Abstract
DNA damage tolerance (DDT) in budding yeast requires Lys-63–linked polyubiquitination of the proliferating cell nuclear antigen. The ubiquitin-conjugating enzyme Ubc13 and the Ubc enzyme variant (Uev) methyl methanesulfonate2 (Mms2) are required for this process. Mms2 homologs have been found in all eukaryotic genomes examined; however, their roles in multicellular eukaryotes have not been elucidated. We report the isolation and characterization of four UEV1 genes from Arabidopsis thaliana. All four Uev1 proteins can form a stable complex with At Ubc13 or with Ubc13 from yeast or human and can promote Ubc13-mediated Lys-63 polyubiquitination. All four Uev1 proteins can replace yeast MMS2 DDT functions in vivo. Although these genes are ubiquitously expressed in most tissues, UEV1D appears to express at a much higher level in germinating seeds and in pollen. We obtained and characterized two uev1d null mutant T-DNA insertion lines. Compared with wild-type plants, seeds from uev1d null plants germinated poorly when treated with a DNA-damaging agent. Those that germinated grew slower, and the majority ceased growth within 2 weeks. Pollen from uev1d plants also displayed a moderate but significant decrease in germination in the presence of DNA damage. This report links Ubc13-Uev with functions in DNA damage response in Arabidopsis.
INTRODUCTION
Cellular DNA is subject to assaults by environmental factors and endogenous metabolites. The alteration of DNA can lead to mutagenesis, genome rearrangements, and cell death (Friedberg et al., 2006). To maintain genome integrity, all living organisms have evolved a variety of DNA repair mechanisms to protect cells from DNA damage. However, despite great advances made during the last decade in the field of DNA repair and mutagenesis, the molecular mechanisms of DNA damage tolerance (DDT) in eukaryotes, especially in multicellular eukaryotes, have not yet been well characterized. In the lower eukaryote Saccharomyces cerevisiae, a DDT process known as DNA postreplication repair (PRR) facilitates DNA synthesis in the presence of replication-blocking lesions in the template. PRR consists of two branches: an error-prone (mutagenesis) branch and an error-free branch. The error-prone branch is mediated by specialized DNA polymerases, including Polζ (Rev3 + Rev7), Polη, and Rev1, which are required for translesion DNA synthesis (TLS). By contrast, the error-free branch is mediated by the ubiquitin (Ub)-conjugating enzyme (Ubc or E2)–Ubc variant (Uev) complex Ubc13-Mms2 (for methyl methanesulfonate2), which acts to prevent spontaneous and DNA damage–induced mutagenesis (Broomfield et al., 2001; Barbour and Xiao, 2003). It is now clear that yeast PRR is accomplished by stepwise covalent modifications of the proliferating cell nuclear antigen (PCNA) encoded by POL30. In response to DNA damage, the Rad6-Rad18 ubiquitination complex monoubiquitinates Pol30 at the Lys-164 residue, then the Mms2-Ubc13-Rad5 complex further polyubiquitinates Pol30 through Lys-63–linked chains (Hoege et al., 2002). It is thus assumed that monoubiquitinated Pol30 promotes error-prone TLS, whereas polyubiquitinated Pol30 promotes error-free bypass of damaged templates (Stelter and Ulrich, 2003; Pastushok and Xiao, 2004). The same Lys-164 residue can also be covalently modified by SUMO (for small ubiquitin-related modifier), which requires the Siz1-Ubc9 complex; sumoylated Pol30 recruits the DNA helicase Srs2 to stalled replication forks to prevent inappropriate recombination (Papouli et al., 2005; Pfander et al., 2005).
Ubc13 and Uev homologs are found in all eukaryotes examined to date (Villalobo et al., 2002; Pastushok and Xiao, 2004), suggesting that their functions are conserved throughout eukaryotes. Despite numerous reports of the isolation and characterization of eukaryotic UBC13 and MMS2/UEV1 genes and indications that they may be involved in DNA repair and damage tolerance, the functions of these genes remain uncharacterized in a multicellular organism. With regard to the fact that the deletion of UBC13 (Yamamoto et al., 2006a, 2006b) and possibly MMS2/UEV1 in mammals may cause embryonic lethality, we turned our attention to Arabidopsis thaliana as an alternative multicellular model organism to study the Lys-63 polyubiquitination and DDT pathway. Analysis of the Arabidopsis genome database indicates that most genes involved in DNA repair are highly conserved between plants and mammals (Tuteja et al., 2001). Most importantly, plants are much more tolerant of genomic instability and chromosome rearrangements, making the plant a suitable model organism to study DNA repair/tolerance defects.
Protein ubiquitination and its role in regulating protein degradation have been extensively studied in plants (Bachmair et al., 2001; Devoto et al., 2003; Vierstra, 2003; Moon et al., 2004). The conventional ubiquitination process is via Lys-48–linked Ub chain formation, which targets these proteins for degradation through 26S proteasomes (Hochstrasser, 1996). By contrast, Ub chains linked to substrates through Ub–Lys-63 regulate diverse functions in a nonproteolytic manner (Pickart, 2001a). To date, Ubc13 is the only known E2 capable of polyubiquitinating target proteins via Lys-63–linked chains, and this activity absolutely requires a Uev as a cofactor (Hofmann and Pickart, 1999; McKenna et al., 2001; Pastushok et al., 2005).
Arabidopsis genes involved in TLS, including REV3 (Sakamoto et al., 2003), REV1, REV7 (Takahashi et al., 2005), POLK (Garcia-Ortiz et al., 2004), and POLH (Curtis and Hays, 2007), have been isolated and characterized. TLS appears to play an important role in the tolerance of DNA damage in plants. We recently showed that two Arabidopsis UBC13 genes could complement a yeast ubc13 null mutant for spontaneous mutagenesis as well as interact with yeast and human Uev proteins (Wen et al., 2006), suggesting that the same pathway for an error-free DDT also exists in plants. However, the roles of UBC13 in DDT have not been directly assessed, and there has been no experimental evidence for plant Ubc13-Uev complex formation or for Lys-63–linked protein ubiquitination. In this report, we describe the molecular cloning and functional characterization of four Arabidopsis UEV1 genes and report a case of mutant phenotypes in DNA damage response when one of the UEV1 genes is inactivated.
RESULTS
Isolation of Arabidopsis UEV1 Genes
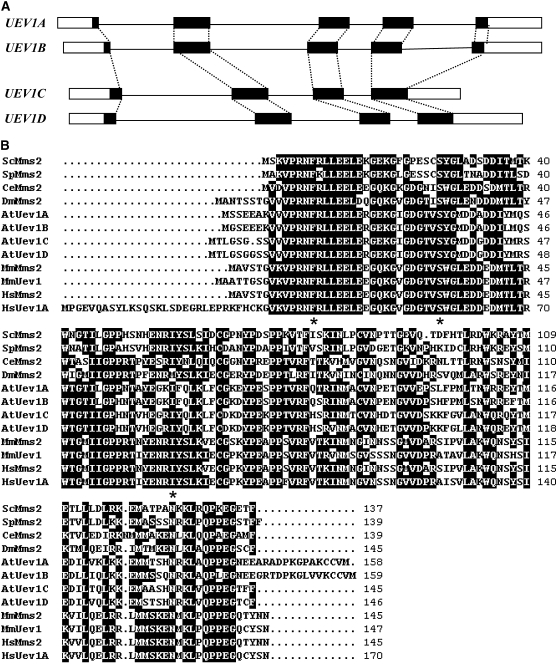
To identify Arabidopsis UEV1 genes, a human Mms2 sequence (Xiao et al., 1998) was used to search for homologs in the Arabidopsis protein database (through The Arabidopsis Information Resource [TAIR]; www.arabidopsis.org). Four hypothetical proteins with a high degree of similarity (E-values < 2e-38) were found and named At Uev1A (At1g23260), At Uev1B (At1g70660), At Uev1C (At2g36060), and At Uev1D (At3g52560).
The genomic structures of these four corresponding UEV1 genes are shown in Figure 1A. UEV1A and UEV1B have the same number of exons and introns, and their exon–intron junctions are identical. UEV1B has shorter intron sequences than UEV1A. Similarly, UEV1C and UEV1D have the same number of exons and introns, the same exon–intron junctions, and all introns in UEV1D are longer than those in UEV1C. Nucleotide sequence alignment of UEV1 open reading frames (ORFs) reveals 86% identity between UEV1A and UEV1B and 88% identity between UEV1C and UEV1D. Based on the above analyses, we predict that the four UEV1 genes resulted from two separate gene duplication events. This agrees with a duplication mapping analysis (http://wolfe.gen.tcd.ie.athal/dup) (Blanc et al., 2003). Further nucleotide sequence analysis of UEV1 promoter and downstream sequences does not reveal significant similarity between the two pairs of duplicated genes, indicating that they were derived from segmental duplications. This is consistent with database analysis (http://www.tigr.org/tdb/e2k1/athl/athl.shtml) and suggests that their expression profiles may be different.
