Abstract
To maintain proper meristem function, cell division and differentiation must be coordinately regulated in distinct subdomains of the meristem. Although a number of regulators necessary for the correct organization of the shoot apical meristem (SAM) have been identified, it is still largely unknown how their function is integrated with the cell cycle machinery to translate domain identity into correct cellular behavior. We show here that the cyclin-dependent kinases CDKB2;1 and CDKB2;2 are required both for normal cell cycle progression and for meristem organization. Consistently, the CDKB2 genes are highly expressed in the SAM in a cell cycle–dependent fashion, and disruption of CDKB2 function leads to severe meristematic defects. In addition, strong alterations in hormone signaling both at the level of active hormones and with respect to transcriptional and physiological outputs were observed in plants with disturbed CDKB2 activity.
INTRODUCTION
All above-ground parts of a plant, such as stem, leaves, flowers, and fruits, are derived from a small number of pluripotent stem cells residing in the shoot apical meristem (SAM). Therefore, plants are absolutely dependent on the presence of these cells to complete their lifecycle. This strict dependence is reflected in the evolution of a robust regulatory system that is able to integrate local meristematic cues with global signals, which relay environmental parameters and the overall growth status of the organism. Forward genetic screens have uncovered some of the key regulators for SAM organization, and their localized expression has helped to identify important functional domains within this tissue. These studies revealed that due to the activity of the homeodomain transcription factor WUSCHEL (WUS), the core of the meristem acts as an organizing center, which is essential for induction of stem cell fate in the overlying cells (Laux et al., 1996; Mayer et al., 1998). The stem cells in turn express CLAVATA3 (CLV3), which codes for a short secreted peptide molecule and acts to restrict WUS expression in deeper layers (Fletcher et al., 1999; Ito et al., 2006; Kondo et al., 2006). Therefore, in clv3 mutants, WUS expression is expanded and stem cells proliferate inappropriately, while in plants with compromised WUS activity, stem cell fate and, thus, meristem function is terminated prematurely. Cells within the central stem cell domain divide slowly, but once they are displaced into the peripheral zone, they divide more rapidly before being incorporated into newly forming organs at the flanks of the meristem. This function is dependent on the activity of another homeodomain transcription factor, SHOOT MERISTEMLESS (STM), which is expressed throughout the meristem and acts to inhibit cell differentiation (Long et al., 1996). The complex pattern of cellular behavior within the meristem highlights the need for careful balancing of cell division and differentiation in the various domains to preserve the structure and function of this tissue. Thus, signals from meristem regulators must be perceived and accurately interpreted by the cell cycle machinery.
The eukaryotic cell cycle is a highly regulated process that relies on a series of discrete checkpoints to ensure proper DNA replication and successful cytokinesis. Progression through the individual steps of the cycle is dependent on the activation of cyclin-dependent kinases (CDKs) by interaction with their specific cyclin partners. While a single CDK is sufficient to execute the cell cycle in yeast, higher eukaryotic organisms, such as plants and animals, have expanded complements of cell cycle regulators. The Arabidopsis thaliana genome contains 38 Cyclin-related genes, many of which are expressed in a cell cycle–specific manner (Menges et al., 2005). Among the 29 CDK-related sequences, CDKA;1 is the archaetypical CDK in Arabidopsis, as demonstrated by its ability to complement the yeast cdc28 mutant (Ferreira et al., 1991). Like most other CDK genes, it is expressed at a similar level throughout the cell cycle, while the plant-specific B-type CDKs are unique in having a strictly cell cycle–dependent transcription profile (Segers et al., 1996; Magyar et al., 1997; Menges and Murray, 2002; Menges et al., 2005). The roles of individual plant cell cycle regulators have been assayed genetically using gain- and loss-of-function strategies. Overexpression of a dominant-negative CDKA;1 allele resulted in plants with fewer but dramatically larger cells (Hemerly et al., 1995), whereas complete loss of CDKA;1 function caused gametophytic lethality, highlighting its essential nature (Iwakawa et al., 2006; Nowack et al., 2006). Plants expressing a dominant-negative allele of CDKB1;1 displayed normal overall morphology but also had fewer and larger cells, which had higher ploidy levels than wild-type controls. In addition, these plants had specific defects in stomatal cell division (Boudolf et al., 2004a, 2004b). Originally identified based on their ability to complement yeast mutants, the D-type cyclins are among the most extensively studied CDK interaction partners in plants (Soni et al., 1995). Significant acceleration of plant growth by CYCLIN D overexpression was accompanied by only subtle changes in SAM structure (Cockcroft et al., 2000; Boucheron et al., 2005); likewise, additional cell divisions could be induced in the SAM without compromising its organization (Dewitte et al., 2003).
These studies have suggested that plant architecture is remarkably resistant to the manipulation of many cell cycle regulators, and it has been proposed that cell divisions are merely subordinate in the execution of developmental programs (Kaplan and Hagemann, 1991; Gutierrez, 2005). However, since meristematic cells proliferate at different rates within the various subdomains, it would appear that the cell cycle machinery is able to integrate organizing signals to maintain a functional stem cell niche. To address this paradox, we have searched for core cell cycle regulators that are active in the meristem and whose expression is dependent on the functional organization of this tissue.
RESULTS
B2-Type CDKs Are Expressed in Functional SAMs
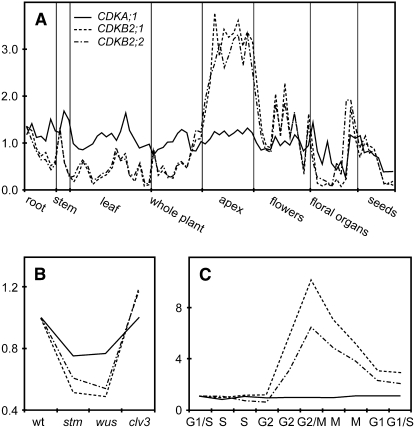
To identify regulators required for both meristem function and cell cycle control, we surveyed Arabidopsis microarray data sets for cell cycle–related genes that show preferential expression in the shoot apex and that are sensitive to disruption of key meristematic regulators. In addition, we scored for cell cycle–dependent mRNA accumulation. Using these criteria, we found a pair of duplicated B-type CDKs, CDKB2;1 and CDKB2;2, that are regarded as core cell cycle regulators and fulfilled our mRNA expression criteria. In the AtGenExpress developmental data set (Schmid et al., 2005), expression of these genes was highest in samples corresponding to the shoot apex (Figure 1A) but was strongly reduced in shoot apices of 3-d-old wus and stm mutant seedlings, which fail to maintain a proper SAM (Figure 1B). By contrast, CDKB2;1 and CDKB2;2 mRNA levels were slightly elevated in clv3 mutants, which have an enlarged meristem (Figure 1B). Furthermore, both genes showed a peak in expression during the G2-to-M phase transition of the cell cycle (Menges et al., 2003) (Figure 1C). Additional information about the presumed phylogenetic relationships and expression patterns of all Arabidopsis CDKs can be found in Supplemental Figure 1 online.
Figure 1.
Relative Expression Levels of CDKA;1, CDKB2;1, and CDKB2;2.
(A) The CDKB2s were highly expressed in the apex, whereas CDKA;1 was expressed at similar levels across all tissues. Expression levels were normalized to the per-gene average across all samples.
(B) CDKB2 expression was reduced in stm and wus mutants, which have impaired meristem function, while their transcript levels were increased in clv3 mutants with enlarged meristems. Expression levels were normalized to the wild-type control.
(C) The CDKB2s showed a peak in expression level at the G2/M transition, while CDKA;1 expression did not change in response to the cell cycle phase. Transcript abundance was normalized to the expression level at the time of cell cycle block release.
Microarray data were extracted from the AtGenExpress compendium (Menges et al., 2003; Schmid et al., 2005) and represent biological triplicates in (A) or biological duplicates in (B) and (C).
