Abstract
Avenacins are antimicrobial triterpene glycosides that are produced by oat (Avena) roots. These compounds confer broad-spectrum resistance to soil pathogens. Avenacin A-1, the major avenacin produced by oats, is strongly UV fluorescent and accumulates in root epidermal cells. We previously defined nine loci required for avenacin synthesis, eight of which are clustered. Mutants affected at seven of these (including Saponin-deficient1 [Sad1], the gene for the first committed enzyme in the pathway) have normal root morphology but reduced root fluorescence. In this study, we focus on mutations at the other two loci, Sad3 (also within the gene cluster) and Sad4 (unlinked), which result in stunted root growth, membrane trafficking defects in the root epidermis, and root hair deficiency. While sad3 and sad4 mutants both accumulate the same intermediate, monodeglucosyl avenacin A-1, the effect on avenacin A-1 glucosylation in sad4 mutants is only partial. sad1/sad1 sad3/sad3 and sad1/sad1 sad4/sad4 double mutants have normal root morphology, implying that the accumulation of incompletely glucosylated avenacin A-1 disrupts membrane trafficking and causes degeneration of the epidermis, with consequential effects on root hair formation. Various lines of evidence indicate that these effects are dosage-dependent. The significance of these data for the evolution and maintenance of the avenacin gene cluster is discussed.
INTRODUCTION
Plants produce a huge array of secondary metabolites. These compounds confer selective advantages by protecting against pathogens, pests, and stress and possibly also by suppressing the growth of neighboring plants (Wink, 1999; Field et al., 2006). The ability of plants to perform in vivo combinatorial chemistry by mixing, matching, and evolving the genes for new natural product pathways is likely to have been critical for survival and diversification. Some plant secondary metabolites (e.g., flavonoids) have also been implicated in primary physiological functions, such as auxin transport, regulation of seed longevity, and dormancy (D'Auria and Gershenzon, 2005).
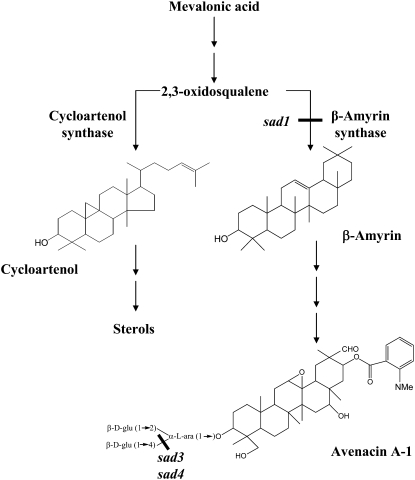
Roots produce a remarkable variety of compounds, including alkaloids, flavonoids, and terpenoids (Flores et al., 1999; Bertin et al., 2003; Bais et al., 2006; Field et al., 2006). These compounds can have marked effects on interactions between plants and other organisms. Oat (Avena) roots synthesize antimicrobial triterpenoid glycosides (saponins) known as avenacins, of which the major component is avenacin A-1 (Figure 1) (Crombie and Crombie, 1986; Crombie et al., 1986). These compounds confer resistance to a range of soil-borne pathogens (Papadopoulou et al., 1999). Avenacins are synthesized from mevalonic acid via the isoprenoid pathway and share a common biogenetic origin with sterols, the two pathways diverging after 2,3-oxidosqualene (Figure 1) (Hostettman and Marston, 1995; Trojanowska et al., 2000; Haralampidis et al., 2001b). The ability to produce avenacins is restricted to the genus Avena, and other cereals and grasses do not make these compounds (Hostettman and Marston, 1995). Therefore, transfer of this pathway into wheat (Triticum aestivum), rice (Oryza sativa), and other plants offers potential for new methods of disease control but is dependent on a better understanding of the biosynthetic process.
Figure 1.
Synthesis of Sterols and Triterpenoid Saponins in Oat.
The first committed step in the avenacin pathway is catalyzed by the oxidosqualene cyclase β-amyrin synthase (encoded by Sad1). sad3 and sad4 mutants accumulate avenacin A-1 lacking the β-1,4–linked d-glucose.
Avenacin A-1 is strongly fluorescent under UV illumination and causes oat roots to fluoresce bright blue (Turner, 1960; Papadopoulou et al., 1999). We have exploited this fluorescence to identify sodium azide–induced mutants of diploid oat (Avena strigosa) that are compromised in avenacin synthesis (Papadopoulou et al., 1999). To date, we have defined nine loci for avenacin biosynthesis, two of which have been cloned (Saponin-deficient1 [Sad1] and Sad2). Sad1 encodes β-amyrin synthase, which catalyzes the first committed step in the pathway (Figure 1) (Haralampidis et al., 2001a), and Sad2 encodes a cytochrome P450 monooxygenase that functions early in biosynthesis (Qi et al., 2006). Intriguingly, these two cloned genes along with five of the six other loci that we have defined genetically (Sad3 and Sad5 to Sad8) are clustered in the oat genome (Papadopoulou et al., 1999; Qi et al., 2004, 2006). Production of secondary metabolites is usually tightly regulated in plants, since the pathway end products and also their biosynthetic intermediates may have phytotoxic activity. Avenacin A-1 accumulates in the epidermal cells of oat roots, and its synthesis is highly localized (Turner, 1960; Osbourn et al., 1994). Expression of Sad1 and Sad2 occurs primarily in these cells and is under strict developmental control (Haralampidis et al., 2001a; Qi et al., 2006). Genes for metabolic pathways in plants are generally unlinked. Although the avenacin pathway has features in common with bacterial operons (gene clustering and coexpression), our evidence indicates that this pathway has evolved from plant components by gene duplication and rapid sequence divergence, and not by horizontal gene transfer (Haralampidis et al., 2001a; Qi et al., 2004, 2006). Therefore, this gene cluster is an example of a rapidly evolving coadapted gene complex.
Proper glycosylation of avenacins is critical for antifungal activity, since removal of a single glucose molecule from the trisaccharide sugar chain leads to a substantial reduction in toxicity (Osbourn et al., 1991). Indeed, some soil fungi produce glycosyl hydrolases that detoxify avenacin A-1 by removing one or more sugars, enabling them to tolerate the compound (Turner, 1961; Bowyer et al., 1995; Carter et al., 1999). In the plant, the addition of sugars to triterpene aglycones occurs late in the biosynthetic pathway and is likely to be important for stability and sequestration within the cell (Hostettman and Marston, 1995; Haralampidis et al., 2001a; Jenner et al., 2005). Mutants affected at seven of the nine loci that we have identified in our screen for avenacin-deficient mutants have normal root morphology apart from reduced root fluorescence (Papadopoulou et al., 1999). These include mutants affected at Sad1, the gene for the first committed enzyme in the pathway (Haralampidis et al., 2001a). Thus, although avenacins confer disease resistance, they are not required for normal root development. However, mutants affected at two other loci, Sad3 and Sad4, have reduced root growth and are root hair–deficient. sad3 and sad4 mutants are both affected in their ability to glucosylate avenacins and accumulate the same intermediate, monodeglucosyl avenacin A-1 (avenacin A-1 lacking the β-1,4–linked d-glucose). These mutants have epidermis-specific membrane trafficking defects that are suppressed by mutations at Sad1, indicating that accumulation of monodeglucosyl avenacin A-1 is the cause rather than a consequence of the observed root defects. Our evidence suggests that the Sad3 gene, which is part of the avenacin gene cluster, has a specific role in avenacin synthesis/sequestration, while Sad4 (which is unlinked to this region) has a broader function.