Figure 1.
Sequence Analysis of At UEV1 Genes and Their Products.
(A) Genomic organization of UEV1. Open boxes, untranslated region; closed boxes, coding regions; solid lines, introns; dotted lines, identical intron–exon alignment between different UEV1 genes.
(B) Amino acid sequence alignment of At Uev1 and Uevs from six other organisms. The sequences were aligned and edited using the BioEdit program version 5.0.9 (Hall, 1999). Residues are highlighted when 50% or more are identical. Critical residues for Mms2/Uev functions are indicated with asterisks underneath the residues.
All four UEV1 ORFs were cloned from Arabidopsis by RT-PCR using gene-specific primers. The nucleotide sequences were identical to the annotated complete coding sequences in the Arabidopsis database. The predicted Uev1A, -B, -C, and -D proteins contain 158, 159, 145, and 146 amino acids, respectively, with differences in length primarily at the N or C terminus. Uev1A and Uev1B contain C-terminal tails not found in other Uevs (Figure 1B). Amino acid sequence alignment (Figure 1B) shows 86% identity between Uev1A and Uev1B and 92% identity between Uev1C and AtUev1D, whereas amino acid sequence identity between the two pairs is ∼75%.
The sequences of At Uev1 proteins were also aligned with those of Uev proteins from six other eukaryotic organisms, including human. As shown in Figure 1B, amino acid sequence identity between At Uev1s and those from other species ranges from 47 to 56%, and similarity ranges from 65 to 75%. Furthermore, several critical residues implicated in Uev activity are also conserved in At Uev1s. These residues include Phe-13 of Hs Mms2 required for physical interaction with Ubc13 (Pastushok et al., 2005) and Ser-32 and Ile-62 of Hs Mms2 (Pastushok et al., 2007) and the corresponding Ser-27 (Eddins et al., 2006) and Ile-57 (Tsui et al., 2005) of Sc Mms2 required for noncovalent interaction with Ub and polyubiquitin chain assembly. It is noted that mammals also contain two Uev proteins, Mms2 and Uev1A, with >91% amino acid sequence identity in their core domains (Xiao et al., 1998; Franko et al., 2001). Amino acid sequence comparison could not assign lineage between the two pairs of At Uev1 proteins and the mammalian Uevs, indicating that plant and animal Uevs evolved independently within their own kingdoms.
Phylogenetic analysis (see Supplemental Figure 1 online) was performed on At Uev1s in relation to Uevs from the above model organisms as well as with other plant species of known genomic sequence. This analysis revealed that plant UEV1 genes evolved from a common UEV1/MMS2 ancestor, which were duplicated and further evolved within each species. Hence, it would be of great interest to examine whether or how functions have evolved in the UEV family of genes.
Physical Interaction of At Uev1 with Ubc13 from Different Species
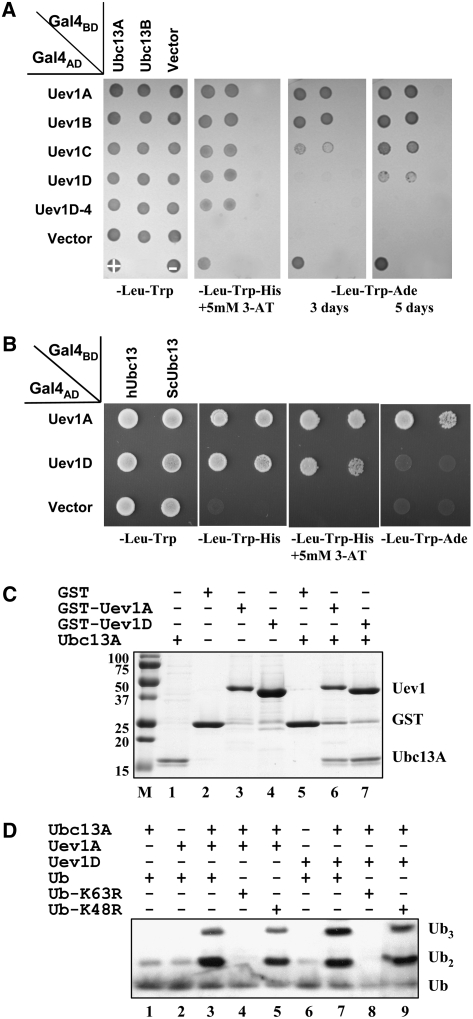
Both yeast and human Uevs play an essential role in Ubc13-mediated Lys-63–linked polyubiquitination; the prerequisite of this activity is that the Uev has to form a stable complex with Ubc13 (Hofmann and Pickart, 1999, 2001; McKenna et al., 2001). In order to detect this interaction, we performed a yeast two-hybrid assay (Fields and Song, 1989) between the cloned At UEV1s and UBC13 genes from different species. All four At Uev1 proteins were able to interact with either At Ubc13A or At Ubc13B; however, the strength of interaction appears to be different. At Uev1A and At Uev1B gave positive results with At Ubc13s under high stringency (SD-Ade for 3 d), but At Uev1C and At Uev1D gave weak and no interaction, respectively, under the same conditions (Figure 2A). Nevertheless, all of the above interactions are robust and deemed strong, as none of the negative controls reveal positive interactions under low stringency and many bona fide positive interactions may not survive as low as 1 mM 1,2,4-aminotriazole concentration under the same experimental conditions. The UEV1D-4 clone was identified among initial UEV1D clones; its ORF contains a three-nucleotide (GTA) insertion at position 175 that would encode the additional amino acid Val. This has been predicted to be a splicing variant of UEV1D (At3g52560.2) in the Arabidopsis genome database. Uev1D-4 appears to be able to interact with Ubc13, albeit at a reduced affinity compared with Uev1D (Figure 2A). The physiological significance of this variant has yet to be investigated. In addition, the yeast two-hybrid analyses showed that all four At Uev1 proteins are able to physically interact with Ubc13 from yeast or human, and the strength of interaction follows the trend Uev1A = Uev1B > Uev1C > Uev1D (Figure 2B; see Supplemental Figure 2A online).
Figure 2.
Biochemical Properties of Uev1.
(A) Physical interaction between Ubc13 and Uev1 in a yeast two-hybrid assay. The PJ69-4A transformants carrying one Gal4AD (from pGAD424) and one Gal4BD (from pGBT9) were replicated onto various plates as indicated and incubated for 3 d or as specified before being photographed. The result is representative of at least five independent transformants from each treatment.
(B) Physical interactions between At Uev1A/D and Ubc13 from yeast or human in a yeast two-hybrid assay. Experimental conditions were the same as in (A).
(C) Protein interactions between Uev1A/D and Ubc13 by an affinity pull-down assay. Purified GST (lane 5), GST-Uev1A (lane 6), or GST-Uev1D (lane 7) was added to GST microspin columns. Following incubation, the columns were spun and washed, and purified Ubc13A was added to the column. After reincubation and washing, the column contents were eluted with reduced glutathione, followed by SDS-PAGE gel analysis. Lanes 1 to 4 contain purified input proteins as indicated at top. Note that spontaneous cleavage occurred in the two GST-Uev1 protein samples (lanes 3 and 4).
(D) Ub conjugation by Ubc13, Uev1A, and Uev1D. An in vitro Ub conjugation assay was performed using purified proteins as indicated. Assay samples were subjected to SDS-PAGE, and a protein gel blot using an anti-Ub antibody was assayed to monitor poly-Ub formation. The low background of spontaneously formed di-Ub in the absence of E2 or Uev (lanes 1, 2, and 6) is commonly observed in these reactions (McKenna et al., 2001).
To further confirm the physical interaction between Uev1 and Ubc13 in vitro, a glutathione S-transferase (GST)–affinity pull-down assay was conducted. As shown in Figure 2C, purified GST-Uev1A (lane 6) and GST-Uev1D (lane 7) are able to specifically interact with Ubc13A. As a negative control, GST alone (lane 5) does not bind to Ubc13A under the same experimental conditions. Similar results were also obtained with Uev1B and Uev1C (see Supplemental Figure 2B online). Hence, all four Uev1 proteins are able to form stable heterodimers with Ubc13.