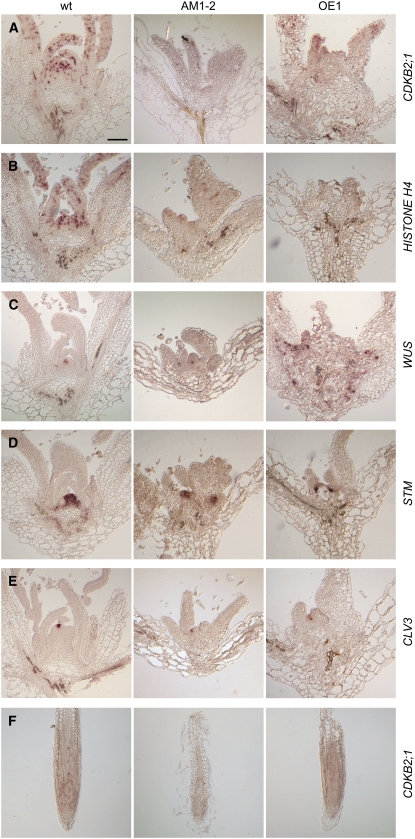
To resolve the localization of CDKB2 mRNAs at a cellular level, we performed in situ hybridizations on 12-d-old seedlings and confirmed strong cell cycle–dependent expression of both transcripts in cells of the SAM and young leaves (Figure 3A). A weaker cell cycle–dependent expression was seen in root tips (Figure 3F; see Supplemental Figure 2A online). Since CDKB2;1 and CDKB2;2 cDNAs share 86% percent identity on the nucleotide level, RNA probes corresponding to the full-length coding sequences likely detect both transcripts simultaneously. To distinguish between the two CDKs, we prepared probes from the untranslated regions of both genes, which are unrelated in sequence. Despite weaker signals, which likely result from short probe lengths, we confirmed that both CDKB2 transcripts are expressed in cells of the SAM (see Supplemental Figure 3 online).
Figure 3.
In Situ Hybridizations.
All images were taken at the same magnification. Bar = 100 μm.
(A) In the wild type, CDKB2;1 was expressed at the apex in a spotty pattern characteristic of a cell cycle–regulated gene. Expression could not be detected in AM1-2 plants, whereas OE1 plants showed a weaker expression in a larger number of cells.
(B) HISTONE H4 was expressed in many cells of wild-type apices, but its expression was attenuated in both AM1-2 and OE1 plants.
(C) to (E) A single center of meristematic activity with WUS, STM, and CLV3 expression was seen in wild-type apices. By contrast, AM1-2 and OE1 plants contained multiple foci of WUS, STM, and CLV3 expression, consistent with the emergence of multiple rosettes observed. See Supplemental Figure 8 online for images of serial sections.
(F) In roots, CDKB2;1 was expressed in a spotty pattern in cells near the root tip. This is consistent with the root digital in situ data available (Brady et al., 2007; see Supplemental Figure 2 online). Like in the shoot, expression could not be detected in AM1-2 plants, whereas OE1 plants showed a weaker expression in a larger number of cells.
Requirement of CDKB2 Function for Proper Organization of the SAM
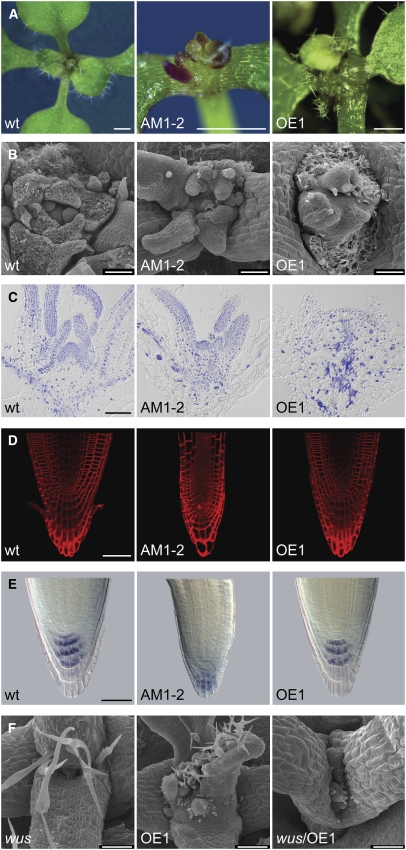
To test whether the identified CDKB2s play a role in the SAM, we analyzed plants in which expression levels of CDKB2;1 and/or CDKB2;2 were modified. As appropriate T-DNA insertion lines were not available for both genes, we used artificial microRNAs (amiRNAs) to assess loss-of-function phenotypes (Schwab et al., 2006; details on amiRNA design can be found in Supplemental Figure 4 online). Plants harboring constructs in which the cauliflower mosaic virus 35S (35S) promoter drove expression of amiRNAs targeting CDKB2;1 (AM1) or CDKB2;2 (AM2) individually did not display aberrant phenotypes. However, when we introduced both constructs simultaneously, we recovered double transformants showing dwarfism, abnormal structure of the shoot meristem, and phyllotaxis defects. To confirm that silencing of both CDKB2 genes caused the observed phenotypes, we designed a third amiRNA that targeted both transcripts for degradation (AM1-2). Thirty-five percent of the resulting T1 transgenic plants recapitulated the phenotype seen in AM1/AM2 double transformants and showed severely disturbed meristem structure and strong overall growth inhibition (Figures 2A to 2C).
Figure 2.
Phenotypes of AM1-2 Double Knockdown and 35S:CDKB2;1 (OE1) Plants.
The genotype is indicated at the lower left of each picture.
(A) Apices of 15-d-old plants. At this age, the emergence of multiple rosettes from the disorganized apex was observed in both AM1-2 and OE1 plants. Bars = 500 μm.
(B) Scanning electron micrographs of apices from 12-d-old seedlings. The strict organization seen at the wild-type apex was disrupted in both AM1-2 and OE1 plants. Bars = 90 μm.
(C) Overlays of differential interference contrast bright-field images and 4′,6-diamidino-2-phenylindole nuclear stainings. Nuclei appear blue. The dome-shaped meristem-like structures found in AM1-2 and OE1 plants contained fewer cells than wild-type meristems. In addition, several nuclei of AM1-2 and OE1 plants were abnormally expanded, and in OE1 plants, these were accompanied by abnormally large cells. Bar = 100 μm.
(D) FM4-64 staining of root tips. The organization of the root meristem is maintained in both AM1-2 and OE1 plants. Bar = 50 μm.
(E) Lugol staining of root tips. Starch grains are deposited normally in the columella cells in both transgenic lines. Bar = 50 μm.
(F) Phenotype of wus/35S:CDKB2;1 (OE1) double mutant plants. Homozygous wus mutant plants produced the first set of fully developed true leaves before terminating in a flat apex with no discernible meristem. Homozygous OE1 plants produced the first set of true leaves, followed by swelling of the apex and initiation of multiple irregularly spaced rosettes. wus/OE1 double homozygous plants displayed developmental arrest and completely failed to produce organs. Bars = 200 μm.
We observed a similar, albeit weaker, phenotype in plants overexpressing either CDKB2;1 or CDKB2;2 cDNAs from the 35S promoter. While meristematic activity was immediately blocked after germination in AM1-2 double knockdown lines, plants carrying CDKB2 overexpression constructs were able to initiate two to four true leaves before the meristem arrested. After a few days of apparent growth arrest, AM1-2 plants started to initiate radialized organs at multiple positions of the meristem, whereas in 35S:CDKB2;1 (OE1) plants, cells of the meristem expanded improperly and formed a bulge before organs emerged at irregularly spaced foci (Figures 2A to 2C). A summary of phenotype frequencies is given in Supplemental Table 1 online.