RESULTS
Mutations at Sad3 and Sad4 Affect Root Morphology
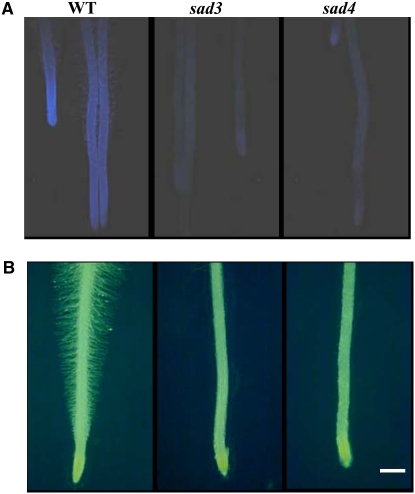
Previously, we screened germinating seedlings of a sodium azide–mutagenized population of diploid oat for reduced root fluorescence to identify mutants that were defective in avenacin biosynthesis (Papadopoulou et al., 1999). Two mutants, #1139 and #9, single mutants representing two different loci—Sad3 and Sad4, respectively—were identified among the mutants isolated during this screen. Both of these mutants accumulated incompletely glucosylated avenacin A-1. Sugar linkage analysis revealed that in both cases, the intermediate that accumulated was avenacin A-1 lacking the β-1,4–linked d-glucose (Figure 1; see Supplemental Table 1 online). These two mutants differed from the other root fluorescence mutants in our collection in that they had reduced numbers of root hairs (Figure 2). They also had reduced root growth (see Supplemental Table 2 online).
Figure 2.
Root Phenotypes of sad3 and sad4 Mutants.
Roots of 3-d-old oat seedlings showing the reduced fluorescence (A) and root hair–deficient (B) phenotypes of sad3 mutant #1139 and sad4 mutant #9. Bar = 2 mm.
F2 populations derived from wild type × mutant crosses were examined to determine whether the reduced root fluorescence phenotype cosegregated with the root morphology defect (short roots and reduced numbers of root hairs). Approximately 1000 F2 progeny were tested for each mutant. These were derived from backcrosses with A. strigosa accession S75 (the parent of the mutants) (Papadopoulou et al., 1999) and additionally from crosses with the two mapping lines CI1994 and CI3814 (Yu and Wise, 2000), both of which also produce avenacins. In all crosses, the reduced fluorescence phenotype cosegregated with both aspects of the root phenotype as single recessive mutations.
Sad3 and/or Sad4 may be required specifically for triterpene glycosylation. Alternatively, they may have functions in root growth and development that indirectly affect the glycosylation process.
Isolation of new Mutant Alleles of Sad3 and Sad4
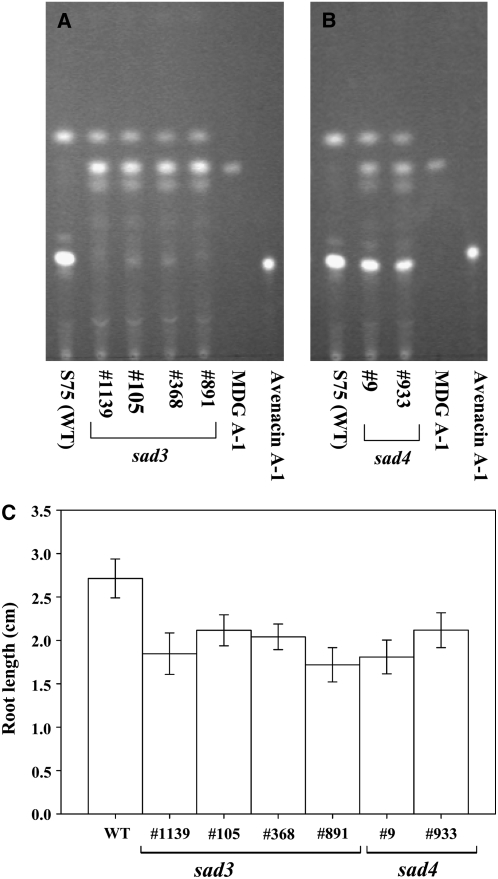
We then examined an extended collection of uncharacterized reduced root fluorescence mutants to identify more mutant alleles of Sad3 and Sad4. We found four mutants with the root morphology defects typical of mutants #1139 and #9. Allelism tests confirmed three of these as new sad3 mutants (#105, 368, and 891) and one as a new sad4 mutant (#933). Root extracts of the original sad3 mutant #1139 contained only trace amounts of avenacin A-1, the bulk of the triterpene accumulating as monodeglucosyl avenacin A-1 (Figure 3A). In contrast, the original sad4 mutant (#9) accumulated a mixture of avenacin A-1 and monodeglucosyl avenacin A-1 (Figure 3B). Root extracts of the new sad3 mutants resembled #1139 in containing monodeglucosyl avenacin A-1 but only trace amounts of avenacin A-1 (Figure 3A), while root extracts of the new sad4 mutant (#933) contained a mixture of avenacin A-1 and monodeglucosyl avenacin A-1 and so resembled mutant #9 (Figure 3B). Thus, the differences in the relative amounts of avenacin A-1 and monodeglycosyl avenacin A-1 in sad3 and sad4 mutants appear to be consistent features of mutations at these loci. Like mutants #1139 and #9, the new sad3 and sad4 mutants all had shorter roots compared with the wild type (Figure 3C) and were deficient in root hairs.
Figure 3.
Characterization of New Mutant Alleles of sad3 and sad4.
(A) and (B) Thin-layer chromatography analysis of root extracts from wild-type oat and from sad3 (A) and sad4 (B) mutant lines. Avenacin A-1 and monodeglucosyl avenacin A-1 (MDG A-1) were visualized under UV illumination.
(C) Mean root lengths (±se) of 3-d-old wild-type and mutant seedlings (sample size, 25).
sad3 and sad4 Mutants Have Root Epidermis Defects
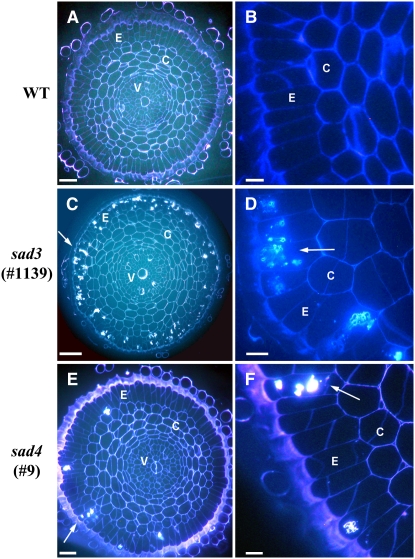
Propidium Iodide Staining
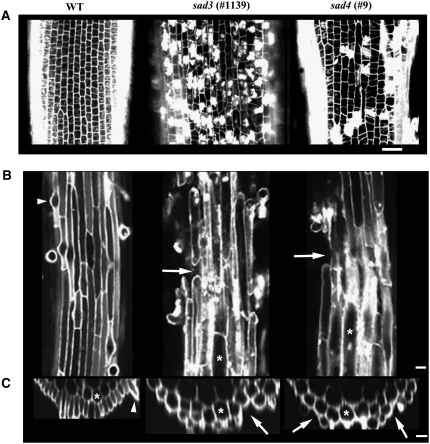
The roots of 2-d-old seedlings of wild-type, sad3, and sad4 lines were stained with propidium iodide (a fluorescent dye that stains nucleic acids and also outlines the periphery of the cells) and imaged by confocal microscopy. Propidium iodide staining of cell contents was restricted to the root epidermis and was not observed in the underlying cell layers. The epidermal cells of sad3 and sad4 mutants in the meristematic region of the root were irregular in shape and size compared with those of the wild type, and many of these cells stained strongly with propidium iodide (Figure 4A). The phenotype of sad3 mutants was more severe than that of sad4 mutants (shown for mutants #1139 and #9, respectively; Figure 4A).
Figure 4.
Degeneration of the Epidermal Layer in sad3 and sad4 Mutants.
(A) Longitudinal optical sections of roots of 2-d-old wild-type and mutant propidium iodide–stained seedlings, showing the epidermal cell layer in the meristematic zone. Bar = 25 μm.
(B) Longitudinal sections through the epidermal layer of roots of 3-d-old wild-type and mutant propidium iodide–stained seedlings, showing the cellular organization at the beginning of the differentiation zone. Bar = 25 μm.