Uev1 Is Required for Ubc13-Mediated Lys-63–Linked Polyubiquitination in Vitro
It has been reported that while yeast and human Ubc13s are bona fide E2 enzymes capable of forming active-site thioesters with Ub, a Uev is absolutely required for Ub chain assembly. Furthermore, these chains are linked through Lys-63 instead of the conventional Lys-48 linkages (Hofmann and Pickart, 1999, 2001; McKenna et al., 2001). We previously reported the cloning and characterization of the At UBC13 genes (Wen et al., 2006). With the cloning of the UEV1 genes in this study, we were able to ask whether Ubc13 requires Uev1 for the assembly of Lys-63–linked poly-Ub chains. As shown in Figure 2D, Ubc13A and Ubc13D alone cannot generate free poly-Ub chains (lanes 2 and 6, respectively). Ubc13A with Uev1A (lane 3) and Uev1D (lane 7) can generate di- and tri-Ub chains. Furthermore, the poly-Ub chains generated are linked through Lys-63, since poly-Ub conjugates were not detected when using a Ub-K63R mutant that lacks Lys-63 (lanes 4 and 8), but were detected when using the Ub-K48R mutant that lacks the predominant Lys-48 residue for conjugation but retains Lys-63 (lanes 5 and 9). Similar results were also obtained with Uev1B and Uev1C (see Supplemental Figure 2C online).
At UEV1 Genes Functionally Complement Yeast mms2 Null Mutants
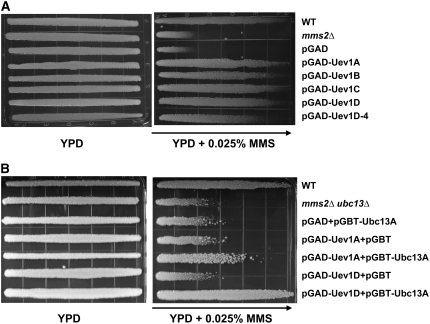
Yeast MMS2 is a member of the error-free DDT pathway and plays an important role in protecting yeast cells from mutagenesis and cell death caused by DNA-damaging agents (Broomfield et al., 1998). We performed yeast killing and spontaneous mutagenesis assays to determine whether At UEV1 could functionally complement the error-free PRR defect in the yeast mms2 null mutant. As shown in Figure 3A, expression of any one of the At UEV1 genes from the yeast two-hybrid plasmid rescued the mms2 mutant from killing by MMS to a level comparable to that in wild-type cells, whereas mms2 mutant cells transformed with the vector alone did not acquire any MMS resistance. It is interesting that the At UEV1D-4 clone also provides protection to mms2 cells, albeit at a slightly reduced level.
Figure 3.
Complementation of Yeast mms2 Mutants by At UEV1.
(A) Complementation of the mms2 single mutant by At UEV1. WXY942 (mms2Δ) transformants were grown overnight and printed onto YPD and YPD + 0.025% MMS gradient plates. The plates were incubated at 30°C for 2 d before being photographed. The arrow indicates higher MMS concentrations. Several transformants of each treatment were tested with the same result, and only one is shown here.
(B) Complementation of the mms2 ubc13 double mutant (WXY955) by At UEV1A, At UEV1D, and At UBC13. Experimental conditions were as in (A).
The complementation of yeast mms2 relies on heterodimer formation between At Uev1 and yeast Ubc13. In order to assess in vivo complex formation and functions between Arabidopsis Uev1 and Ubc13, we created a yeast mms2 ubc13 double mutant and cotransformed it with At UEV1 and At UBC13. When the double mutant cells were transformed with only At UBC13 or At UEV1, the transformed cells did not display enhanced resistance to MMS (Figure 3B), indicating that both Ubc13 and a Uev are required for the DDT function. Interestingly, when combined with At UBC13, UEV1C and UEV1D completely restored the MMS resistance to the wild-type level, whereas UEV1A and UEV1B barely rescued the host cells (Figure 3B; see Supplemental Figure 2D online). This result is in sharp contrast with the observations that all At UEV1s functioned equally well in the complementation of yeast mms2 single mutant (Figure 3A) and that Uev1A/B displayed higher binding capacity than Uev1C/D in yeast two-hybrid assays (Figures 2A and 2B; see Supplemental Figure 2A online).
One of the most astonishing phenotypes of a yeast mms2 (Broomfield et al., 1998) or ubc13 (Brusky et al., 2000) mutant is its massive increase in spontaneous mutagenesis, indicating that these genes play an important role in protecting cells from genome instability. Indeed, in this experiment, the mms2 mutant strain showed an increase of >20-fold in spontaneous mutagenesis compared with wild-type cells (Table 1). When the same mms2 mutant was transformed with a plasmid expressing an At UEV1, the spontaneous mutation rate was reduced to a level similar to that of the wild-type cells (Table 1). Again, UEV1C and UEV1D appear to be more effective than UEV1A and UEV1B in limiting mutagenesis. Collectively, the results obtained from the yeast complementation experiments suggest that At UEV1 genes are able to replace the PRR function of yeast MMS2 and that perhaps UEV1C and UEV1D are more efficient than UEV1A and UEV1B in such a function in Arabidopsis.
Table 1.
Effects of At UEV1 on the Spontaneous Mutation Rate of the S. cerevisiae mms2 Mutant
| Straina | Key Alleles | Rate (×10−8)b | Foldc |
|---|---|---|---|
| DBY747 | Wild type | 3.2 ± 0.18 | 1.00 |
| WXY642/pGAD424 | mms2Δ | 70.2 ± 7.96 | 22.10 |
| WXY642/At UEV1A | mms2Δ At UEV1A | 8.1 ± 0.16 | 2.53 |
| WXY642/At UEV1B | mms2Δ At UEV1B | 7.1 ± 0.13 | 2.22 |
| WXY642/At UEV1C | mms2Δ At UEV1C | 5.3 ± 0.98 | 1.66 |
| WXY642/At UEV1D | mms2Δ At UEV1D | 4.9 ± 0.56 | 1.53 |
All strains are isogenic derivatives of DBY747.
The spontaneous mutation rates are the average of three independent experiments with standard deviations.
Relative to the wild-type mutation rate.
At UEV1 Expression in Different Tissues and under Stresses
Since UEV1 is presumed to be involved in DDT and the ubiquitination process is often involved in stress responses, we analyzed UEV1 expression under various stress conditions. Arabidopsis cell suspension culture was subjected to treatments as indicated and total RNA was isolated for RNA gel blot hybridization. The results from samples of 24-h treatments are presented in Supplemental Figure 3A online. It appears that UEV1 expression is slightly decreased after treatment with MMS or H2O2 and slightly increased after treatment with abscisic acid or mannitol, although for the latter two treatments the transcript level of the control UBQ11 was also higher.
Since all four UEV1 genes share >72% nucleotide sequence identity in their core coding region and all four predicted transcripts are similar in size, we suspect that the UEV1C probe used for RNA gel blot hybridization actually detected all four UEV1 transcripts. Given the fact that the two human Uev homologs (UEV1A and hMMS2) play distinct roles in cellular metabolism (Andersen et al., 2005) and our observation in this study that the two pairs of UEV1 genes may function differently, it is important to assess the expression of individual UEV1 genes. To fulfill this objective, we analyzed the existing microarray data (available from www.Arabidopsis.org) for individual UEV1 gene expression profiles and found no evidence of strong stress responses after treatment of Arabidopsis plants (see Supplemental Figure 3B online). This analysis suggests that various environmental stresses used in this study have little effect on the expression of UEV1 genes at the transcriptional level.
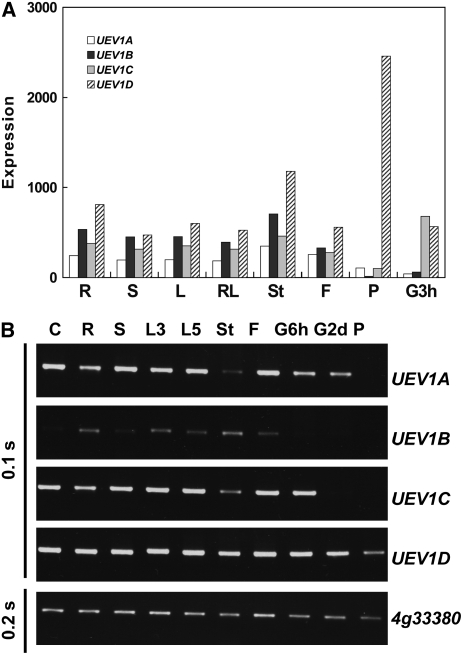
The expression of UEV1 genes in different tissues was also determined by RNA gel blot hybridization (see Supplemental Figure 3C online) and by analyzing the microarray data (Figure 4A; see Supplemental Figure 4 online). While most tissues express variable levels of each UEV1 transcript, UEV1D appears to show a higher level of expression than the other three UEV1 genes in most tissues examined (Figure 4A; see Supplemental Figure 4 online). Greater differences in transcript levels of the UEV1 genes were found in samples from pollen and geminating seeds. The microarray data indicate that 3 h after seed germination, the expression of UEV1C and UEV1D is much higher than that of UEV1A and UEV1B and that UEV1D is essentially the only UEV1 transcript detected from pollen. To validate the microarray data, we performed RT-PCR with various tissues, including germinating seeds and pollen. Under the conditions used, the amount of PCR product was not excessive and was deemed to reflect the amount of cDNA template. Representative results are shown in Figure 4B and summarized as follows. First, all four UEV1 genes are indeed expressed in most common tissues, such as root, shoot, leaf, and stem. Second, only UEV1D transcript is detectable in pollen under our experimental conditions, consistent with the microarray data. Third, 6 h after seed germination, all transcripts except UEV1B are detected, while after 2 d of seed germination, only UEV1A and UEV1D transcripts are found, with UEV1D at a clearly higher level than UEV1A. Microarray data show little expression of UEV1A in 3-h germinating seeds, but we consistently observed UEV1A transcript by PT-PCR in the sample we used. These differences may be due to the conditions used in the microarray experiments and in this study.