In contrast with AM1-2 lines, in which development irreversibly terminated, mildly affected 35S:CDKB2;1 plants were able to make the transition to flowering and produced seeds, which allowed us to establish a stable single insertion transgenic line. The offspring from these plants segregated for mild and severe phenotypes dependent on the copy number of the transgene. Hemizygous plants showed defects in phyllotaxis and organ spacing, while homozygous plants recapitulated the phenotypes of strong T1 lines described above (Figures 2A to 2C; see Supplemental Figure 5 online).
To determine whether the defects in meristematic organization are restricted to the SAM, we investigated the structure of the root apical meristem (RAM) in overexpression and double knockdown lines. Roots of 35S:CDKB2;1 plants were indistinguishable from wild-type roots in terms of root length and RAM organization, while the number of lateral roots was reduced (Figures 2D and 2E; see Supplemental Figure 6 online). Meristem organization was also largely unaffected in roots of AM1-2 plants, despite the fact that they consisted of fewer cells. By comparison with control lines, we found that kanamycin selection contributed little to the smaller root size of AM1-2 plants, indicating that it, like the shoot phenotype, was caused by the transgene. Due to the early manifestation of the shoot phenotype in AM1-2 plants, we were unable to distinguish whether the root phenotype was caused directly by aberrant CDKB2 activity in the root or whether it was an indirect effect of the reduced shoot size. Thus, we cannot rule out that CDKB2 disruption has a negative impact on root growth. However, since the structure of the root meristem was maintained in both CDKB2 overexpressors and double knockdown plants, it appears that the RAM was much more resistant to perturbations in CDKB2 activity than the SAM.
To investigate the basis for the similar phenotypes in overexpression and double knockdown plants, we analyzed CDKB2 expression by quantitative real-time RT-PCR (q-RT-PCR) and in situ hybridization. As expected, mRNA abundance of both CDKB2 transcripts in the shoot and root was reduced in AM1-2 double knockdown plants (Figures 3A and 3F; see Supplemental Figure 7 online). In 35S:CDKB2;1 plants, overall CDKB2;1 mRNA levels were increased in both root and shoot, although endogenous CDKB2;1 and CDKB2;2 expression was reduced (see Supplemental Figure 7 online). In situ hybridization revealed that the strong cell cycle–dependent expression of CDKB2;1 in discrete cells of the meristem was lost in these plants and was replaced by a uniform but weaker expression in all cells (Figures 3A and 3F). These findings suggest that strong, cell cycle–dependent expression of CDKB2;1 is required for its full function and that the similar phenotypes of knockdown and overexpression lines are likely caused by a disruption of this pattern.
CDKB2s Play a Role in Both Cell Cycle Regulation and Meristem Organization
Our analysis of tissue sections not only allowed us to asses the changes in expression levels of CDKB2;1 but also revealed abnormal cellular organization and morphology within the apices of both double knockdown and overexpression lines. While the basic dome shape of the meristem was maintained in our transgenic lines, the strict organization into three distinct tissue layers was disrupted (Figure 2C). In addition, cell number was significantly reduced in the meristems of knockdown and overexpression lines, and cells were dramatically enlarged in apices of 35S:CDKB2 plants. Using 4′,6-diamidino-2-phenylindole staining, we also observed abnormally large nuclei in double knockdown and overexpression lines (Figure 2C).
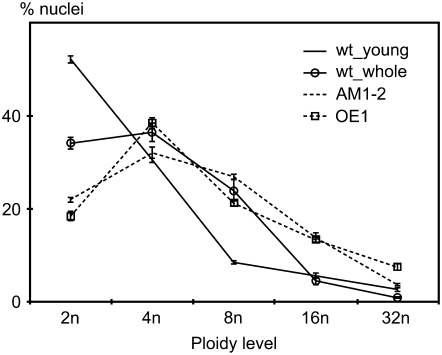
To asses whether the cell cycle was affected in these apices, we analyzed HISTONE H4 expression as a marker for cells in S-phase and found that while HISTONE H4 mRNA could be detected in a pattern similar to CDKB2 transcripts in the wild type, expression was strongly reduced in overexpression and double knockdown plants (Figure 3B). We then measured nuclear DNA content as an independent marker for cell cycle activity, since nonproliferating plant cells that undergo differentiation expand their DNA content by endoreduplication. To control for potential differences in leaf age and size, wild-type seedlings with and without the oldest set of leaves were included. Nuclei from both overexpression and double knockdown lines had significantly higher nuclear DNA content than either wild-type sample (Figure 4), consistent with a misregulation of the G2–M transition. The reduced cell cycle activity within the meristem and the increase in nuclear DNA content demonstrated that disruption of CDKB2 function causes defects in cell division control.
Figure 4.
Nuclear DNA Content of AM1-2 Double Knockdown and 35S:CDKB2;1 (OE1) Plants.
Ploidy of AM1-2 and OE1 plants was compared with wild-type control seedlings with (wt_whole) and without (wt_young) the oldest set of leaves. The horizontal axis indicates the genome copy number, and the vertical axis shows the percentage of nuclei counted. Error bars represent se. In both AM1-2 and OE1 plants, the relative number of 2n nuclei was reduced, while the number of 16n nuclei was increased compared with either wild-type sample, indicating a shift toward higher genome copy numbers in plants with altered CDKB2 activity.
Since both knockdown and overexpression lines had multiple centers of organ initiation, we asked whether meristem-organizing genes are ectopically activated in these functional niches. In situ hybridization on overexpression and knockdown plants showed that expression of WUS, STM, and CLV3 colocalized with multiple spots of apparent organ initiation within a single meristem (Figures 3C to 3E; see Supplemental Figure 8 online). This observation suggests that there might be feedback from the cell cycle to the network of meristem organizers and that ectopic activation of meristem regulators could be required for the formation of functional centers of activity. To test this hypothesis, we crossed the CDKB2;1 overexpression line to wus mutants and observed segregation of three distinct phenotypes in the F2 generation. The first phenotypic class corresponded to wus mutants (Laux et al., 1996), in which meristematic activity ceases after the formation of the first true leaves, resulting in a flat apex without a discernible SAM (Figure 2F). The second phenotypic class displayed all features of severely affected CDKB2;1 overexpression plants, which produce one set of leaves followed by disorganization of the meristem and reinitiation of organ formation from multiple sites (Figure 2F). Intriguingly, the third class exhibited a new phenotype. In these plants, the SAM never acquired activity and development was terminated after expansion of the cotelydons (Figure 2F). Genotyping showed that these plants were wus mutants and contained two copies of the 35S:CDKB2;1 transgene, while wus mutants hemizygous for the transgene were similar to nontransgenic mutants (see Supplemental Figure 9 online). Consistently, 12 (expected 14) out of 56 wus mutants displayed the completely arrested phenotype. Taken together, these results demonstrate that ectopic activation of WUS is required for formation of functional meristematic centers in plants with compromised CDKB2 activity and conversely that CDKB2s are required for the remaining organ forming capacity of wus mutants.
CDKB2 Activity Is Required for Proper Plant Hormone Signaling
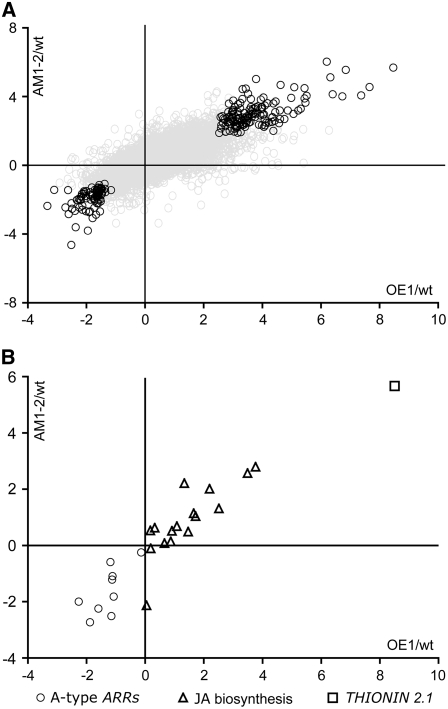
To elucidate the molecular basis for the phenotypes observed in lines with disrupted CDKB2 function, we performed expression-profiling experiments using Affymetrix Ath1 microarrays. Double knockdown and overexpression seedlings were compared with wild-type controls 10 d after germination in two biological replicates consisting of pools of six to 15 individuals each. The overall molecular signature of the two lines was similar, consistent with our findings at the phenotypic and cellular levels, where both lines showed meristems with fewer cells, increased ploidy levels, and multiple centers of meristematic activity. Accordingly, statistical testing using the Rank products algorithm (Breitling et al., 2004) identified 219 genes whose expression changed significantly in both of the CDKB2 transgenic lines (Figure 5A; see Supplemental Data Set 1 online). The direction of change for all these genes was the same in double knockdown and overexpression lines (Figure 5A), confirming that 35S:CDKB2;1 plants can be regarded as mild CDKB2 loss-of-function lines.