(C) Optical cross sections of roots shown in (B). Gaps in the epidermis of sad3 and sad4 are indicated by arrows, and exposed cortical cells are indicated by asterisks. Bar = 25 μm.
We also examined the differentiation zone of the roots following propidium iodide staining. In 2-d-old seedlings, the appearance of the root epidermis in the differentiation zone of the mutants was similar to that of the wild type. However, in roots of 3-d-old seedlings, the epidermis of the mutants was defective (Figure 4B). Optical cross sections through the roots of sad3 and sad4 mutants confirmed that the epidermis was discontinuous and that cortical cells were often in direct contact with the environment (Figure 4C).
These experiments indicate that the epidermal cells of roots of sad3 and sad4 seedlings degenerate. This process progresses as the seedlings develop. The obvious explanation for the reduced fluorescence and the root hair deficiency phenotypes of sad3 and sad4 mutants is the degeneration of the epidermis.
Saponin Localization
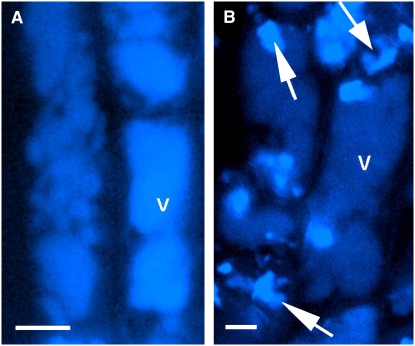
Previously, we showed that avenacin A-1 is the major UV fluorescent compound in wild-type oat roots (Osbourn et al., 1994). Confocal microscopy using excitation and emission conditions that detect avenacin A-1 revealed that the fluorescence associated with this saponin appears to be localized in the vacuoles of the root epidermal cells (Figure 5A). The root epidermal cells of sad1 mutants did not fluoresce under these conditions, confirming that the fluorescence observed in the wild type was due to the presence of avenacin A-1 (data not shown). sad3 mutants accumulate monodeglucosyl avenacin A-1 as the major fluorescent compound and contain only trace amounts of fully glycosylated avenacin A-1 (Papadopoulou et al., 1999) (Figure 3A). Therefore, the fluorescent patches seen in roots of these mutants are likely to be primarily due to this compound. Epidermal cells of roots of sad3 mutants have overall reduced fluorescence compared with the wild type, with patches of more intense fluorescence (shown for mutant #1139 in Figure 5B and in Supplemental Figure 1D online). These patches appeared to be either inside or just outside the vacuole. Irregular patches of fluorescence were also visible in the epidermal cells of roots of sad4 mutants (see Supplemental Figure 1F online). These data imply that sad3 and sad4 mutations directly or indirectly affect the appropriate sequestration and/or distribution of triterpene glycosides.
Figure 5.
Localization of UV Fluorescent Material in Wild-Type and Mutant Oat Root Epidermal Cells.
UV confocal microscopy of oat root epidermal cells showing uniform distribution of avenacin A-1 in vacuoles of wild-type roots (A) and patchy distribution of monodeglucosyl avenacin A-1 in sad3 roots (B) (arrow). v, vacuole. Bars = 100 μm.
Calcofluor
Further cytological analysis revealed the presence of Calcofluor-staining aggregates in the epidermal cells of roots of sad3 and sad4 mutants but not in the wild type (arrows in Figure 6). Calcofluor stains both cellulose and callose. This effect was more severe for sad3 than for sad4, as was the case with propidium iodide staining. At higher resolution, these aggregates often had a spaghetti-like appearance (Figures 6D and 6F). Autofluorescence associated with avenacin A-1 was lost during the fixation and embedding of the material, and fluorescence was not observed in the absence of Calcofluor staining. Staining with aniline blue, which detects callose but not cellulose, indicated that the aggregates contained callose (see Supplemental Figure 2 online), although they may also contain primary cell wall material as well.
Figure 6.
Epidermal Cells of Roots of sad3 and sad4 Mutants Contain Aggregates That Stain with Calcofluor.
Cross sections of roots of 2-d-old wild-type ([A] and [B]) and mutant ([C] to [F]) seedlings (meristematic zone), showing Calcofluor-staining aggregates (arrows). C, cortex; E, epidermis; V, vasculature. Bars = 50 μm ([A], [C], and [E]), 16 μm (B), 12 μm (D), and 17 μm (F).
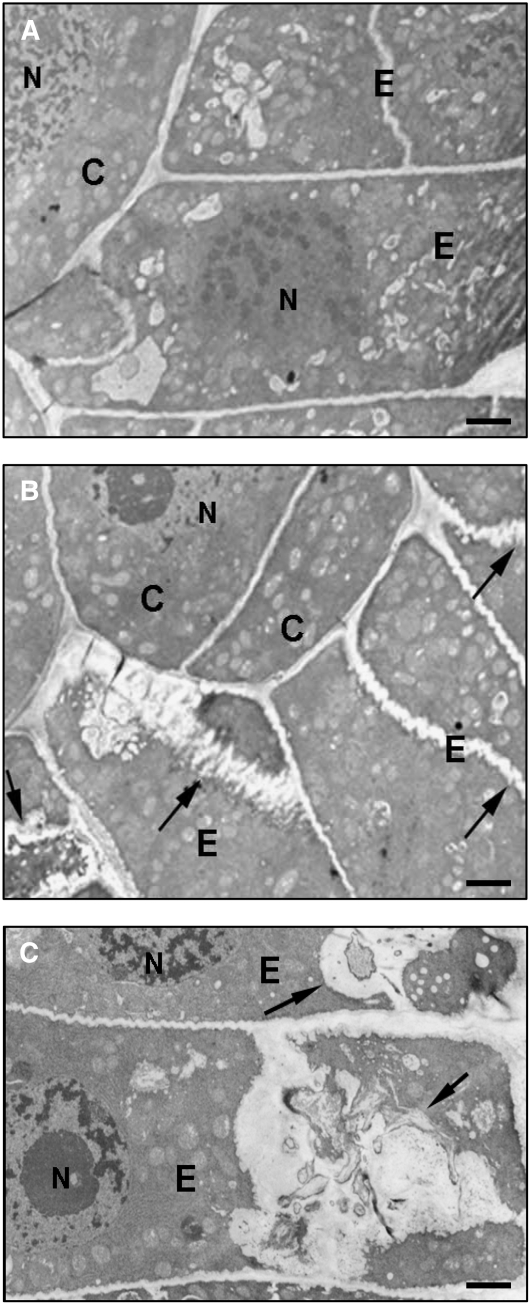
Transmission Electron Microscopy
We then investigated the nature of the epidermal defects of roots of sad3 and sad4 at the ultrastructural level using transmission electron microscopy (shown for the sad3 mutant #1139 in Figure 7). The margins of sad3 epidermal cells had a wavy appearance and appeared to be thickened in places compared with the wild type, suggestive of a cell wall and/or membrane defect (arrows in Figure 7B). In some epidermal cells, the infolding of the plasma membrane extended deep into the cytoplasm, forming sac-like structures (arrows in Figure 7C). The normal morphology of the nucleus and the endoplasmic reticulum of the epidermal cells containing these sacs indicates that these structures are not likely to be due to autolysis or an artifact of fixation. Similar defects were seen in occasional epidermal cells in roots of sad4 mutants (see Supplemental Figure 3 online). For both sad3 and sad4 mutants, the observed membrane and cell wall defects were restricted to the epidermal cells of the roots. Our transmission electron microscopy data suggest that sad3 and sad4 mutants have membrane trafficking defects.
Figure 7.
sad3 and sad4 Mutants Have Membrane Trafficking Defects.
Transmission electron micrographs of cross sections of the epidermal cells of roots of 2-d-old wild-type (A) and sad3 mutant ([B] and [C]) seedlings in the meristematic zone. C, cortex; E, epidermis; N, nucleus. Arrows indicate wavy/thickened appearance of cell margins (B) and sac-like structures (C) in sad3 mutants. Bars = 2 μm (A), 6 μm (B), and 1.8 μm (C).