Figure 4.
Tissue Distribution of UEV1 Expression.
(A) Relative expression of UEV1 transcripts in different tissues was determined using data from the Arabidopsis NASCArrays microarray database (http://affymetrix.Arabidopsis.info/narrays/experimentbrowse.pl) (Craigon et al., 2004). R, roots of 17-d plants; S, shoots of 8-d seedlings; L, rosette leaf 2 of 17-d plants; RL, mature rosette leaves of 23-d plants; St, second internode of 21-d plants; F, stage 12 flowers of 21-d plants; P, mature pollen; G3h, seed germinating for 3 h. The original microarray data are from AtGenExpress: Expression Atlas of Arabidopsis Development (TAIR accession number 1006710873: ATGE_9, ATGE_12, ATGE_24, ATGE_27, ATGE_33, ATGE_73, and ATGE_96 samples) (Schmid et al., 2005), except for G3h data, which are from AtGenExpress: Expression Profiling of Early Germinating Seeds (TAIR accession number 1007966994: RIKEN-PRESTON2 sample).
(B) Expression of UEV1 transcripts in different tissues analyzed by RT-PCR. The At4g33380 gene was assayed as an input control (Czechowski et al., 2005). The exposure time of the gels is shown at left (BioDoc-It System; UVP). C, cell suspension; R, roots of 13-d seedlings; S, shoots of 13-d seedlings; L3, leaves of 3-week plants; L5, leaves of 5-week plants; St, stems of 5-week plants; F, floral tissues of 5-week plants; G6h and G2d, seeds germinating on Petri dishes for 6 h and 2 d, respectively; P, pollen.
uev1d Mutant Plants Are Sensitive to the DNA-Damaging Agent MMS
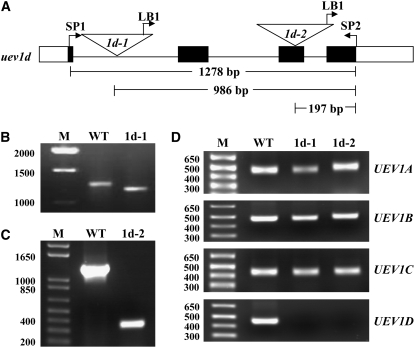
The analysis of UEV1 expression as well as the observation that in combination with UBC13, UEV1D but not UEV1A could completely rescue the yeast ubc13 mms2 double mutant, prompted us to focus our attention on UEV1D. We reasoned that uev1d mutant plants may display compromised tolerance to DNA damage in pollen and during seed germination. The UEV1D T-DNA insertion line SALK_064912 was obtained from the ABRC (www.arabidopsis.org), and the allele was named uev1d-1. Sequence analysis revealed that the T-DNA was inserted in the first intron of UEV1D, with the left border oriented toward the 3′ end of the gene (Figure 5A). The gene-specific primers (SP1 and SP2) and a primer specific to the left border sequence (LB1) were used to confirm the insertion of T-DNA (Figure 5B). To further confirm that UEV1D expression was abolished by this T-DNA insertion, total RNA was extracted from seedlings of wild-type and homozygous uev1d-1 plants and analyzed by RT-PCR for the expression of four UEV1 genes. As shown in Figure 5D, a fragment corresponding to the UEV1D ORF could be amplified from wild-type plants but not from the uev1d-1 line, while the expression of the other three At UEV1 genes remained unaltered.
Figure 5.
Confirmation of Two uev1d T-DNA Insertion Mutants.
(A) Genomic structure showing the positions of two T-DNA insertions in UEV1D. Open boxes, exons; closed boxes, UEV1D ORF; lines, introns. SP1, 5′ gene-specific primer AtUEV1D-1; SP2, 3′ gene-specific primer AtUEV1D-2; LB1, T-DNA left border primer.
(B) and (C) Genomic DNA PCR to confirm uev1d-1 (1d-1) (B) and uev1d-2 (1d-2) (C). The fragment was amplified using three primers (SP1, SP2, and LB1) in each reaction and genomic DNA from Columbia (WT), 1d-1 (B), or 1d-2 (C) as a template.
(D) RT-PCR detection of the UEV1 transcripts. UEV1 gene-specific primers were used for RT-PCR against total RNA extracted from Columbia (WT), uev1d-1, and uev1d-2 lines. Total RNA was extracted from flowers.
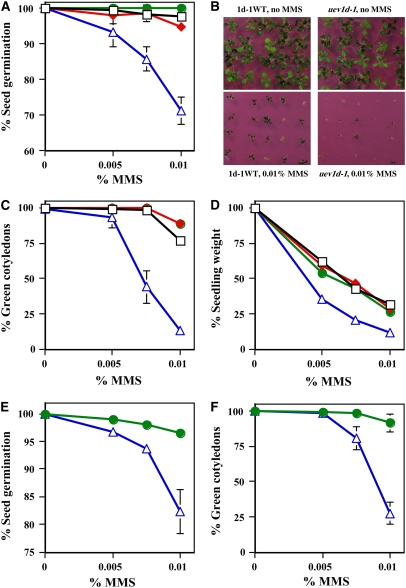
The homozygous uev1d-1 plants do not display apparent morphological variations. In order to investigate the possible role of UEV1D in protecting cells from DNA damage, we analyzed the effect of MMS on seed germination, considering that UEV1D is strongly expressed during seed germination. We examined three controls—wild-type Arabidopsis ecotype Columbia, a T-DNA insertion line (SALK_042050) not affecting UEV1 genes, and a wild type segregant line derived from the initial SALK_064912 seeds (1d-1WT)—along with the homozygous uev1d-1 T-DNA insertion line. Three parameters related to seed germination were surveyed. First, the percentage of seeds that germinated in the presence of various concentrations of MMS was scored after a 5-d incubation. Seeds from uev1d-1 plants were much more sensitive to MMS treatment than any of the three control plants, and this response was dose-dependent (Figure 6A). By contrast, in the absence of MMS, the uev1d-1 seeds did not show a noticeable difference from controls in the percentage of seed germination. Second, it was observed that the homozygous uev1d-1 seedlings were dying relatively quickly in the presence of MMS and displayed bleached pale cotyledons rather than the normal green cotyledons. Thus, the percentage of germinated seeds with green cotyledons was scored after 13 d. The data clearly indicate that the uev1d-1 line had reduced numbers of viable seedlings in the presence of MMS. In particular, in the presence of 0.01% MMS, 75 to 90% of control seedlings were viable, as judged by green seedlings, compared with <15% viable uev1d-1 seedlings under the same growth condition (Figures 6B and 6C). Finally, the average fresh weight of 13-d uev1d-1 mutant seedlings was reduced compared with that in control seedlings after MMS treatments. More specifically, with 0.005% MMS treatment, even though almost all uev1d-1 seedlings remained green, they only had half the fresh weight of the wild-type seedlings (Figure 6D).
Figure 6.
Phenotypic Analysis of DNA Damage Response during Seed Germination.
(A) to (D) The homozygous uev1d-1 mutant (open triangles) is compared with three controls: Columbia (open squares), an unrelated T-DNA insertion line, SALK_042051 (closed diamonds), and a wild-type segregant line from the same SALK_064912 seeds (1d-1WT; closed circles). Synchronized seeds were sown on half-strength Murashige and Skoog agar plates with or without MMS as indicated and incubated for the given period, and phenotypes were quantitatively assessed.
(A) Percentage of seed germination after 5 d.
(B) Representative photographs after 13 d.
(C) Percentage of seedlings with green cotyledons after 13 d.
(D) Relative fresh weight of seedlings with green cotyledons after 13 d.
(E) and (F) The homozygous uev1d-2 mutant (open triangles) is compared with 1d-2WT, a wild-type segregant from the same T-DNA insertion line SALK_052144 (closed circles).
(E) Percentage of seed germination after 5 d.
(F) Percentage of seedlings with green cotyledons after 13 d.
All data are averages of three independent experiments with sd.