Figure 5.
Molecular Phenotypes of AM1-2 Double Knockdown and 35S:CDKB2;1 (OE1) Plants by Global Expression Analysis.
The horizontal axis shows the log2-transformed OE1/wild type expression ratio, and the vertical axis indicates the log2-transformed AM1-2/wild type expression ratio.
(A) Expression ratios of all genes (gray circles). The 219 genes that changed significantly in both conditions (percentage of false positives < 10%) are highlighted in black.
(B) Expression ratios of A-type ARRs, JA biosynthesis genes (JA biosynthesis), and THIONIN 2.1.
Analyzing the functional categories of the differentially expressed genes, we found many transcripts of the plant hormone signaling and response class among the top ranks. THIONIN 2.1 (AT1G72260) stood out as the most highly induced gene, changing 50- and 350-fold in knockdown and overexpression plants, respectively (Figure 5B). THIONIN 2.1 has been identified as a jasmonate (JA)–induced transcript (Bohlmann et al., 1998), and several transcripts coding for enzymes involved in JA synthesis were also elevated in both CDKB2 lines (Figure 5B). JA plays important roles in stress and pathogen signaling as well as in the regulation of growth and development (Farmer and Ryan, 1992; Sanders et al., 2000; Stintzi and Browse, 2000; Turner et al., 2002). It has also been shown that JA signaling is modulated by the cell wall status (Ellis et al., 2002), suggesting the changes in cell morphology observed in the CDKB2 disruption lines might trigger JA responses.
At the other end of the spectrum, the primary cytokinin response genes were most prominent among the genes expressed at lower than wild-type levels, and three A-type ARRs (ARR6, ARR15, and ARR16) were found among the 32 most reduced transcripts (Figure 5B; see Supplemental Data Set 1 online). Cytokinin is essential for meristem function, since it is required for cell cycle progression at multiple checkpoints (Zhang et al., 1996; Riou-Khamlichi et al., 1999), and removal of active cytokinin from the plant by overexpression of a cyctokinin oxidase leads to meristem termination (Werner et al., 2003). A-type ARRs have been implicated in a negative feedback regulation of cytokinin response (Kiba et al., 2003; To et al., 2004), and we have recently shown that ARR7 and ARR15 play prominent roles in meristem regulation (Leibfried et al., 2005).
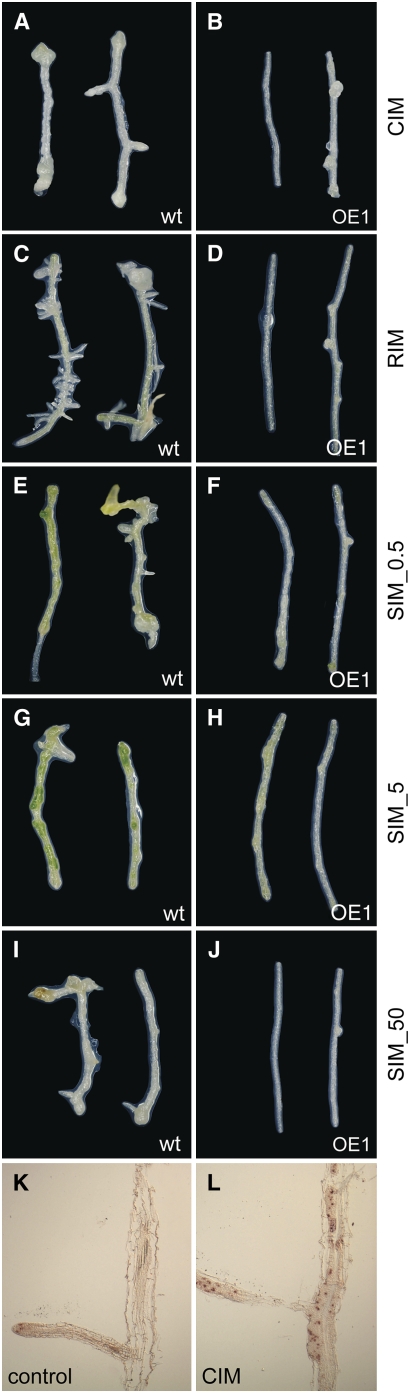
To directly test whether hormone-dependent developmental programs are affected in our CDKB2 lines, we performed tissue regeneration assays. Wild-type root explants can be induced to form callus, root, or shoot in vitro, depending on the relative levels of auxin and cytokinin in the regeneration medium. This response is dependent on the proper function of hormone signaling pathways and on developmental regulators (Reinert and Bajaj, 1977; Endrizzi et al., 1996; Kakimoto, 1996). While roots of CDKB2;1 overexpression plants were largely normal at the time of explantation, they were not able to respond properly to the hormone stimuli leading to regeneration and differentiation in wild-type roots. On callus induction medium, all wild-type root explants responded with formation of calli at the wounding sites. By contrast, explants from overexpression plants did not form calli but produced irregular outgrowths from young lateral roots (Figures 6A and 6B). The induction of roots by auxin was completely inhibited in these lines (Figures 6C and 6D). Explants growing on shoot induction medium with 0.5 to 5 μM cytokinin (2-iP) responded by limited cell proliferation and weak tissue greening, whereas wild-type explants formed clearly discernible green foci at 5 μM (Figures 6E to 6J). Increasing the cytokinin concentration in the shoot induction medium further to 50 μM 2-iP caused wild-type explants to produce callus, whereas overexpression explants showed no response (Figures 6I and 6J). We detected CDKB2 transcripts in proliferating calli by in situ hybridization, consistent with a role for these genes in tissue regeneration (Figures 6K and 6L). Bearing in mind that CDKB2 activity is not necessary for cell divisions in the root in general, these results demonstrate that normal CDKB2 activity is essential for the initiation of coordinated developmental programs in response to hormonal stimuli and suggest that the phenotypes observed in plants with disrupted CDKB2 function could at least in part be caused by the inability of cells to properly interpret hormone signals.
Figure 6.
Hormone Response of Wild-Type and 35S:CDKB2;1 (OE1) Tissue.
(A) to (J) Wild-type and OE1 root explants were subjected to hormone treatments to assess their regeneration capacity. Plant genotype and growth medium are indicated. CIM, callus induction medium; RIM, root induction medium; SIM_0.5, shoot induction medium with 0.5 μM 2-iP; SIM_5, shoot induction medium with 5 μM 2-iP; SIM_50, shoot induction medium with 50 μM 2-iP. OE1 roots were recalcitrant to tissue regeneration and, in contrast with wild-type explants, did not form calli, roots, or green foci in response to hormone treatment.
(K) and (L) Wild-type root explants after 5 d of incubation on either hormone-free (control) or CIM plates. Whereas CDKB2 expression in the control explants could only be detected in root tips, CDKB2 expression was detected throughout proliferating callus tissue in the explants incubated on CIM.