Thus, sad3 and sad4 mutants have root epidermal cells with aberrant morphology that are defective in the glycosylation and sequestration of avenacin A-1, that accumulate callose, and that have membrane trafficking defects. These effects are more severe in sad3 mutants than in sad4 mutants. sad1 mutants had normal root morphology (apart from a lack of fluorescence), indicating that these effects were not due to a lack of avenacins. Our mRNA in situ data suggest very weak expression of avenacin biosynthesis genes in the root cap cells (Qi et al., 2006), and wild-type root cap cells are UV fluorescent, suggestive of the presence of avenacin A-1 (see Supplemental Figure 1 online). sad3 and sad4 mutants exhibit a distinct root cap defect: root cap cells form a thick ring of dark brown cells that adhere strongly to the root tip. They also have a patchy fluorescent phenotype resembling that seen in sad3 and sad4 mutant root epidermal cells (see Supplemental Figure 1 online).
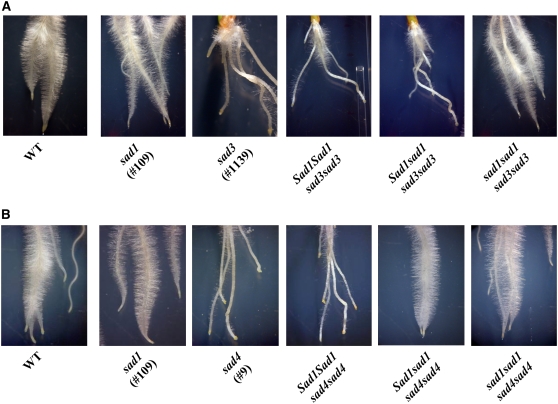
Characterization of sad1/sad1 sad3/sad3 and sad1/sad1 sad4/sad4 Double Mutants
The root epidermis defects that we observed in sad3 and sad4 mutants could be caused by the accumulation of monodeglucosyl avenacin A-1. Alternatively, they could be the result of mutations that affect root growth and development and that have an indirect effect on avenacin glucosylation. sad1 mutants are blocked in the first committed step in the avenacin pathway (Figure 1) and so do not accumulate avenacin A-1 or other pathway intermediates. These mutants have normal root morphology. We generated double mutants in which the homozygous sad3 or sad4 mutations were introduced into a sad1/sad1 mutant background to establish whether the accumulation of monodeglucosyl avenacin A-1 was the cause or the consequence of the root morphology defects.
Sad3 is part of the avenacin biosynthetic gene cluster and maps 3.6 centimorgan from Sad1 (Qi et al., 2004). Since Sad1 and Sad3 are linked, recombinants between these loci are rare. Therefore, we screened F2 progeny derived from a sad1 sad1 × sad3 sad3 cross for seedlings with the genotype Sad1/sad1 sad3/sad3, so that we could self these and follow segregation of the Sad1 locus in a sad3/sad3 mutant background. The F2 progeny derived from the sad1/sad1 × sad3/sad3 cross were genotyped for Sad1/sad1 by DNA sequence analysis (Haralampidis et al. 2001a). Sad3 has not yet been cloned. However, homozygous mutations at this locus give a clear biochemical phenotype (accumulation of monodeglucosyl avenacin A-1) in the Sad1/Sad1 and Sad1/sad1 backgrounds, enabling Sad1/Sad1 sad3/sad3 and Sad1/sad1 sad3/sad3 seedlings to be readily identified. Therefore, we were able to use a combination of genotyping and chemotyping to identify a single seedling with the genotype Sad1/sad1 sad3/sad3 among 305 F2 seedlings. This seedling had the root morphology phenotype typical of sad3 mutants, as expected. The progeny derived by selfing this plant were also analyzed by genotyping and chemotyping as described above, and the expected segregation patterns were observed (see Supplemental Table 3 online). Progeny with the genotypes Sad1/Sad1 sad3/sad3 and Sad1/sad1 sad3/sad3 accumulated monodeglucosyl avenacin A-1 and had root morphology defects (Figure 8A; see Supplemental Table 3 online). Progeny with the genotypes sad1/sad1 sad3/sad3 did not accumulate monodeglucosyl avenacin A-1 (see Supplemental Table 3 online), since the pathway is blocked in the first committed step (Figure 1). These progeny had normal roots (Figure 8A), indicating that accumulation of monodeglucosyl avenacin A-1 is the cause of the root morphology defects. Propidium iodide and Calcofluor staining confirmed that the epidermal cells of these double mutants were normal in appearance (apart from the reduced fluorescence phenotype).
Figure 8.
Root Phenotypes of sad1/sad1 sad3/sad3 and sad1/sad1 sad4/sad4 Double Mutants.
(A) sad1/sad1 sad3/sad3 double mutants have normal root morphology.
(B) The sad4 root morphology phenotype is dependent on the dosage of Sad1. Sad1/sad1 sad4/sad4 and sad1/sad1 sad4/sad4 mutants both have normal root morphology.
Four-day-old seedlings are shown.
Sad4 is unlinked to Sad1, so segregation of Sad1/sad1 in a sad4 mutant background can be followed in F2 progeny derived by crossing a homozygous sad1 mutant with a homozygous sad4 mutant and then selfing. Again, seedlings were genotyped for Sad1/sad1 and also chemotyped to identify Sad1/Sad1 sad4/sad4 and Sad1/sad1 sad4/sad4 lines using the methods described above. The segregation patterns of genotypes were consistent with a 9:3:4, ratio as expected for two unlinked loci (see Supplemental Table 4 online). Progeny with the genotypes Sad1_ Sad4_ (i.e., at least one Sad1 allele together with one Sad4 allele) accumulated avenacin A-1 (not monodeglucosyl avenacin A-1) and had wild-type root morphology, as expected. Progeny that were homozygous mutant for sad1 did not accumulate avenacin A-1 or monodeglucosyl avenacin A-1 and had normal root morphology (although these seedlings had reduced root fluorescence under UV illumination). Since sad1/sad1 sad4/sad4 double mutants must be within this group, this implies that these double mutants have normal root morphology, as is the case for sad1/sad1 sad3/sad3 double mutants. We expected all progeny with a Sad1_ sad4/sad4 genotype to have the root morphology phenotype typical of homozygous sad4 mutants. However, we found that this group of seedlings consisted of two subgroups: those with sad4 mutant root morphology and those with normal root morphology. Genotyping indicated that the seedlings within the former subgroup were all homozygous wild type for Sad1, while those in the latter were all Sad1/sad1 heterozygotes (see Supplemental Table 4 online). These data indicate that the manifestation of the sad4 root morphology phenotype is dependent on the dosage of Sad1.
We then went on to verify the phenotypes of Sad1/Sad1 sad4/sad4, Sad1/sad1 sad4/sad4, and sad1/sad1 sad4/sad4 seedlings by selfing plants with the genotype Sad1/sad1 sad4/sad4 and following the segregation of Sad1 and sad1 in a sad4/sad4 background. These experiments confirmed that Sad1/Sad1 sad4/sad4 seedlings have the root morphology defect and that sad1/sad1 sad4/sad4 seedlings have wild-type root morphology, as expected. They also verified that seedlings with the genotype Sad1/sad1 sad4/sad4 accumulate monodeglucosyl avenacin A-1 but have normal root morphology (Figure 8B). sad1/sad1 sad4/sad4 and Sad1/sad1 sad4/sad4 lines also had normal root epidermal cells, as assessed by propidium iodide and Calcofluor staining (data not shown).