To ensure that the above observations were specific to the T-DNA insertion at UEV1D, we obtained the second UEV1D T-DNA insertion line SALK_052144 from the ABRC, in which the T-DNA was inserted in the third exon of UEV1D (Figure 5A). We confirmed the T-DNA insertion by genomic PCR (Figure 5C) and named it uev1d-2. RT-PCR analysis demonstrated that the UEV1D mRNA is absent in the homozygous uev1d-2 line (Figure 5D). Phenotypic analyses showed that, like uev1d-1, the uev1d-2 mutant is hypersensitive to MMS treatment during seed germination (Figures 6E and 6F). From these results, we conclude that UEV1D is required for tolerance to DNA damage during seed germination.
To assess whether the observed uev1d mutant phenotype is specific to DNA damage or is triggered by general stress, we measured seed germination in the presence of up to 0.2 M NaCl. There was no significant difference in the response of wild-type and uev1d plants to the salt stress (data not shown).
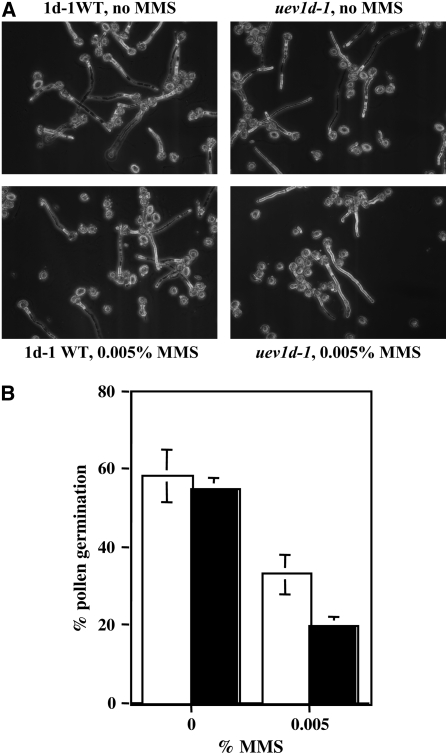
We also attempted to assess the role of UEV1D in pollen germination by measuring the percentage of pollen germination in the presence of MMS. As shown in Figure 7, inactivation of UEV1D resulted in a moderate but significant decrease in pollen germination. In the presence of 0.005% MMS, 33% of wild-type pollens germinated, while only 20% of uev1d-1 pollens germinated after 8 h of incubation, indicating that UEV1D also plays a critical role in protecting pollen from environmental DNA damage.
Figure 7.
Phenotypic Analysis of DNA Damage Response during in Vitro Pollen Germination.
(A) Representative in vitro pollen germination images of 1d-1WT and uev1d-1 with or without MMS treatment as indicated.
(B) Summary of the pollen germination results. Data presented are averages of three independent experiments with sd. Open bars, 1d-1WT; closed bars, uev1d-1.
uev1a Mutant Plants Do Not Display MMS Sensitivity
Since UEV1A is the only other UEV1 gene expressed during seed germination, we were interested in the phenotypes of this mutant plant. Unfortunately, a uev1a T-DNA insertion mutant line is not available from the ABRC; instead, we found a line (FLAG_128G02) with a T-DNA insertion at the fourth exon (see Supplemental Figure 5A online) from the Institut Jean-Pierre Bourgin collection. We obtained this line, screened the segregants, and confirmed the homozygous uev1a mutant (uev1a-1) by both genomic PCR (see Supplemental Figure 5B online) and RT-PCR (see Supplemental Figure 5C online). Seed germination assays were performed under the same experimental conditions described above. uev1a-1 mutant plants did not display enhanced MMS sensitivity compared with its wild-type segregants or with the parental strain Ws-4 (data not shown). This result indicates that inactivation of UEV1A does not alter DDT during Arabidopsis seed germination.
DISCUSSION
Lys-63–linked polyubiquitination of target proteins is considered to be a fundamentally different process from conventional Lys-48–linked polyubiquitination, which targets proteins for degradation via the 26S proteasome (Hochstrasser, 1996; Pickart, 2001b). Instead, it is deemed analogous to other posttranslational regulatory processes, such as phosphorylation and sumoylation, that alter target protein activities. Although Lys-63–linked polyubiquitination has been reported to be involved in several cellular processes, including stress response (Arnason and Ellison, 1994), mitochondrial inheritance (Fisk and Yaffe, 1999), plasma membrane protein endocytosis (Galan and Haguenauer-Tsapis, 1997), ribosome function (Spence et al., 2000), innate immunity (Deng et al., 2000; Wang et al., 2001; Zhou et al., 2004), mitotic cell cycle checkpoint (Bothos et al., 2003), and DNA repair (Hofmann and Pickart, 1999; Hoege et al., 2002), to date Ubc13 is the only known E2 capable of mediating Lys-63–linked Ub chain assembly. Interestingly, genetic analyses to date define only three functions of Ubc13, namely PRR in yeast (Hoege et al., 2002) and possibly mammals (Andersen et al., 2005) through covalent modification of PCNA, activation of NF-κB by polyubiquitinating NEMO/IKKγ in mammals (Zhou et al., 2004), and involvement in synaptic connectivity between the giant fiber and a motor neuron in Drosophila. Drosophila ubc13 is also known as bendless (Muralidhar and Thomas, 1993; Oh et al., 1994), although the molecular mechanism of Ubc13 activity in this case remains unclear. The unique feature of Ubc13 compared with other Ubcs is its ability to form a stable complex with a Uev, which is homologous to other Ubcs but lacks the active Cys residue (Broomfield et al., 1998; Sancho et al., 1998). This family of Uevs engages a noncovalent interaction with Ub (McKenna et al., 2001) and orients this acceptor Ub to allow its Lys-63 residue to be exposed to the C terminus of donor Ub covalently bound to Ubc13 (McKenna et al., 2003). It appears that in mammals, Uevs not only facilitate polyubiquitination but also serve as a regulatory subunit to promote the ubiquitination of different targets (Andersen et al., 2005). In this study, we identified four Arabidopsis genes that meet the criteria of encoding a Uev and found by sequence analysis that they are two pairs of duplicated genes. We argue that these UEV1 genes are paralogs and that they evolved from a common ancestor within plants, since sequence alignment could not distinguish which Uev1 pair is more related to one of the two mammalian Uevs (Uev1A or Mms2). This observation raises two major questions. First, do Ubc13-Uev complexes from different organisms confer a conserved function? Second, if gene duplications occurred after animal–plant separation, how are their functions preserved? In this study, we demonstrated that all At Uev1 proteins are able to form stable complexes with At Ubc13 and to promote Ubc13-mediated Lys-63–linked polyubiquitination. The only other known Arabidopsis Uev found to date is Cop10, which was identified as a negative regulator of photomorphogenesis and functions by promoting target protein degradation (Suzuki et al., 2002).
The observation that At UBC13 (Wen et al., 2006) or At UEV1 (this study) is able to functionally complement the corresponding yeast mutants defective in DDT is insufficient to claim that they also play the same role in their own host. A good example is human UEV1A, which confers a similar DDT function in yeast but is exclusively involved in NF-κB activation instead of DNA repair (Andersen et al., 2005). In this study, we took advantage of the fact that UEV1D is the predominant UEV1 gene expressed in germinating seeds and in pollen and characterized the sensitivity of uev1d mutant plants to a DNA-damaging agent in these tissues. Our results clearly show that in the presence of DNA damage, lack of Uev1D activity compromises seed germination, seedling survival, and growth. Based on the following observations, we argue that the above phenotypic effects are specific to the inactivation of UEV1D. First, two independent UEV1D T-DNA insertion lines displayed similar phenotypes, while none of three control lines showed these phenotypes. Second, a uev1a mutant line also did not show such phenotypes compared with its wild-type controls. This study, along with previous studies in yeast and mammalian cells, supports the notion that error-free DDT promoted by Lys-63–linked polyubiquitination via Ubc13-Uev is an evolutionarily conserved function throughout eukaryotes.
Despite its predominant expression among the four UEV1 genes, inactivation of UEV1D caused only a very moderate compromise in pollen tube growth in the presence of a DNA-damaging agent. This probably reflects the lack of cell division during pollen germination, whereas DDT is expected to operate only on replicated DNA (Barbour and Xiao, 2003). It is of great interest that in contrast with many other plant species, it has been observed that DNA synthesis in the Arabidopsis sperm nuclei is initiated prior to anthesis and continues as the pollen tube develops (Friedman, 1999). Thus, the Arabidopsis sperm nuclei are essentially in a prolonged S phase at the time of anthesis, in preparation for eventual double fertilization. Our observation that in the presence of replication-blocking lesions induced by MMS, pollen tubes from the uev1d mutant plants did not develop as well as those from wild-type segregant lines is consistent with the notion that UEV1D plays a more active role than other UEV1 genes when DNA synthesis occurs in the presence of DNA damage, which is essentially a DDT activity. We wish to stress that our analysis does not rule out the possibility that MMS-induced DNA lesions may inhibit transcription and that MMS can also directly methylate and damage RNA (Friedberg et al., 2006), which could contribute to the observed phenotypes, although Lys-63 ubiquitination has not been linked to these processes.