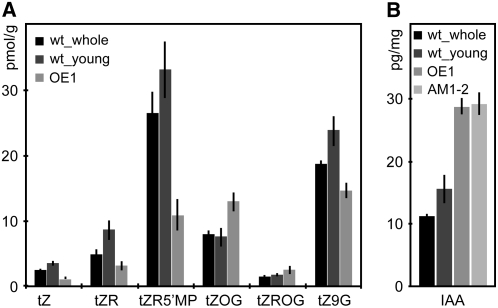
To distinguish whether the defects in hormone signaling we observed on the functional and transcriptional levels are due to changes in hormone levels or if they are brought about by a direct transcriptional modulation of response genes, we quantified hormone content in our CDKB2 disruption lines. Consistent with the former explanation, we found that auxin levels were significantly elevated in overexpression and knockdown lines (Figure 7). By contrast, the bioactive cytokinins, trans-zeatin (tZ), tZ-riboside, and tZ-5′monophosphate, were reduced, whereas the levels of inactive zeatin-O-glucosides were increased in overexpression seedlings (Figure 7). These results are consistent with the observed changes in gene expression and demonstrate that CDKB2 disruption plants suffer from a severe perturbation of the level of bioactive hormones.
Figure 7.
Hormone Content of 35S:CDKB2;1 (OE1) and AM1-2 Double Knockdown Plants.
Hormone levels of AM1-2 and OE1 plants were compared with wild-type control seedlings with (wt_whole) and without (wt_young) the oldest set of leaves.
(A) Quantification of trans-zeatin (tZ), its precursors tZ riboside (tZR) and tZ riboside 5′ monophospate (tZR5′MP), and conjugates tZ O-glucoside (tZOG), tZ riboside O-glucoside (tZROG), and tZ 9-glucoside (tZ9G). The bioactive nonglycosylated forms tZ, tZR, and tZR5′MP were present at lower levels in OE1 plants than in the wild-type controls, whereas the inactive O-glycosylated forms tZOG and tZROG were increased.
(B) Quantification of indole-3-acetic-acid (IAA). Both AM1-2 and OE1 plants contained significantly more IAA than either wild-type control. Error bars indicate sd. Cytokinin quantifications were performed on biological quadruplicates, and biological triplicates were used for IAA measurements.
DISCUSSION
In the course of evolution, the machinery controlling the eukaryotic cell cycle has undergone substantial changes. While unicellular organisms, such as yeast, rely on the activity of a singe CDK, higher land plants contain a plethora of CDKs. Among those, the B-type CDKs appear to be plant specific, and their origin can be traced back to the unicellular green algae Ostreococcus tauri. This member of a lineage that is ancestral to vascular plants has a single B-type CDK, which is regulated in a cell cycle–dependent manner similar to that of Arabidopsis CDKB1s and is able to complement the yeast cdc28 mutant (Corellou et al., 2005; Robbens et al., 2005). This function has been assumed by CDKA;1 in Arabidopsis, but B-type CDKs are still integral parts of the Arabidopsis cell cycle machinery as evidenced by the sharing of interaction partners, such as CYCD4;1, CDK inhibitors, and CKS1 by A- and B-type CDKs (Ferreira et al., 1991; De Veylder et al., 1997; Boudolf et al., 2001; Kono et al., 2003; Nakai et al., 2006). The duplication of B-type CDKs could have allowed them to take on more specific roles, such as those related to multicellularity. Consistent with such a hypothesis, CDKB1;1 is highly expressed in specialized cells that form stomatal complexes and is required for their development (Boudolf et al., 2004a), while we have now shown that CDKB2s are active in the SAM and necessary for its function.
Coordinated cell divisions are a prerequisite for the formation of organized multicellular tissues, but how much cell divisions actively contribute to the shaping of an organism is still a matter of debate (Gutierrez, 2005; Ramirez-Parra et al., 2005; Fleming, 2006; Inzé and De Veylder, 2006). The cellular theory claims that cell divisions are the main driving force of development, whereas the organismal theory proposes that cells are merely slaves to a higher-level developmental plan (Kaplan and Hagemann, 1991). To elucidate which of the principles underlie plant growth and development, cell division rates have been altered through manipulation of cell cycle regulator activity. Unfortunately, lethality has precluded the use of knockout alleles for many of the core regulators, such as CDKA;1 and RBR (Ebel et al., 2004; Iwakawa et al., 2006; Nowack et al., 2006). However, overexpression of functional and/or dominant-negative alleles of core cell cycle regulators has allowed cell division rates to be modified in planta. The effects of these manipulations on overall plant morphology reported so far have been mild, supporting the organismal theory (Hemerly et al., 1995; Doerner et al., 1996; Cockcroft et al., 2000; Boudolf et al., 2004b). For example, manipulation of CDKB1s and CDKA;1, the regulators most closely related to the CDKB2s, by overexpression of dominant-negative alleles did not lead to disturbed meristem structure (Hemerly et al., 1995; Boudolf et al., 2004b). By contrast, we now show that interfering with CDKB2 function not only causes abnormalities in cell cycle progression, but also in meristem organization. These phenotypes were not only observed in plants in which endogenous expression is knocked down by amiRNAs, a method new to the study of cell cycle regulators, but also in overexpression lines. This suggests that the defects are not merely a consequence of our knockdown strategy. While we certainly cannot rule out that knocking down other cell cycle regulators by amiRNAs would produce phenotypes similar to those seen for the CDKB2s, the results from interfering with the related CDKB1s and CDKA;1 by dominant-negative strategies suggest that the function in meristem organization is not shared by all cell cycle regulators.
The organizational activity of the CDKB2s is tightly integrated with the function of classical meristem regulators. Consistently, a strong additive effect was seen upon introducing the wus mutant allele into plants with reduced CDKB2 activity. These plants maintain some meristematic activity despite the perturbations in cell cycle control; however, when remaining organizational cues were removed by a mutation in WUS, the meristem was permanently arrested. This result demonstrates that meristem organization and cell division rates need to be coordinated for proper meristem function. Thus, our results are in line with the view that cell cycle regulators may serve as targets for the integration of cell proliferation with differentiation, morphogenesis, and growth (Gutierrez, 2005) and highlight the CDKB2s as such potential targets within the shoot meristem. The observed resistance of the root meristem to CDKB2 disruption could be explained by the activity of redundant regulators in the root, which are absent from the shoot. Alternatively, the regulatory machinery of the root meristem could rely on factors different from the CDKB2s to integrate meristem organization and cell cycle control, in line with the differences observed in meristem development between root and shoot. Variations in root and shoot responses have also been observed in overexpression of the CDK inhibitor KRP2, where leaves and lateral roots were severely affected, whereas main root growth was uninhibited (De Veylder et al., 2001; Himanen et al., 2002).
Further support for a tight integration of cell cycle control and developmental programs comes from the finding that plant hormone signaling is strongly modulated at multiple levels in plants with disrupted CDKB2 function. Auxin and cytokinin levels are known to control the cell cycle and many developmental programs throughout the plant (Skoog and Miller, 1957; Riou-Khamlichi et al., 1999; Himanen et al., 2002), and the levels of both hormones were changed in response to loss-of-CDKB2 function. A striking example for the action of auxin and cytokinin is tissue regeneration, where the auxin/cytokinin ratio not only determines cell division rates, but also the developmental fate of the tissue (Skoog and Miller, 1957). We have found that tissue with disrupted CDKB2 function cannot translate these stimuli into a proper developmental output, despite the fact that it was normal at the time of explantation. Thus, the developmental block observed is not merely a consequence of an inability of cells to proliferate, but rather underlines the importance for a coordination of cell cycle and developmental programs by hormonal signals. In the SAM, the central role of plant hormones in these processes has been highlighted by the identification of the pasticcino and tumorous shoot development mutants, which have disorganized meristems and altered cytokinin sensitivity (Frank et al., 2002; Harrar et al., 2003). In addition, cytokinin biosynthesis in the meristem is stimulated by STM, whereas WUS is thought to modulate cyctokinin response by directly repressing a subset of the A-type ARR primary cytokinin response genes (Leibfried et al., 2005; Yanai et al., 2005). Both the hormonal requirements and the meristem organizing factors differ between shoot and root meristems, and this could explain why disruption of CDKB2 activity had such a severe effect on the SAM, while RAM structure remained intact.