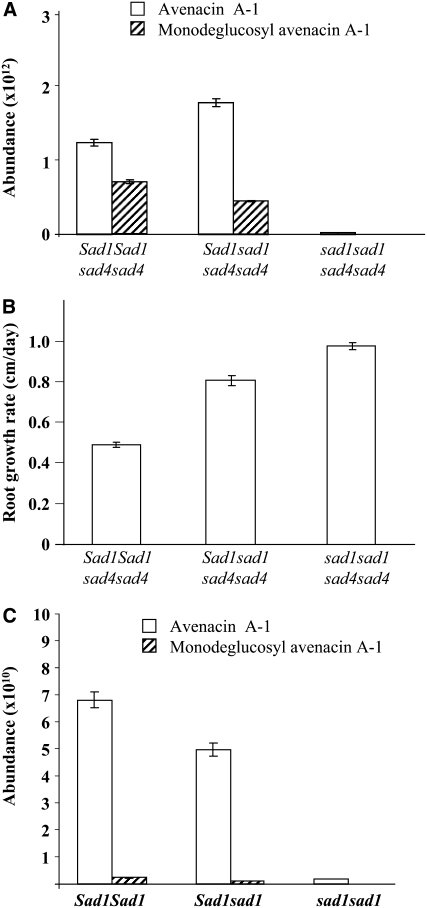
The ratios of avenacin A-1 to monodeglucosyl avenacin A-1 were significantly different when root extracts from seedlings with the genotypes Sad1/Sad1 sad4/sad4 and Sad1/sad1 sad4/sad4 were compared (∼2:1 and 4:1, respectively; Figure 9A). We also noted a small but significant reduction in the growth rates of the roots of Sad1/sad1 sad4/sad4 seedlings compared with sad1/sad1 sad4/sad4 seedlings, but this effect was far less severe than that observed for Sad1/Sad1 sad4/sad4 seedlings (Figure 9B). These data suggest that the severity of the root morphology defects may depend on the amounts of monodeglucosyl avenacin A-1 that accumulate. Direct comparison of the metabolite content of Sad1/Sad1 sad4/sad4 and Sad1/sad1 sad4/sad4 is complicated by the fact that Sad1/Sad1 sad4/sad4 seedlings have substantial root morphology defects while Sad1/sad1 sad4/sad4 seedlings do not. Therefore, we analyzed the avenacin A-1 content of F2 progeny derived from a cross between the wild-type oat line and a homozygous sad1 mutant (in the absence of sad3 or sad4 mutations) to establish whether the dosage effect of the Sad1 gene affected avenacin A-1 levels and found that this was indeed the case. Avenacin A-1 levels were higher in Sad1/Sad1 homozygotes than in Sad1/sad1 lines (Figure 9C). Collectively, these data suggest that Sad1/sad1 sad4/sad4 seedlings accumulate less monodeglucosyl avenacin A-1 than Sad1/Sad1 sad4/sad4, because the β-amyrin synthase enzyme is limiting in seedlings with the Sad1/sad1 genotype.
Figure 9.
Effects of Sad1 Gene Dosage on Avenacin A-1 Content.
(A) Liquid chromatography–mass spectrometry (LC-MS) analysis of the avenacin A-1 and monodeglucosyl avenacin A-1 content of root extracts of F3 progeny derived from a cross between sad1 and sad4 mutants. Values are means ± se of 25 seedlings.
(B) Growth rates (measured over a 4-d period) of roots of seedlings with the genotypes shown in (A). Values are means ± se for 25 seedlings.
(C) LC-MS analysis of root extracts of F2 seedlings derived from a cross between wild-type oat and a homozygous sad1 mutant (in the absence of sad3 or sad4 mutations). Values are means ± se of 25 seedlings.
DISCUSSION
sad3 and sad4 mutants are compromised in avenacin synthesis and sequestration. The roots of these mutants are stunted and have reduced numbers of root hairs. Cytological examination indicates that the reduced root hair phenotype is caused by defects in the root epidermal cells. Ultrastructural studies indicate that membrane trafficking is disrupted in these cells. The root epidermal cells of sad3 and sad4 mutants accumulate Calcofluor-staining material that contains callose and probably also primary cell wall components. Callose deposition in plants can be induced by wounding or pathogen attack, but it has also been observed in Arabidopsis thaliana mutants that have defects in cell wall biogenesis and cell expansion (Lukowitz et al., 2001; Ko et al., 2006). The accumulation of callose in sad3 and sad4 mutants may be a stress response caused by cytotoxic effects of monodeglucosyl avenacin A-1 and/or disruption of cell wall deposition/cell expansion. One of the manifestations of disruption of the epidermal cell layer is the root hair deficiency phenotype.
The morphological defects that we observed in sad3 and sad4 mutants could in principle be due to mutations that affect root growth and development and that have an indirect secondary effect on avenacin glucosylation. Mutations in genes that affect membrane trafficking in plants have been shown to affect cell wall metabolism, growth, and development (Jürgens, 2004; Johansen et al., 2006). In this case, effects on the synthesis and sequestration of avenacin A-1 would be an indirect consequence of defects in fundamental cellular processes. However, we have shown that the root morphology defects in both sad3 and sad4 mutants are suppressed in a sad1/sad1 mutant background. This implies that the accumulation of monodeglucosyl avenacin A-1 is the cause, rather than the consequence, of the reduced root growth and root hair deficiency phenotypes. Homozygous sad1 mutants lacking the entire avenacin biosynthetic pathway have no obvious defects in root development and morphology, although they are compromised in disease resistance (Papadopoulou et al., 1999; Haralampidis et al., 2001a). The effects that we observed in sad3 and sad4 mutants, therefore, are not due to an inability to synthesize avenacin A-1. Exogenous application of monodeglucosyl avenacin A-1 to roots of wild-type oat seedlings and also to tobacco (Nicotiana tabacum) BY2 cell cultures did not have any obvious effects on morphology or growth (data not shown). However, we were limited in the amounts of compound that we could apply due to availability (the maximum concentrations used were 5 to 10 μM); also, we cannot exclude the possibility that the compound may not have been taken up or may have been modified by endogenous glucosyltransferases. Interestingly, the pea (Pisum sativum) triterpene glycoside chromosaponin I stimulates Arabidopsis root growth when applied exogenously and also causes a reduction in root hair length and number. Chromosaponin I is believed to act through the regulation of auxin influx to affect various developmental processes in Arabidopsis, including ethylene-mediated responses and gravitropism (Rahman et al., 2000, 2001, 2002). However the significance of chromosaponin I for growth and development in pea is not known.
There are numerous reports that support the importance of glycosylation in cell division, growth, and morphogenesis both in animals and in plants. More specifically, glycosylation of secondary metabolites is a common mechanism of self-protection against compounds that are phytotoxic (von Rad et al., 2001; Bowles et al., 2005, 2006). A root-expressed pea UDP-glycosyltransferase, Ps UGT1, that glycosylates flavonoids has been shown to be essential for plant development, possibly via regulation of the cell cycle (Woo et al., 1999). Downregulation of the Arabidopsis ortholog of Ps UGT1 resulted in altered root growth, development, and gravity response (Woo et al., 2003, 2007). Similarly, loss-of-function mutations in an Arabidopsis glucosyltransferase required for glucosinolate biosynthesis gave a leaf chlorosis phenotype that has been ascribed to the accumulation of toxic levels of thiohydroximate, the substrate for this enzyme (Grubb et al., 2004). Other aspects of plant physiology have been reported to be affected by the glycosylation status of secondary metabolites. For example, partially glycosylated triterpenes and their aglycones can suppress induced defense responses in solanaceous plants (Bouarab et al., 2002; Ito et al., 2004).