In our opinion, this study provides an important step toward understanding Ubc13-Uev–mediated Lys-63 polyubiquitination in general and the mechanisms of DDT in particular in plants. Several questions remain to be addressed. First, is UEV1D the only UEV1 gene involved in error-free DDT? Second, is UEV1D also involved in other cellular processes? Third, what are the other cellular processes that also require Ubc13-Uev–mediated polyubiquitination? We feel that given the near identity in amino acid sequence and similar complementation phenotypes in yeast, Uev1C is likely involved in the same cellular processes as Uev1D. This could explain our failure to detect a DNA repair/tolerance defect in uev1d plants in a root growth assay (data not shown). Indeed, the phenotypes of uev1d mutant plants may be considered to be moderate, which is due to either the backup or residual expression of other UEV1 genes or the nature of the error-free DDT defect in plants. One important aspect of future work will be to identify a uev1c null mutant, combine this mutation with uev1d, examine various tissues for a DDT defect, and relate the results to the UEV1 expression profile. It is also interesting that the yeast mms2/ubc13 or rev1/rev3/rev7 single mutants are moderately sensitive to killing by DNA-damaging agents, but the combination of any two mutations from different pathways results in strong synergistic interactions (Broomfield et al., 1998; Xiao et al., 1999). It would be of great interest to determine whether the combination of uev1d and rev mutations also results in a synergistic sensitivity to DNA damage in Arabidopsis.
The two pairs of UEV1s may be involved in different functions in plants. Although it remains possible that Uev1A and Uev1B are involved in DDT as well, our observations favor the suggestion that the Uev1A/B pair is probably involved in other cellular processes unrelated to DNA damage response. First, although UEV1A is expressed during seed germination, inactivation of this gene does not result in compromised seed germination in the presence of MMS, in sharp contrast with the uev1d mutant lines. Second, in the presence of UBC13, the DDT activities of UEV1A/B are much lower than those of UEV1C/D in yeast cells, despite the fact that Uev1A/B interact with At Ubc13 and Ubc13 from other species very well and are able to fully complement the yeast mms2 mutant when yeast Ubc13 is present. This result also effectively rules out the possibility that partial complementation by UEV1A/B was due to their poor expression in yeast cells. Third, it is interesting that human Uev1A contains an additional N-terminal 25 amino acid residues and plays a distinct role from that of hMms2; it may be reverted to play a role in DDT when its N-terminal sequence is experimentally deleted (Andersen et al., 2005). Similarly, At Uev1A and At Uev1B contain a unique C-terminal tail that may be critical for their functions other than DDT. It is difficult at this stage to predict what type of activity it may be, given the fact that Uev1 appears to have evolved independently of vertebrate Uev paralogs, that Drosophila ubc13/bendless confers a very different function than its mammalian counterpart, and that other reports have claimed additional Lys-63–mediated cellular processes and some of them have also been linked to Ubc13-Uev (Bothos et al., 2003; Doss-Pepe et al., 2005; Laine et al., 2006). What we can predict is that additional Arabidopsis Ubc13-Uev1 functions should be mediated by its Lys-63–linked poly-Ub chains and that different Uev1 proteins may serve as cofactors and critical regulators in these processes. In this regard, future research may focus on the search for Ubc13-Uev ubiquitination targets and cognate E3s through bioinformatics and proteomic approaches as well as through genomic approaches such as microarray analysis of the At uev1 mutants reported in this study.
METHODS
Plant and Yeast Cell Cultures
Arabidopsis thaliana ecotypes Columbia and Ws and their mutant derivatives were used in this study. The conditions for plant growth and maintenance of Arabidopsis Columbia cell suspension culture have been described previously (Wen et al., 2006).
The haploid yeast strains used in this study are listed in Supplemental Table 1 online. Yeast cells were grown at 30°C in either rich YPD or in a synthetic dextrose (SD) medium (0.67% Bacto-yeast nitrogen base without amino acids, 2% glucose) supplemented with necessary nutrients as recommended (Sherman et al., 1983). For solid plates, 2% agar was added to either YPD or SD medium prior to autoclaving. Yeast cells were transformed using a LiAc method as described (Ito et al., 1983). The sources and preparation of ubc13Δ∷hisG-URA3-hisG (Brusky et al., 2000) and mms2Δ∷HIS3 (Xiao et al., 1999) cassettes was as described previously.
Cloning Arabidopsis UEV1 cDNAs and Plasmid Construction
To clone At UEV1s, total RNA was isolated from Arabidopsis seedlings using TRIzol reagent (Invitrogen) for RT-PCR with the ThermoScript RT-PCR kit (Invitrogen) according to the manufacturer's instructions. Each At UEV1 ORF was amplified by PCR from the above cDNA preparation using gene-specific primers (see Supplemental Table 2 online), and the flanking SalI restriction site was used to clone the PCR products into the yeast two-hybrid vector pGAD424E (for Gal4AD fusion), which was derived from pGAD424 (Bartel and Fields, 1995), with a 1-bp frameshift at the multiple cloning site. The identity of each cloned ORF was verified by sequencing.
Yeast Two-Hybrid Analysis
The yeast two-hybrid strain PJ69-4A (James et al., 1996) was cotransformed with different combinations of Gal4BD and Gal4AD constructs. The construction of pGBT-At Ubc13A, pGBT-At Ubc13B (Wen et al., 2006), pGBT-Sc Ubc13 (Brown et al., 2002), and pGBT-Hs Ubc13 (Pastushok et al., 2005) has been described previously. The cotransformed colonies were initially selected on SD-Leu-Trp plates. For each transformation, at least five independent colonies were plated onto SD-Leu-Trp-His with various concentrations of 1,2,4-aminotriazole to test the activation of the GAL1-HIS3 gene and onto SD-Leu-Trp-Ade to detect the activation of the GAL2-ADE2 reporter gene.
Protein Expression, Purification, and GST Pull-Down Assay
The SalI fragments containing UEV1 ORFs were isolated from the pGAD424E plasmids and cloned into pGEX6p-2 (Amersham Biosciences). The resulting pGEX-Uev1s were transformed into Escherichia coli strain BL21 (DE3)-RIL (Stratagene). The GST-Uev1 fusion proteins were produced and purified as described previously (McKenna et al., 2001). The source and preparation of the Ubc13A fusion protein and the protocol for the GST pull-down assay were as described previously (Wen et al., 2006).
Ub Conjugation Reaction
In vitro Ub conjugation reactions were performed using the purified Ubc13A and GST-Uev1A proteins as described above, and Ub thioester/conjugation initiation reagents were purchased from Boston Biochem. Unless noted otherwise, the 20-μL reaction mixture contained 225 nM E1 enzyme, 450 μM Ub, 1 mM MgATP, 1 μM Ubc13, and 1 μM Uev1 in the supplied reaction buffer. The K63R and K48R mutant Ub proteins were purchased from Boston Biochem (UM-K63R and UM-K48R). The conjugation reactions were performed at 37°C for 2 h. Samples were subjected to SDS-PAGE (12%), and Ub and poly-Ub were detected through protein gel blots using polyclonal rabbit anti-Ub antibodies (Sigma-Aldrich).
Yeast Killing and Spontaneous Mutagenesis Assay
Yeast strain HK580-10D and its isogenic mms2Δ single or ubc13Δ mms2Δ double mutants were either singly transformed with pGAD-Uev1 or cotransformed with pGAD-Uev1 and pGBT-Ubc13A, and transformants were selected on SD-Leu or SD-Leu-Trp plates, respectively. The gradient plate assay was performed as described previously (Pastushok et al., 2005).
Yeast strain DBY747 and its mms2Δ derivative WXY642 bear a trp1-289 amber mutation that can be reverted to Trp+ by several different mutation events (Xiao and Samson, 1993). WXY642 was transformed with pGAD-UEV1 or vector pGAD424E, and transformants were selected on SD-Leu plates. The spontaneous mutagenesis assay was performed as described previously (Wen et al., 2006).
Expression Analysis by RNA Gel Blot and RT-PCR
To determine the expression of UEV1 genes under different stress conditions, Arabidopsis cell suspension culture was used. The culture was maintained for 5 d after subculture and then subjected to various treatments as specified. After a 24-h treatment, total RNA was isolated and 15 μg of RNA from each sample was used for RNA gel blot analysis as described (Wang et al., 1995). The DNA fragment containing the UEV1C ORF was isolated from an agarose gel after restriction enzyme digestion and electrophoresis, and the UBQ11 gene probe was amplified using gene-specific primers (see Supplemental Table 2 online). The DNA fragment was labeled with a 32P-dCTP using the Random Primer labeling kit from Invitrogen. The membrane containing total RNA was hybridized with the UEV1C probe, stripped, and hybridized with the UBQ11 probe.