Our results demonstrate that cell cycle control by CDKB2s is essential for proper shoot meristem development and that hormones such as auxin and cytokinin might play an important role in relaying information between nodes of the regulatory network. Whether the CDKB2s act at the level of signal interpretation or in translating developmental programs into cellular behavior and whether there are other cell cycle regulators with similar functions represent intriguing questions for future study.
METHODS
Cloning of CDKB2 amiRNA and Overexpression Constructs
To create the 35S:CDKB2;1 and 35S:CDKB2;2 constructs, CDKB2;1 and CDKB2;2 cDNAs were PCR amplified with primers adding Gateway B1 and B2 sites to their open reading frames. The resulting PCR products were recombined into the pDONR221 vector and subsequently into a vector containing the 35S promoter in front of a Gateway cassette. The amiRNAs were created as previously described (Schwab et al., 2006) using Gateway tailed primers. All primer sequences are listed in Supplemental Table 2 online.
Plant Material
All plants were in the Columbia (Col-0) background and were grown in continuous light at 23°C and 65% relative humidity.
RNA Extraction and Expression Quantification
Double knockdown and overexpression seedlings showing the phenotypes described were compared with the wild type 10 d after germination in two biological replicates consisting of pools of 6 to 15 individuals each. For the double knockdown plants, pools of T1 plants grown on soil and selected with Basta were used, whereas pools of plants from a segregating T2 population grown on soil without selection were used for the overexpression line. RNA extraction and microarray analyses using the Affymetrix ATH1 platform were performed as described (Schmid et al., 2003). The data have been deposited at ArrayExpress (EMBL-EBI) under accession number E-MEXP-1100. Expression estimates were derived by GC-RMA at standard settings implemented in R. We determined significant changes on a per-gene level by applying the Rank products algorithm (Breitling et al., 2004) using 1000 permutations and a false discovery rate cutoff of 10%. For q-RT-PCR, total RNA was extracted using RNeasy Mini columns (Qiagen), and reverse transcription was performed with 1 μg of total RNA using the RevertAid first-strand cDNA synthesis kit (Fermentas). q-RT-PCR amplification was performed in the presence of the double-stranded DNA binding dye SYBR Green (Molecular Probes) and monitored in real time with the Opticon continuous fluorescence detection system (MJ Research). Amplification of TUBULIN BETA-2 served as cDNA loading control. Primer sequences are listed in Supplemental Table 3 online.
Genotyping
The wus mutants were genotyped using a derived cleaved amplified polymorphic sequence marker. Oligos 5′-GTAGTAAAGTTCTTTGAGGATTTTGGTTT-3′ and 5′-AGTCGAATCAAACACACATG-3′ were used for amplification, and the PCR product was then digested with MjaIV (Hpy8I). The 35S:CDKB2;1 transgene copy number was measured by q-RT-PCR. To amplify the internal genomic control, oligos 5′-GCTATCCACAGGTTAGATAAAGGAG-3′ and 5′-GAGAAAGATTGTGTGAGAATGAAA-3′ were used. The 35S:CDKB2;1 transgene was quantified using oligos 5′-CACGAGGAGCATCGTGGAAA-3′ and 5′-GTGAGGATCACGAGCGAGCA-3′.
In Situ Hybridizations and Microscopy
Scanning electron microscopy was performed as previously described (Schmid et al., 2003). In situ hybridization was done in accordance with standard protocols (Weigel and Glazebrook, 2002). FM4-64 (Molecular Probes) staining of roots was done by mounting roots directly in FM4-64 working solution (4 μg/mL) and viewing the stained samples with a confocal microscope. Starch granules in the columella root cap were visualized with 1% Lugol solution. Seedlings were stained for 1 min, rinsed with water, cleared with chloral hydrate, and photographed.
Ploidy Measurements
Nuclei from whole seedlings or from seedlings without the oldest set of true leaves were extracted and labeled using the CyStain PI Absolute P (05-5022; Partec) kit according to the manufacturer's instructions. Stained samples (biological triplicates) were analyzed with the BD LSRII flow cytometer, and the data were acquired with BD FACS DiVA software (BDBiosciences). More than 3800 nuclei were counted per sample.
Hormone Measurements
Quantification of IAA
Seedlings were pooled in triplicates, weighed, and frozen in liquid nitrogen for quantification of free IAA content. The frozen sample containing between 13 and 54 mg of tissue (fresh weight) was homogenized in 0.5 mL of 50 mM sodium phosphate buffer, pH 7.0, containing 0.02% diethyldithiocarbamic acid (antioxidant) and 500 pg 13C6-IAA internal standard, using the Retsch vibration mill (Retsch) and a 3-mm tungsten carbide bead at a frequency of 30 Hz for 2 min. The sample was then incubated for 15 min at +4°C with continuous shaking. The pH was adjusted to 2.7 with 1 M HCl, and the sample was purified by solid phase extraction on a 500-mg Isolute C8-EC column (International Sorbent Technology) conditioned with 2 mL of methanol and 2 mL of 1% acetic acid. The column was washed with 2 mL of 10% methanol in 1% acetic acid and eluted with 2 mL 70% methanol in 1% acetic acid, and the sample was evaporated to dryness. The sample was dissolved in 0.2 mL of 2-propanol and 1 mL of dichloromethane, and IAA was methylated by adding 5 μL 2 M trimethylsilyl-diazomethane in hexane (Aldrich) and incubating the sample at room temperature for 30 min. Five microliters of 2 M acetic acid in hexane was added to destroy excess diazomethane, and the sample was then evaporated to dryness. The methylated sample was then trimethyl-silylated and analyzed by gas chromatography–selected reaction monitoring–mass spectrometry as described (Edlund et al., 1995).
Quantification of Cytokinin
The procedure used for cytokinin analysis was a modification of a previously described method (Novak et al., 2003). Frozen plant material (∼0.1 g fresh weight) was extracted in Bieleski buffer using a vibration mill combined with ultrasonication. Deuterium-labeled CK internal standards (Olchemim) were added, each at 5 pmol per sample to check the recovery during purification and to validate the determination. The extracts were purified using a combined cation (SCX-cartridge) and anion (DEAE-Sephadex-C18-cartridge) exchanger and immunoaffinity chromatography based on wide-range specific monoclonal antibodies against cytokinins (Faiss et al., 1997). This resulted in three fractions: (1) the free bases, ribosides, and N-glucosides (fraction B), (2) a nucleotide fraction, and (3) an O-glucoside fraction. The metabolic eluates were evaporated to dryness and stored at −20°C until further analyses. CK fractions were quantified by ultra performance liquid chromatography (Acquity UPLC; Waters) coupled to a Quatro micro API triple quadrupole mass spectrometer (Waters) equipped with an electrospray interface. The purified samples were injected onto a C18 reversed-phase column (BEH C18; 1.7 μm; 2.1 ± 50 mm; Waters). The column was eluted with a linear gradient (0 min, 10% B; 0 to 8 min, 50% B; flow rate of 0.25 mL/min) of 15 mM ammonium formate, pH 4.0, (A) and methanol (B). Quantitation was obtained by multiple reaction monitoring of [M+H]+ and the appropriate product ion. In MRM mode, the limit of detection for most of cytokinins was below 5.0 fmol, and the linear range was at least five orders of magnitude.