Sad3 and/or Sad4 may encode glucosyltransferases that are required for the addition of the β-1,4–linked d-glucose molecule to the triterpene backbone of avenacin A-1. Alternative possibilities are that the Sad3 and/or Sad4 gene products are involved in the regulation of avenacin A-1 glucosylation or in avenacin transport/sequestration. Glycosylation is the final step in the synthesis of many plant secondary metabolites and occurs prior to entry into the vacuole (Hostettman and Marston, 1995; Wink, 1999; Frangne et al., 2002; Richman et al., 2005; Bowles et al., 2006; Martinoia et al., 2007), although there are examples of glucosylated compounds that cannot enter the vacuole unless they are further modified by acylation (Matern et al., 1986; Hopp and Seitz, 1987). The process by which avenacin A-1 is transported to the vacuole is not known. Glycosides, including endogenous secondary metabolites, may be transported into the vacuole by ATP binding cassette or multidrug and toxic compound extrusion transporters (Klein et al., 1996; Debeaujon et al., 2001; Bartholomew et al., 2002; Goodman et al., 2004; Yazaki, 2005). Alternatively, they may be delivered to the vacuole through the fusion of endoplasmic reticulum–derived vesicles (Poustka et al., 2007) or membrane-bound biosynthetic organelles (Bouvier et al., 2003).
Since sad3 mutants accumulate monodeglucosyl avenacin A-1 and contain only trace amounts of avenacin A-1, it is clear that the Sad4 gene product is unable to compensate for mutations at Sad3. By contrast, root extracts of sad4 mutants contain a mixture of avenacin A-1 and monodeglucosyl avenacin A-1. This is not likely to be attributable to weak mutant alleles, since both sad4 mutants show this effect. Previously, we showed that sad4 mutant #9 is also affected in the glucosylation of avenacosides, which are steroidal compounds found in oat leaves, while the sad3 mutant #1139 is wild type in this regard (Papadopoulou et al., 1999). Several lines of evidence suggest that the Sad3 gene product has a specific role in avenacin synthesis/sequestration, while the Sad4 gene product has a broader function. First, sad4 mutants are only partially affected in avenacin glucosylation, while sad3 mutants are fully compromised. Second, sad4 mutant #9 has been shown to be defective in glucosylation of avenacosides, implying that the Sad4 gene product has functions beyond avenacin synthesis and is not restricted to the roots. Third, Sad3 is part of the avenacin biosynthetic gene cluster, while Sad4 is unlinked to this region (Papadopoulou et al., 1999; Qi et al., 2004). In filamentous fungi, gene clusters for the production of specialized metabolites (e.g., toxins) are common, and it is often the case that genes that are important for detoxification and storage of the pathway end product (sugar transferases and transporters) are contained within the cluster (Yu and Keller, 2005). The maintenance of such genes within the clusters is presumably favored by selection, since disruption of the cluster will lead to the generation of deleterious intermediates. The fact that the Sad3 gene is genetically linked to the avenacin biosynthetic gene cluster further implicates Sad3 as a dedicated cluster component. Homozygous sad3 mutations have clear detrimental effects on plant growth in both Sad1/Sad1 and Sad1/sad1 backgrounds, indicating that the gene cluster is highly sensitive to perturbation. By contrast, homozygous mutations at the Sad4 locus (which segregates independently of Sad1) are detrimental in a Sad1/Sad1 background but can be tolerated in a Sad1/sad1 background. A future priority is to clone and characterize Sad3 and Sad4, since this should give insights into their function. This will also enable us to generate lines that are homozygous mutant for both sad3 and sad4 in order to carry out epistasy analysis.
To date, the only other well-characterized example of a gene cluster for a metabolic pathway in plants is that of the benzoxazinoids, a different class of defense-related compounds produced by maize (Zea mays) (Frey et al., 1997; Gierl and Frey, 2001). Interestingly, a putative biosynthetic gene cluster was also recently reported for momilactone (diterpene) synthesis in rice (Shimura et al., 2007). Thus, gene clusters for natural product pathways may be more common in plants than previously anticipated. Mechanisms that act to disperse genes (translocation, inversion, and unequal crossing over) are common in eukaryotes, and genes for most well-studied metabolic pathways are not clustered in plants. This raises some intriguing questions about why the genes for the avenacin, benzoxazinoid, and momilactone pathways are maintained as clusters. Clustering of genes has obvious advantages for coordinated regulation at the higher order levels of chromatin and nuclear organization. Clustering can also facilitate the inheritance of the genes as a functional unit and is likely to be selected for because the end products of these pathways confer an advantage (pest and pathogen resistance). Interference with the integrity of such clusters could result not only in loss of the ability to produce the protective pathway end product but also in the accumulation of deleterious intermediates (Gierl and Frey, 2001; Qi et al., 2004). Our experiments demonstrate that this is indeed the case.
METHODS
Plant Material
Wild-type and mutant oat (Avena strigosa) lines have been described previously (Papadopoulou et al., 1999; Yu and Wise, 2000; Qi et al., 2004). Methods for the isolation of new mutants and subsequent biochemical analysis were as described by Papadopoulou et al. (1999). For growth assessment and cytological analysis of mutants, seeds were dehusked, surface-sterilized in 5% sodium hypochlorite solution, and placed on 0.8% water agar. They were then stratified at 4°C for 1 to 2 d before being transferred to a growth chamber (16 h of light and 8 h of dark at 22°C). The agar plates were maintained at an angle of 25°.
Confocal Microscopy
Two- to 5-d-old seedlings were stained with 0.1 mg/mL propidium iodide solution for 15 to 25 min. The roots were imaged with a Leica SP confocal microscope (488 nm excitation, 598 to 651 nm emission). Images were processed with NIH Image (http://rsb.info.hih.gov/nih-image) and assembled using Adobe Photoshop 7. A MRC500 Bio-Rad confocal laser scanning microscope was used for imaging avenacin A-1 and monodeglucosyl avenacin A-1 (363 nm excitation, 450 to 465 nm emission) (Osbourn et al., 1994). Approximately 50 seedlings of each line were imaged per time point.
Fluorescence and Transmission Electron Microscopy
Roots were fixed in 2.5% (v/v) glutaraldehyde, 0.05 M sodium cacodylate, pH 7.3, and embedded in LR White resin (London Resin) as described by Lodwig et al. (2005). The material was sectioned with a glass knife using a Reichert ultramicrotome (Leica). For fluorescence microscopy, 0.5-μm-thick sections were dried onto glass slides and stained with Calcofluor (Sigma-Aldrich; 0.1% in water; 395 to 415 nm excitation and 455 nm emission) or with aniline blue (Biosupplies; 0.1% in water; 390 nm excitation and 480 nm emission) and viewed with a Nikon Eclipse E800 fluorescence microscope with a 4′,6-diamidino-2-phenylindole filter set. Photographs were taken with a Nikon Coolpix digital camera. For electron microscopy, ultrathin sections of ∼90 nm were picked up on 200 mesh copper grids that had been pyroxylin- and carbon-coated. The sections were stained with 2% (w/v) uranyl acetate and 2% lead citrate. The grids were viewed with a Jeol 1200 EX transmission electron microscope at 80 kV, and photographs were taken on Kodak electron image film.
Analysis of Segregating Progeny
Seeds were dehusked and surface-sterilized as described above. They were then stratified on moist filter paper in the dark at 4°C for 2 d before incubation at 22°C (16 h of light, 8 h of dark) for 3 d. Individual seedlings were assessed as follows. For root fluorescence, seedlings were scored for root fluorescence as described by Papadopoulou et al. (1999). For metabolite analysis, individual root tips (1-cm sections) were immersed in 75% methanol and left at 4°C overnight. The methanol extract was then dried down in a speed vacuum and resuspended in 10 μL of absolute methanol before being subjected to thin-layer chromatography analysis as described by Papadopoulou et al. (1999). Avenacin A-1, monodeglucosyl avenacin A-1, and other fluorescent components were visualized under UV illumination. For genotyping, DNA extracts of seedlings were genotyped for a characterized single nucleotide polymorphism at Sad1 by sequence analysis (Haralampidis et al., 2001a; Qi et al., 2001).