For RT-PCR analysis, total RNA from various tissues was isolated using TRIzol and treated with DNaseI (Promega). Total RNA from mature pollen was extracted as described (Fei et al., 2004), and total RNA from germinating seeds was extracted as described (Vicient and Delseny, 1999). Reverse transcript synthesis of the first-strand cDNA was performed with the SuperScript RT-PCR III system (Invitrogen) using the protocol as described (Karsai et al., 2002). Briefly, 2 to 4 μg of total RNA for each sample were treated with DNase I (Roche Diagnostics) and reverse-transcribed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and d(T)18. The final input amount of cDNA used for RT-PCR was adjusted by analyzing the expression of the At4g33380 control gene (Czechowski et al., 2005). Experiments were performed using UEV1 gene-specific primer pairs with different cycle conditions (22, 28, and 35 cycles) to make sure that the amount of PCR product was not excessive and that the differences among different tissue samples were not disturbed by saturation of PCR amplification. Eight microliters of each reaction was used for agarose gel electrophoresis. All RT-PCR series were assayed at least twice with highly consistent results.
Analysis of Microarray Expression Data
Expression values of UEV1 genes were obtained from publicly available Affymetrix ATH1 array data available up to June 2006 (http://affymetrix.Arabidopsis.info/) with a total of 2392 arrays. A subset of 237 arrays was used to calculate the average expression levels and to compare the expression levels that correspond with (1) 63 conditions from the Developmental Affymetrix Gene Expression Atlas: AtGenExpress (Schmid et al., 2005), (2) three root stages (Birnbaum et al., 2003), (3) five root layer tissues (Nawy et al., 2005), (4) four pollen stages (Honys and Twell, 2004), (5) four flower induction treatments (Schmid et al., 2003), and (6) three germination stages (http://www.weigelworld.org/resources/microarray/AtGenExpress/). Heat maps representing the expression levels of At UEV1 genes were produced with the Genesis 1.6.0 Beta 1 program (Sturn et al., 2002) (http://genome.tugraz.at/) using absolute expression values as standards in the Nottingham Arabidopsis Stock Centre's International Affymetrix Service.
Seed Germination Assays
The homozygous UEV1D T-DNA insertion lines SALK_064912 (uev1d-1) and SALK_052144 (uev1d-2) were used in the sensitivity assay to the DNA-damaging agent MMS. To exclude any possible nonspecific effect, we used three controls, the wild-type Arabidopsis Columbia, a T-DNA insertion line not related to UEV1 genes (SALK_042050), and a homozygous wild-type segregant line (1d-1WT or 1d-2WT) derived from the initial mutant seeds received. For the uev1a-1 mutant, the parental wild-type line Ws-4 and a homozygous wild-type (1a-1WT) segregant from FLAG_128G02 were used as controls. The identity of the wild-type and mutant segregants was determined by genomic PCR and RT-PCR. In addition, to minimize the effect of individual plants, seeds of three homozygous mutant plants were pooled and used for the assay. Seeds were surface-sterilized with 20% bleach and 0.1% Triton X-100 for 20 min, followed by three rinses in sterile water. After sterilization, seeds were suspended in 0.1% agarose and stored at 4°C in the dark for 3 d to synchronize germination. Three days later, the seeds were removed from the dark and sown on half-strength Murashige and Skoog agar plates supplemented with different concentrations of MMS. Each plate was planted with 50 seeds, and at least three plates (150 seeds) were used for each treatment. After a 5-d incubation in a growth chamber, the germination of the seeds was surveyed. After a 13-d incubation, the color of the cotyledons was rated (green versus nongreen) while the first pair of true leaves was still small. Since seedlings with nongreen cotyledons at this stage were dead or dying, the percentage of seedlings with green cotyledons was an indicator of seedling viability. The fresh weight of seedlings was also determined and was used as an indicator of seedling growth.
In Vitro Pollen Germination Assay
To quantitatively measure pollen germination efficiency in the presence of MMS, 2 mL of the germination medium (Fan et al., 2001) containing 1% agar with or without 0.005% MMS was poured into a 35-mm Petri dish to form a thin layer. Freshly anther-dehisced flowers at stage 13 or 14 (Smyth et al., 1990) were randomly picked and used to carefully touch the central area of the agar plate in order to spread pollen grains. The Petri dishes were incubated in a humid chamber at 26°C for 8 h without light before counting pollen germination and photographing. For each plate, >400 pollen grains were counted using a phase-contrast microscope for each plate, and three plates were used for each treatment.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: NP_011428.1 (Saccharomyces cerevisiae); NP_588162.1 (Schizosaccharomyces pombe); NP_647959.1 (Drosophila melanogaster); NP_076074.2 (Mus musculus); At Uev1A = NP_565834.1 (At1g23260), At Uev1B = NP_564994.1 (At1g70660), At Uev1C = NP_850259.1 (At2g36060), and At Uev1D = NP_566968.1 (At3g52560). (Arabidopsis thaliana); NP_493578.1 (Caenorhabditis elegans); and Hs Mms2 = NP_003341.1 and Hs Uev1A = NP_068823.2 (Homo sapiens).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Analyses of Hypothetical Uev Family Proteins from Different Organisms.
Supplemental Figure 2. Characterization of Arabidopsis UEV1B and UEV1C.
Supplemental Figure 3. UEV1 Expression Profiles.
Supplemental Figure 4. Heat Map Showing the Transcript Levels of Arabidopsis UEV1 Genes in Different Tissues and Developmental Stages.
Supplemental Figure 5. Confirmation of the uev1a-1 T-DNA Insertion Mutant.
Supplemental Table 1. S. cerevisiae Strains.
Supplemental Table 2. Oligonucleotide Sequences.
Supplementary Material
Acknowledgments
We thank Vipon Sawhney for the protocol on collecting pollen samples, Gordon Gray for the Arabidopsis cell suspension culture, Bernard Kunz for critical comments on the manuscript, and Michelle Hanna for proofreading the manuscript. This work was supported by Canadian Institutes of Health Research Operating Grant MOP-53240 to W.X. and a Natural Sciences and Engineering Research Council of Canada Discovery Grant to H.W.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Hong Wang (hong.wang@usask.ca) and Wei Xiao (wei.xiao@usask.ca).
Online version contains Web-only data.