Tissue Regeneration
Tissue regeneration was done using a modified procedure based on previously described protocols (Ezura and Harberd, 1995; Catterou et al., 2002; Che et al., 2002). Plants were grown for 15 d on germination plates (half strength Murashige and Skoog [1962] supplemented with 1% saccharose, 0.5 g/L MES, and 0.8% agar). Root explants of 5 to 10 mm in length were then moved to CIM (Gamborg's B5 medium [Gamborg et al., 1968] with 0.5 g/L MES, 2% glucose, 0.2 μM kinetin, and 2.2 μM 2,4-D, 0.8% agar) and incubated for 5 d in continuous light. Finally, explants were either left on CIM or moved to root-inducing medium (Gamborg's B5 medium with 0.5 g/L MES, 2% glucose, and 0.9 μM 3-indoleacetic acid) or shoot-inducing medium (Gamborg's B5 medium with 0.5 g/L MES, 2% glucose, 0.9 μM 3-indoleacetic acid, and 0.5, 5, or 50 μM 2-isopentenyladenine) and incubated for 10 d in continuous light.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers are as follows: CDKA;1 (AT3G48750), CDKB1;1 (AT3G54180), CDKB2;1 (AT1G76540), CDKB2;2 (AT1G20930), CKS1 (AT2G27960), CLV3 (AT2G27250), CYCD4;1 (AT5G65420), STM (AT1G62360), THIONIN 2.1 (AT1G72260), TUBULIN BETA-2 (AT5G62690), and WUS (AT2G17950). The microarray data set has been deposited at ArrayExpress (EMBL-EBI) under accession number E-MEXP-1100.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogeny and Expression Patterns of Arabidopsis CDKs.
Supplemental Figure 2. Expression of CDKB2s in Roots and during Callus Induction.
Supplemental Figure 3. In Situ Hybridizations with CDKB2;1 and CDKB2;2 3′ Untranslated Region Probes.
Supplemental Figure 4. Alignment of CDKB2 Sequences with Indication of amiRNA Target Sites.
Supplemental Figure 5. Phenotype of Hemizygous 35S:CDKB2;1 (OE1) Plants.
Supplemental Figure 6. Root Phenotypes of Wild-Type and 35S:CDKB2;1 (OE1) Plants.
Supplemental Figure 7. CDKB2;1 and CDKB2;2 Expression Levels in Transgenic Lines.
Supplemental Figure 8. Serial Sections through Apices of AM1-2 Plants Showing Multiple Expression Foci of Meristem Regulator Genes.
Supplemental Figure 9. Genotypes of F2 WUS/35S:CDKB2;1 Plants.
Supplemental Table 1. Characterization of T1 CDKB2 Transgenic Lines.
Supplemental Table 2. Primers Used to Amplify Open Reading Frames and amiRNAs.
Supplemental Table 3. Primers Used in q-RT-PCR Quantifications.
Supplemental Data Set 1. Genes Showing Significant Change in Expression Level between Wild-Type and Transgenic Lines.
Supplementary Material
Acknowledgments
We thank Jürgen Berger for assistance with electron microscopy and Richard Clark, Kay Schneitz, Karin Schumacher, and Detlef Weigel for critically reading the manuscript. In addition, we thank Petra Amakorová for skillful technical assistance. This work was supported by the Max Planck Society, the SFB 446 funded by the Deutsch Forschungsgemeinschaft (J.U.L.); a Career Development Award of the Human Frontier Science Program and an EMBO Young Investigator Programme award to J.U.L.; a Carlsberg Foundation fellowship to S.U.A.; a fellowship of the Cusanuswerk to W.B.; and Grant MSM6198959216 of the Ministry of Education, Youth, and Sports of the Czech Republic to O.N.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jan U. Lohmann (jlohmann@meristemania.org).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bohlmann, H., Vignutelli, A., Hilpert, B., Miersch, O., Wasternack, C., and Apel, K. (1998). Wounding and chemicals induce expression of the Arabidopsis thaliana gene Thi2.1, encoding a fungal defense thionin, via the octadecanoid pathway. FEBS Lett. 437 281–286. [DOI] [PubMed] [Google Scholar]
- Boucheron, E., Healy, J.H., Bajon, C., Sauvanet, A., Rembur, J., Noin, M., Sekine, M., Riou Khamlichi, C., Murray, J.A., Van Onckelen, H., and Chriqui, D. (2005). Ectopic expression of Arabidopsis CYCD2 and CYCD3 in tobacco has distinct effects on the structural organization of the shoot apical meristem. J. Exp. Bot. 56 123–134. [DOI] [PubMed] [Google Scholar]
- Boudolf, V., Barroco, R., Engler Jde, A., Verkest, A., Beeckman, T., Naudts, M., Inzé, D., and De Veylder, L. (2004. a). B1-type cyclin-dependent kinases are essential for the formation of stomatal complexes in Arabidopsis thaliana. Plant Cell 16 945–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf, V., Rombauts, S., Naudts, M., Inzé, D., and De Veylder, L. (2001). Identification of novel cyclin-dependent kinases interacting with the CKS1 protein of Arabidopsis. J. Exp. Bot. 52 1381–1382. [PubMed] [Google Scholar]
- Boudolf, V., Vlieghe, K., Beemster, G.T., Magyar, Z., Torres Acosta, J.A., Maes, S., Van Der Schueren, E., Inzé, D., and De Veylder, L. (2004. b). The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16 2683–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady, S.M., Orlando, D.A., Lee, J., Wang, J.Y., Koch, J., Dinneny, J.R., Mace, D., Ohler, U., and Benfey, P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318 801–806. [DOI] [PubMed] [Google Scholar]
- Breitling, R., Armengaud, P., Amtmann, A., and Herzyk, P. (2004). Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573 83–92. [DOI] [PubMed] [Google Scholar]
- Catterou, M., Dubois, F., Smets, R., Vaniet, S., Kichey, T., Van Onckelen, H., Sangwan-Norreel, B.S., and Sangwan, R.S. (2002). hoc: An Arabidopsis mutant overproducing cytokinins and expressing high in vitro organogenic capacity. Plant J. 30 273–287. [DOI] [PubMed] [Google Scholar]
- Che, P., Gingerich, D.J., Lall, S., and Howell, S.H. (2002). Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 14 2771–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockcroft, C.E., den Boer, B.G., Healy, J.M., and Murray, J.A. (2000). Cyclin D control of growth rate in plants. Nature 405 575–579. [DOI] [PubMed] [Google Scholar]
- Corellou, F., Camasses, A., Ligat, L., Peaucellier, G., and Bouget, F.Y. (2005). Atypical regulation of a green lineage-specific B-type cyclin-dependent kinase. Plant Physiol. 138 1627–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., Krols, L., Terras, F., Landrieu, I., van der Schueren, E., Maes, S., Naudts, M., and Inzé, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Segers, G., Glab, N., Casteels, P., Van Montagu, M., and Inzé, D. (1997). The Arabidopsis Cks1At protein binds the cyclin-dependent kinases Cdc2aAt and Cdc2bAt. FEBS Lett. 412 446–452. [DOI] [PubMed] [Google Scholar]
- Dewitte, W., Riou-Khamlichi, C., Scofield, S., Healy, J.M., Jacqmard, A., Kilby, N.J., and Murray, J.A. (2003). Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner, P., Jorgensen, J.E., You, R., Steppuhn, J., and Lamb, C. (1996). Control of root growth and development by cyclin expression. Nature 380 520–523. [DOI] [PubMed] [Google Scholar]
- Ebel, C., Mariconti, L., and Gruissem, W. (2004). Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429 776–780. [DOI] [PubMed] [Google Scholar]
- Edlund, A., Eklof, S., Sundberg, B., Moritz, T., and Sandberg, G. (1995). A microscale technique for gas chromatography-mass spectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissues. Plant Physiol. 108 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, C., Karafyllidis, I., Wasternack, C., and Turner, J.G. (2002). The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi, K., Moussian, B., Haecker, A., Levin, J.Z., and Laux, T. (1996). The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10 967–979. [DOI] [PubMed] [Google Scholar]
- Ezura, H., and Harberd, N.P. (1995). Endogenous gibberellin levels influence in-vitro shoot regeneration in Arabidopsis thaliana (L.) Heynh. Planta 197 301–305. [DOI] [PubMed] [Google Scholar]