LC-MS Analysis
Semiquantitative analysis of avenacins was performed by LC-MS using a Thermo Finnigan Surveyor HPLC system (Thermo Scientific) equipped with a diode array detector and a DecaXPplus ion trap mass spectrometer (Thermo Scientific). Avenacins were separated on a 3 μm, 100 × 2 mm Luna C18(2) column (Phenomenex) at 30°C and a flow rate of 300 μL/min using the following gradient of increasing acetonitrile versus water:formic acid (99.9:0.1): 0 min, 20% acetonitrile; 3 min, 25%; 20 min, 50%; 30 min, 80%; 32 min, 80%. Detection was by positive ion electrospray mass spectrometry. Spray chamber conditions were 50 units of sheath gas, 5 units of auxiliary gas, 5.2 kV spray voltage, and 350°C capillary temperature. Quantitation was by integration of extracted ion chromatograms for the parent masses of the avenacins, whose identities were confirmed by tandem mass spectrometry at 35% collision energy and isolation width of 3 m/z.
Accession Numbers
Sequence data from this article can be found in the GenBank data library under accession numbers AY618699 (Sad1) and DQ680852 (Sad2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. sad3 and sad4 Mutants Have Root Cap Defects.
Supplemental Figure 2. Aniline Blue Staining of sad3 and sad4 Mutants.
Supplemental Figure 3. Epidermal Defects in sad4 Mutants.
Supplemental Table 1. Sugar Linkage Analysis of Monodeglucosyl Avenacin A-1 Intermediates Accumulating in sad3 and sad4 Mutants.
Supplemental Table 2. Reduced Growth Rate of Roots of sad3 and sad4 Mutants.
Supplemental Table 3. Analysis of F2 Progeny Derived by Selfing an Oat Plant with the Genotype Sad1/sad1 sad3/sad3.
Supplemental Table 4. Analysis of F2 Progeny Derived from a Cross between sad1 Mutant #109 and sad4 Mutant #9.
Supplementary Material
Acknowledgments
We thank Mike Leggett (Institute of Grasslands and Environmental Research, Aberystwyth, Wales) for carrying out oat crosses, Susan Bunnewell (John Innes Centre) for technical assistance and photography, and Rob Field (John Innes Centre) for valuable comments on the manuscript. This work was supported by Dupont Agricultural Products (P.M., H.J.), the Royal Thai Government (A.O.), the Biotechnology and Biological Sciences Research Council (X.Q., S.B.), the European Union (Marie Curie Fellowship to K.P.), and the Gatsby Charitable Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Anne Osbourn (anne.osbourn@bbsrc.ac.uk).
Online version contains Web-only data.
References
- Bais, H.P., Weir, T.L., Perry, L.G., Gilroy, S., and Vivanco, J.M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57 233–266. [DOI] [PubMed] [Google Scholar]
- Bartholomew, D.M., Van Dyk, D.E., Lau, S.M.C., O'Keefe, D.P., Rea, P.A., and Viitanen, P.V. (2002). Alternate energy-dependent pathways for the vacuolar uptake of glucose and glutathione conjugates. Plant Physiol. 130 1562–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin, C., Yang, X., and Weston, L.A. (2003). The role of root exudates and allelochemicals in the rhizosphere. Plant Soil 256 67–83. [Google Scholar]
- Bouarab, K., Melton, R., Peart, J., Baulcombe, D., and Osbourn, A. (2002). A saponin-detoxifying enzyme mediates suppression of plant defences. Nature 418 889–892. [DOI] [PubMed] [Google Scholar]
- Bouvier, F., Suire, C., Mutterer, J., and Camara, B. (2003). Oxidative remodeling of chromoplast carotenoids: Identification of the carotenoid dioxygenase CsCCD and CsZCD genes involved in crocus secondary metabolite biogenesis. Plant Cell 15 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles, D., Isayenkova, J., Lim, E.-K., and Poppenberger, B. (2005). Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 8 254–263. [DOI] [PubMed] [Google Scholar]
- Bowles, D., Lim, E.-K., Poppenberger, B., and Vaistij, E. (2006). Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 57 567–597. [DOI] [PubMed] [Google Scholar]
- Bowyer, P., Clarke, B.R., Lunness, P., Daniels, M.J., and Osbourn, A.E. (1995). Host range of a plant pathogenic fungus determined by a saponin detoxifying enzyme. Science 267 371–384. [DOI] [PubMed] [Google Scholar]
- Carter, J.P., Spink, J., Cannon, P.F., Daniels, M.J., and Osbourn, A.E. (1999). Isolation, characterization, and avenacin sensitivity of a diverse collection of cereal-root-colonizing fungi. Appl. Environ. Microbiol. 65 3364–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombie, W.M.L., and Crombie, L. (1986). Distribution of avenacins A-1, A-2, B-1 and B-2 in oat roots—Their fungicidal activity towards take-all fungus. Phytochemistry 25 2069–2073. [Google Scholar]
- Crombie, W.M.L., Crombie, L., Green, J.B., and Lucas, J.A. (1986). Pathogenicity of take-all fungus to oats—Its relationship to the concentration and detoxification of the 4 avenacins. Phytochemistry 25 2075–2083. [Google Scholar]
- D'Auria, J.C., and Gershenzon, J. (2005). The secondary metabolism of Arabidopsis thaliana. Curr. Opin. Plant Biol. 8 308–316. [DOI] [PubMed] [Google Scholar]
- Debeaujon, I., Peeters, A.J.M., Léon-Kloosterziel, K.M., and Korneef, M. (2001). The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13 853–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field, B., Jordán, F., and Osbourn, A. (2006). First encounters—Deployment of defence-related natural products by plants. New Phytol. 172 193–207. [DOI] [PubMed] [Google Scholar]
- Flores, H.E., Vivanco, J.M., and Loyola-Vargas, V.M. (1999). ‘Radicle’ biochemistry: The biology of root-specific metabolism. Trends Plant Sci. 4 220–226. [DOI] [PubMed] [Google Scholar]
- Frangne, N., Eggmann, T., Koblischke, C., Weissenböck, G., Martinoia, E., and Klein, M. (2002). Flavone glucoside uptake into barley mesophyll and Arabidopsis cell culture vacuoles. Energization occurs by H+-antiport and ATP-binding cassette-type mechanisms. Plant Physiol. 128 726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, M., Chomet, P., Glawischnig, E., Stettner, C., Grun, S., Winklmair, A., Eisenreich, W., Bacher, A., Meeley, R.B., Briggs, S.P., Simcox, K., and Gierl, A. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277 696–699. [DOI] [PubMed] [Google Scholar]
- Gierl, A., and Frey, M. (2001). Evolution of benzoxazinone biosynthesis and indole production in maize. Planta 213 493–498. [DOI] [PubMed] [Google Scholar]
- Goodman, C.D., Casati, P., and Walbot, V. (2004). A multidrug resistance-associated protein involved in anthocyanin transport in Zea mays. Plant Cell 16 1812–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb, C.D., Zipp, B.J., Ludiwig-Mueller, J., Masuno, M.N., Molinski, T.F., and Abel, S. (2004). Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 40 893–908. [DOI] [PubMed] [Google Scholar]
- Haralampidis, K., Bryan, G., Qi, X., Papadopoulou, K., Bakht, S., Melton, R., and Osbourn, A.E. (2001. a). A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc. Natl. Acad. Sci. USA 98 13431–13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haralampidis, K., Trojanowska, M., and Osbourn, A.E. (2001. b). Biosynthesis of triterpenoid saponins in plants. In Advances in Biochemical Engineering/Biotechnology, T. Scheper, ed (Berlin: Springer-Verlag), pp. 31–50. [DOI] [PubMed]
- Hopp, W., and Seitz, H.U. (1987). The uptake of acylated anthocyanin into isolated vacuoles from cell-suspension culture of Daucus carota. Planta 170 74–85. [DOI] [PubMed] [Google Scholar]
- Hostettmann, K.A., and Marston, A. (1995). Saponins. Chemistry and Pharmacology of Natural Products. (Cambridge, UK: Cambridge University Press).
