Cartagena, J.S., Robles, P., Ponce, M.R., and Micol, J.L. (1999). Genetic analysis of leaf form mutants from the Arabidopsis information service collection. Mol. Gen. Genet. 261 725–739. [DOI] [PubMed] [Google Scholar]
- Sharp, J.A., Fouts, E.T., Krawitz, D.C., and Kaufman, P.D. (2001). Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11 463–473. [DOI] [PubMed] [Google Scholar]
- Shuai, B., Reynaga-Pena, C.G., and Springer, P.S. (2002). The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 129 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, N.R., Williams, R.E., and Hake, S. (1993). Overexpression of the maize homeobox gene, knotted-1, causes a switch from determinate to indeterminate cell fates. Genes Dev. 7 787–795. [DOI] [PubMed] [Google Scholar]
- Spector, M., Raff, A., DeSilva, H., Lee, K., and Osley, M. (1997). Hir1p and hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 17 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 447–456. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9 1797–1810. [DOI] [PubMed] [Google Scholar]
- Tagami, H., Ray-Gallet, D., Almouzni, G., and Nakatani, Y. (2004). Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116 51–61. [DOI] [PubMed] [Google Scholar]
- Theodoris, G., Inada, N., and Freeling, M. (2003). Conservation and molecular dissection of ROUGH SHEATH2 and ASYMMETRIC LEAVES1 function in leaf development. Proc. Natl. Acad. Sci. USA 100 6837–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans, M.C.P., Hudson, A., Becraft, P.W., and Nelson, T. (1999). ROUGH SHEATH2: A myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284 151–153. [DOI] [PubMed] [Google Scholar]
- Truernit, E., Siemering, K.R., Hodge, S., Grbic, V., and Haseloff, J. (2006). A map of KNAT gene expression in the Arabidopsis root. Plant Mol. Biol. 60 1–20. [DOI] [PubMed] [Google Scholar]
- Tsiantis, M., Schneeberger, R., Golz, J.F., Freeling, M., and Langdale, J.A. (1999). The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284 154–156. [DOI] [PubMed] [Google Scholar]
- Ueno, Y., Ishikawa, T., Watanabe, K., Terakura, S., Iwakawa, H., Okada, K., Machida, C., and Machida, Y. (2007). Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. Plant Cell 19 445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollbrecht, E., Reiser, L., and Hake, S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127 3161–3172. [DOI] [PubMed] [Google Scholar]
- Waites, R., Selvadurai, H.R.N., Oliver, I.R., and Hudson, A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93 779–789. [DOI] [PubMed] [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69 843–859. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Nilsson, O. (1995). A developmental switch sufficient for flower initiation in diverse plants. Nature 377 495–500. [DOI] [PubMed] [Google Scholar]
- Xu, L., Xu, Y., Dong, A.W., Sun, Y., Pi, L.M., Xu, Y.Q., and Huang, H. (2003). Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying adaxial identity. Development 130 4097–4107. [DOI] [PubMed] [Google Scholar]
- Zhang, R., et al. (2005). Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8 19–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.