- Faiss, M., Zalubilova, J., Strnad, M., and Schmulling, T. (1997). Conditional transgenic expression of the ipt gene indicates a function for cytokinins in paracrine signaling in whole tobacco plants. Plant J. 12 401–415. [DOI] [PubMed] [Google Scholar]
- Farmer, E.E., and Ryan, C.A. (1992). Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell 4 129–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, P.C., Hemerly, A.S., Villarroel, R., Van Montagu, M., and Inzé, D. (1991). The Arabidopsis functional homolog of the p34cdc2 protein kinase. Plant Cell 3 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming, A.J. (2006). The co-ordination of cell division, differentiation and morphogenesis in the shoot apical meristem: A perspective. J. Exp. Bot. 57 25–32. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 1911–1914. [DOI] [PubMed] [Google Scholar]
- Frank, M., Guivarc'h, A., Krupkova, E., Lorenz-Meyer, I., Chriqui, D., and Schmulling, T. (2002). Tumorous shoot development (TSD) genes are required for co-ordinated plant shoot development. Plant J. 29 73–85. [DOI] [PubMed] [Google Scholar]
- Gamborg, O.L., Miller, R.A., and Ojima, K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50 151–158. [DOI] [PubMed] [Google Scholar]
- Gutierrez, C. (2005). Coupling cell proliferation and development in plants. Nat. Cell Biol. 7 535–541. [DOI] [PubMed] [Google Scholar]
- Harrar, Y., Bellec, Y., Bellini, C., and Faure, J.D. (2003). Hormonal control of cell proliferation requires PASTICCINO genes. Plant Physiol. 132 1217–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly, A., Engler Jde, A., Bergounioux, C., Van Montagu, M., Engler, G., Inzé, D., and Ferreira, P. (1995). Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 14 3925–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen, K., Boucheron, E., Vanneste, S., de Almeida Engler, J., Inzé, D., and Beeckman, T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14 2339–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé, D., and De Veylder, L. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40 77–105. [DOI] [PubMed] [Google Scholar]
- Ito, Y., Nakanomyo, I., Motose, H., Iwamoto, K., Sawa, S., Dohmae, N., and Fukuda, H. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313 842–845. [DOI] [PubMed] [Google Scholar]
- Iwakawa, H., Shinmyo, A., and Sekine, M. (2006). Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 45 819–831. [DOI] [PubMed] [Google Scholar]
- Kakimoto, T. (1996). CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274 982–985. [DOI] [PubMed] [Google Scholar]
- Kaplan, D.R., and Hagemann, W. (1991). The relationship of cell and organism in vacscular plants - Are cells the building-blocks of plant form? Bioscience 41 693–703. [Google Scholar]
- Kiba, T., Yamada, H., Sato, S., Kato, T., Tabata, S., Yamashino, T., and Mizuno, T. (2003). The type-A response regulator, ARR15, acts as a negative regulator in the cytokinin-mediated signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 44 868–874. [DOI] [PubMed] [Google Scholar]
- Kondo, T., Sawa, S., Kinoshita, A., Mizuno, S., Kakimoto, T., Fukuda, H., and Sakagami, Y. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313 845–848. [DOI] [PubMed] [Google Scholar]
- Kono, A., Umeda-Hara, C., Lee, J., Ito, M., Uchimiya, H., and Umeda, M. (2003). Arabidopsis D-type cyclin CYCD4;1 is a novel cyclin partner of B2-type cyclin-dependent kinase. Plant Physiol. 132 1315–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122 87–96. [DOI] [PubMed] [Google Scholar]
- Leibfried, A., To, J.P., Busch, W., Stehling, S., Kehle, A., Demar, M., Kieber, J.J., and Lohmann, J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438 1172–1175. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Moan, E.I., Medford, J.I., and Barton, M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379 66–69. [DOI] [PubMed] [Google Scholar]
- Magyar, Z., et al. (1997). Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 9 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K.F., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815. [DOI] [PubMed] [Google Scholar]
- Menges, M., de Jager, S.M., Gruissem, W., and Murray, J.A. (2005). Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 41 546–566. [DOI] [PubMed] [Google Scholar]
- Menges, M., Hennig, L., Gruissem, W., and Murray, J.A. (2003). Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53 423–442. [DOI] [PubMed] [Google Scholar]
- Menges, M., and Murray, J.A. (2002). Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30 203–212. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Nakai, T., Kato, K., Shinmyo, A., and Sekine, M. (2006). Arabidopsis KRPs have distinct inhibitory activity toward cyclin D2-associated kinases, including plant-specific B-type cyclin-dependent kinase. FEBS Lett. 580 336–340. [DOI] [PubMed] [Google Scholar]
- Novak, O., Tarkowski, P., Tarkowska, D., Dolezal, K., Lenobel, R., and Strnad, M. (2003). Quantitative analysis of cytokinins in plants by liquid chromatography-single-quadrupole mass spectrometry. Anal. Chim. Acta 480 207–218. [Google Scholar]
- Nowack, M.K., Grini, P.E., Jakoby, M.J., Lafos, M., Koncz, C., and Schnittger, A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38 63–67. [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra, E., Desvoyes, B., and Gutierrez, C. (2005). Balance between cell division and differentiation during plant development. Int. J. Dev. Biol. 49 467–477. [DOI] [PubMed] [Google Scholar]
- Reinert, J., and Bajaj, Y.P.S. (1977). Plant Cell, Tissue, and Organ Culture. (New York: Springer-Verlag).
- Riou-Khamlichi, C., Huntley, R., Jacqmard, A., and Murray, J.A. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283 1541–1544. [DOI] [PubMed] [Google Scholar]
- Robbens, S., Khadaroo, B., Camasses, A., Derelle, E., Ferraz, C., Inzé, D., Van de Peer, Y., and Moreau, H. (2005). Genome-wide analysis of core cell cycle genes in the unicellular green alga Ostreococcus tauri. Mol. Biol. Evol. 22 589–597. [DOI] [PubMed] [Google Scholar]
- Sanders, P.M., Lee, P.Y., Biesgen, C., Boone, J.D., Beals, T.P., Weiler, E.W., and Goldberg, R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Uhlenhaut, N.H., Godard, F., Demar, M., Bressan, R., Weigel, D., and Lohmann, J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130 6001–6012. [DOI] [PubMed] [Google Scholar]
- Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers, G., Gadisseur, I., Bergounioux, C., de Almeida Engler, J., Jacqmard, A., Van Montagu, M., and Inzé, D. (1996). The Arabidopsis cyclin-dependent kinase gene cdc2bAt is preferentially expressed during S and G2 phases of the cell cycle. Plant J. 10 601–612. [DOI] [PubMed] [Google Scholar]
- Skoog, F., and Miller, C.O. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 54 118–130. [PubMed] [Google Scholar]
- Soni, R., Carmichael, J.P., Shah, Z.H., and Murray, J.A. (1995). A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 7 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To, J.P., Haberer, G., Ferreira, F.J., Deruere, J., Mason, M.G., Schaller, G.E., Alonso, J.M., Ecker, J.R., and Kieber, J.J. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, J.G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14 (suppl.): S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmulling, T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai, O., Shani, E., Dolezal, K., Tarkowski, P., Sablowski, R., Sandberg, G., Samach, A., and Ori, N. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15 1566–1571. [DOI] [PubMed] [Google Scholar]
- Zhang, K., Letham, D.S., and John, P.C. (1996). Cytokinin controls the cell cycle at mitosis by stimulating the tyrosine dephosphorylation and activation of p34cdc2-like H1 histone kinase. Planta 200 2–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.