- Ito, S.-I., Eto, T., Tanaka, S., Yamauchi, N., Takahara, H., and Ikeda, T. (2004). Tomatidine and lycotetraose, hydrolysis products of α-tomatine by Fusarium oxysporum tomatinase, suppress induced defence responses in tomato cells. FEBS Lett. 57 31–34. [DOI] [PubMed] [Google Scholar]
- Jenner, H., Townsend, B., and Osbourn, A.E. (2005). Unravelling triterpene glycoside synthesis in plants: Phytochemistry and functional genomics join forces. Planta 220 503–506. [DOI] [PubMed] [Google Scholar]
- Johansen, J.N., Vernhettes, S., and Höfte, H. (2006). The ins and outs of cell walls. Curr. Opin. Plant Biol. 9 616–620. [DOI] [PubMed] [Google Scholar]
- Jürgens, G. (2004). Membrane trafficking in plants. Annu. Rev. Cell Dev. Biol. 20 481–504. [DOI] [PubMed] [Google Scholar]
- Klein, M., Weisseböck, G., Dufaud, A., Gaillard, C., Kreuz, K., and Martinoia, E. (1996). Different energization mechanisms drive the vacuolar uptake of a flavonoid glucoside and a herbicide glucoside. J. Biol. Chem. 271 29666–29671. [DOI] [PubMed] [Google Scholar]
- Ko, J.-H., Kim, J.H., Jayanti, S.S., Howe, G.A., and Han, K.-H. (2006). Loss of function of COBRA, a determinant of oriented cell expansion, invokes cellular defense responses in Arabidopsis thaliana. J. Exp. Bot. 57 2923–2936. [DOI] [PubMed] [Google Scholar]
- Lodwig, E., Leonard, M., Marroqui, S., Wheeler, T., Findlay, K., Downie, J., and Poole, P. (2005). Role of polyhydroxybutyrate and glycogen as carbon storage compounds in pea and bean bacteroids. Mol. Plant Microbe Interact. 18 67–74. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Nickle, T.C., Meinke, D.W., Last, R.L., Conklin, P.L., and Somerville, C.R. (2001). Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 98 2262–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia, E., Maeshima, M., and Neuhuas, H.E. (2007). Vacuolar transporters and their essential role in plant metabolism. J. Exp. Bot. 58 83–102. [DOI] [PubMed] [Google Scholar]
- Matern, U., Reichenbach, C., and Heller, W. (1986). Efficient uptake of flavonoids into parsley (Petroselinum hortense) vacuoles requires acylated glycosides. Planta 167 183–189. [DOI] [PubMed] [Google Scholar]
- Osbourn, A.E., Clarke, B.R., Dow, J.M., and Daniels, M.J. (1991). Partial characterization of avenacinase from Gaeumannomyces graminis var. avenae. Physiol. Mol. Plant Pathol. 38 301–312. [Google Scholar]
- Osbourn, A.E., Clarke, B.R., Lunness, P., Scott, P.R., and Daniels, M.J. (1994). An oat species lacking avenacin is susceptible to infection by Gaeumannomyces graminis var. tritici. Physiol. Mol. Plant Pathol. 45 457–467. [Google Scholar]
- Papadopoulou, K., Melton, R.E., Leggett, M., Daniels, M.J., and Osbourn, A.E. (1999). Compromised disease resistance in saponin-deficient mutants. Proc. Natl. Acad. Sci. USA 96 12923–12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka, F., Irani, N.G., Feller, A., Lu, Y., Pourcel, L., Frame, K., and Grotewald, E. (2007). A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiol. 145 1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, X., Bakht, S., Devos, K.M., Gale, M.D., and Osbourn, A. (2001). L-RCA (ligation-rolling circle amplification): A general method for genotyping of single nucleotide polymorphisms (SNPs). Nucleic Acids Res. 29 E116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, X., Bakht, S., Leggett, M., Maxwell, C., Melton, R., and Osbourn, A. (2004). A gene cluster for secondary metabolism in oat—Implications for the evolution of metabolic diversity in plants. Proc. Natl. Acad. Sci. USA 101 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, X., Bakht, S., Qin, B., Leggett, M., Hemmings, A., Mellon, F., Eagles, J., Werck-Reichardt, D., Schaller, H., Lesot, A., Melton, R., and Osbourn, A. (2006). A different function for a member of an ancient and highly conserved cytochrome P450 family: From essential sterols to plant defense. Proc. Natl. Acad. Sci. USA 103 18848–18853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, A., Ahamed, A., Goto, N., and Tsurumi, S. (2001). Chromosaponin I specifically interacts with AUX1 protein in regulating the gravitropic response of Arabidopsis roots. Plant Physiol. 125 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, A., Hosokawa, S., Amakawa, T., Goto, N., and Tsurumi, S. (2002). Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiol. 130 1908–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, A., Tsurumi, S., Amakawa, T., Soga, K., Hoson, T., Goto, N., and Kamisaka, S. (2000). Involvement of ethylene and gibberellin signalings in chromosaponin I-induced cell division and cell elongation in the roots of Arabidopsis seedlings. Plant Cell Physiol. 41 1–9. [DOI] [PubMed] [Google Scholar]
- Richman, A., Swanson, A., Humphrey, T., Chapman, R., McGarvey, B., Pocs, R., and Brandle, J. (2005). Functional genomics uncovers three glucosyltransferases involved in the synthesis of the major glucosides of Stevia rebaudiana. Plant J. 41 56–67. [DOI] [PubMed] [Google Scholar]
- Shimura, K., et al. (2007). Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282 34013–34018. [DOI] [PubMed] [Google Scholar]
- Trojanowska, M.R., Osbourn, A.E., Daniels, M.J., and Threlfall, D.R. (2000). Biosynthesis of avenacins and phytosterols in roots of Avena sativa cv. Image. Phytochemistry 54 153–164. [DOI] [PubMed] [Google Scholar]
- Turner, E.M. (1960). The nature of resistance of oats to the take-all fungus. III. Distribution of the inhibitor in oat seedlings. J. Exp. Bot. 11 403–412. [Google Scholar]
- Turner, E.M. (1961). An enzymatic basis for pathogen specificity in Ophiobolus graminis. J. Exp. Bot. 12 169–175. [Google Scholar]
- von Rad, U., Huttl, R., Lottspeich, F., Gierl, A., and Frey, M. (2001). Two glucosyltransferases are involved in detoxification of benzoxazinoids in maize. Plant J. 28 633–642. [DOI] [PubMed] [Google Scholar]
- Wink, M. (1999). Biochemistry, role and biotechnology of secondary metabolites. Annu. Plant Rev. 3 1–16. [Google Scholar]
- Woo, H.H., Faull, K.F., Hirsch, A.M., and Hawes, M.C. (2003). Altered life cycle in Arabidopsis thaliana plants expressing PsUGT1, a UDP-gluconosyltransferase encoding gene from Pisum sativum. Plant Physiol. 133 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, H.-H., Jeong, B.R., Koo, K.B., Choi, J.W., Hirsch, A.M., and Hawes, M.C. (2007). Modifying expression of closely related UDP-glycosyltransferases from pea and Arabidopsis results in altered root development and function. Physiol. Plant. 130 250–260. [Google Scholar]
- Woo, H.H., Orbach, M., Hirsch, A.M., and Hawes, M.C. (1999). Meristem-localized inducible expression of a UDP-glycosyltransferase gene is essential for growth and development in pea and alfalfa. Plant Cell 11 2303–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki, K. (2005). Transporters of secondary metabolites. Curr. Opin. Plant Biol. 8 301–307. [DOI] [PubMed] [Google Scholar]
- Yu, G.X., and Wise, R.P. (2000). An anchored AFLP- and retrotransposon-based map of diploid Avena. Genome 43 736–749. [PubMed] [Google Scholar]
- Yu, J.H., and Keller, N. (2005). Regulation of secondary metabolism in filamentous fungi. Annu. Rev. Phytopathol. 43 437–458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.