References
- Andersen, P.L., Zhou, H., Pastushok, L., Moraes, T., McKenna, S., Ziola, B., Ellison, M.J., Dixit, V.M., and Xiao, W. (2005). Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J. Cell Biol. 170 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnason, T., and Ellison, M.J. (1994). Stress resistance in Saccharomyces cerevisiae is strongly correlated with assembly of a novel type of multiubiquitin chain. Mol. Cell. Biol. 14 7876–7883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair, A., Novatchkova, M., Potuschak, T., and Eisenhaber, F. (2001). Ubiquitylation in plants: A post-genomic look at a post-translational modification. Trends Plant Sci. 6 463–470. [DOI] [PubMed] [Google Scholar]
- Barbour, L., and Xiao, W. (2003). Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: A yeast model. Mutat. Res. 532 137–155. [DOI] [PubMed] [Google Scholar]
- Bartel, P.L., and Fields, S. (1995). Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 254 241–263. [DOI] [PubMed] [Google Scholar]
- Birnbaum, K., Shasha, D.E., Wang, J.Y., Jung, J.W., Lambert, G.M., Galbraith, D.W., and Benfey, P.N. (2003). A gene expression map of the Arabidopsis root. Science 302 1956–1960. [DOI] [PubMed] [Google Scholar]
- Blanc, G., Hokamp, K., and Wolfe, K.H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothos, J., Summers, M.K., Venere, M., Scolnick, D.M., and Halazonetis, T.D. (2003). The Chfr mitotic checkpoint protein functions with Ubc13-Mms2 to form Lys-63-linked polyubiquitin chains. Oncogene 22 7101–7107. [DOI] [PubMed] [Google Scholar]
- Broomfield, S., Chow, B.L., and Xiao, W. (1998). MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl. Acad. Sci. USA 95 5678–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broomfield, S., Hryciw, T., and Xiao, W. (2001). DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 486 167–184. [DOI] [PubMed] [Google Scholar]
- Brown, M., Zhu, Y., Hemmingsen, S.M., and Xiao, W. (2002). Structural and functional conservation of error-free DNA postreplication repair in Schizosaccharomyces pombe. DNA Repair (Amst.) 1 869–880. [DOI] [PubMed] [Google Scholar]
- Brusky, J., Zhu, Y., and Xiao, W. (2000). UBC13, a DNA-damage-inducible gene, is a member of the error-free postreplication repair pathway in Saccharomyces cerevisiae. Curr. Genet. 37 168–174. [DOI] [PubMed] [Google Scholar]
- Craigon, D.J., James, N., Okyere, J., Higgins, J., Jotham, J., and May, S. (2004). NASCArrays: A repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res. 32 D575–D577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M.J., and Hays, J.B. (2007). Tolerance of dividing cells to replication stress in UVB-irradiated Arabidopsis roots: Requirements for DNA translesion polymerases η and ζ. DNA Repair (Amst.) 6 1341–1358. [DOI] [PubMed] [Google Scholar]
- Czechowski, T., Stitt, M., Altmann, T., Udvardi, M.K., and Scheible, W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, L., Wang, C., Spencer, E., Yang, L., Braun, A., You, J., Slaughter, C., Pickart, C., and Chen, Z.J. (2000). Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103 351–361. [DOI] [PubMed] [Google Scholar]
- Devoto, A., Muskett, P.R., and Shirasu, K. (2003). Role of ubiquitination in the regulation of plant defence against pathogens. Curr. Opin. Plant Biol. 6 307–311. [DOI] [PubMed] [Google Scholar]
- Doss-Pepe, E.W., Chen, L., and Madura, K. (2005). α-Synuclein and parkin contribute to the assembly of ubiquitin lysine 63-linked multiubiquitin chains. J. Biol. Chem. 280 16619–16624. [DOI] [PubMed] [Google Scholar]
- Eddins, M.J., Carlile, C.M., Gomez, K.M., Pickart, C.M., and Wolberger, C. (2006). Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat. Struct. Mol. Biol. 13 915–920. [DOI] [PubMed] [Google Scholar]
- Fan, L.M., Wang, Y.F., Wang, H., and Wu, W.H. (2001). In vitro Arabidopsis pollen germination and characterization of the inward potassium currents in Arabidopsis pollen grain protoplasts. J. Exp. Bot. 52 1603–1614. [PubMed] [Google Scholar]
- Fei, H., Zhang, R., Pharis, R.P., and Sawhney, V.K. (2004). Pleiotropic effects of the male sterile33 (ms33) mutation in Arabidopsis are associated with modifications in endogenous gibberellins, indole-3-acetic acid and abscisic acid. Planta 219 649–660. [DOI] [PubMed] [Google Scholar]
- Fields, S., and Song, O. (1989). A novel genetic system to detect protein-protein interactions. Nature 340 245–246. [DOI] [PubMed] [Google Scholar]
- Fisk, H.A., and Yaffe, M.P. (1999). A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 145 1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franko, J., Ashley, C., and Xiao, W. (2001). Molecular cloning and functional characterization of two murine cDNAs which encode Ubc variants involved in DNA repair and mutagenesis. Biochim. Biophys. Acta 1519 70–77. [DOI] [PubMed] [Google Scholar]
- Friedberg, E.C., Walker, G.C., Wolfram, S., Wood, R.D., Schultz, R.A., and Ellenberger, T. (2006). DNA Repair and Mutagenesis, 2nd ed. (Washington, DC: ASM Press).
- Friedman, W.E. (1999). Expression of the cell cycle in sperm of Arabidopsis: Implications for understanding patterns of gametogenesis and fertilization in plants and other eukaryotes. Development 126 1065–1075. [DOI] [PubMed] [Google Scholar]
- Galan, J.M., and Haguenauer-Tsapis, R. (1997). Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 16 5847–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Ortiz, M.V., Ariza, R.R., Hoffman, P.D., Hays, J.B., and Roldan-Arjona, T. (2004). Arabidopsis thaliana AtPOLK encodes a DinB-like DNA polymerase that extends mispaired primer termini and is highly expressed in a variety of tissues. Plant J. 39 84–97. [DOI] [PubMed] [Google Scholar]
- Hall, T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41 95–98. [Google Scholar]
- Hochstrasser, M. (1996). Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30 405–439. [DOI] [PubMed] [Google Scholar]
- Hoege, C., Pfander, B., Moldovan, G.L., Pyrowolakis, G., and Jentsch, S. (2002). RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419 135–141. [DOI] [PubMed] [Google Scholar]
- Hofmann, R.M., and Pickart, C.M. (1999). Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96 645–653. [DOI] [PubMed] [Google Scholar]
- Hofmann, R.M., and Pickart, C.M. (2001). In vitro assembly and recognition of Lys-63 polyubiquitin chains. J. Biol. Chem. 276 27936–27943. [DOI] [PubMed] [Google Scholar]
- Honys, D., and Twell, D. (2004). Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 5 R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsai, A., Muller, S., Platz, S., and Hauser, M.T. (2002). Evaluation of a homemade SYBR green I reaction mixture for real-time PCR quantification of gene expression. Biotechniques 32 790–792, 794–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine, A., Topisirovic, I., Zhai, D., Reed, J.C., Borden, K.L., and Ronai, Z. (2006). Regulation of p53 localization and activity by Ubc13. Mol. Cell. Biol. 26 8901–8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, S., Moraes, T., Pastushok, L., Ptak, C., Xiao, W., Spyracopoulos, L., and Ellison, M.J. (2003). An NMR-based model of the ubiquitin-bound human ubiquitin conjugation complex Mms2.Ubc13. The structural basis for lysine 63 chain catalysis. J. Biol. Chem. 278 13151–13158. [DOI] [PubMed] [Google Scholar]
- McKenna, S., Spyracopoulos, L., Moraes, T., Pastushok, L., Ptak, C., Xiao, W., and Ellison, M.J. (2001). Noncovalent interaction between ubiquitin and the human DNA repair protein Mms2 is required for Ubc13-mediated polyubiquitination. J. Biol. Chem. 276 40120–40126. [DOI] [PubMed] [Google Scholar]
- Moon, J., Parry, G., and Estelle, M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidhar, M.G., and Thomas, J.B. (1993). The Drosophila bendless gene encodes a neural protein related to ubiquitin-conjugating enzymes. Neuron 11 253–266. [DOI] [PubMed] [Google Scholar]
- Nawy, T., Lee, J.Y., Colinas, J., Wang, J.Y., Thongrod, S.C., Malamy, J.E., Birnbaum, K., and Benfey, P.N. (2005). Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17 1908–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, C.E., McMahon, R., Benzer, S., and Tanouye, M.A. (1994). bendless, a Drosophila gene affecting neuronal connectivity, encodes a ubiquitin-conjugating enzyme homolog. J. Neurosci. 14 3166–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli, E., Chen, S., Davies, A.A., Huttner, D., Krejci, L., Sung, P., and Ulrich, H.D. (2005). Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19 123–133. [DOI] [PubMed] [Google Scholar]
- Pastushok, L., Moraes, T.F., Ellison, M.J., and Xiao, W. (2005). A single Mms2 “key” residue insertion into a Ubc13 pocket determines the interface specificity of a human Lys-63 ubiquitin conjugation complex. J. Biol. Chem. 280 17891–17900. [DOI] [PubMed] [Google Scholar]
- Pastushok, L., Spyracopoulos, L., and Xiao, W. (2007). Two Mms2 residues that cooperatively interact with ubiquitin are critical for Lys-63-linked polyubiquitination. FEBS Lett. 581 5343–5348. [DOI] [PubMed] [Google Scholar]
- Pastushok, L., and Xiao, W. (2004). DNA postreplication repair modulated by ubiquitination and sumoylation. Adv. Protein Chem. 69 279–306. [DOI] [PubMed] [Google Scholar]
- Pfander, B., Moldovan, G.L., Sacher, M., Hoege, C., and Jentsch, S. (2005). SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436 428–433. [DOI] [PubMed] [Google Scholar]
- Pickart, C.M. (2001. a). Ubiquitin enters the new millennium. Mol. Cell 8 499–504. [DOI] [PubMed] [Google Scholar]
- Pickart, C.M. (2001. b). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70 503–533. [DOI] [PubMed] [Google Scholar]
- Sakamoto, A., Lan, V.T., Hase, Y., Shikazono, N., Matsunaga, T., and Tanaka, A. (2003). Disruption of the AtREV3 gene causes hypersensitivity to ultraviolet B light and γ-rays in Arabidopsis: Implication of the presence of a translesion synthesis mechanism in plants. Plant Cell 15 2042–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho, E., et al. (1998). Role of UEV-1, an inactive variant of the E2 ubiquitin-conjugating enzymes, in in vitro differentiation and cell cycle behavior of HT-29-M6 intestinal mucosecretory cells. Mol. Cell. Biol. 18 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., Davison, T.S., Henz, S.R., Pape, U.J., Demar, M., Vingron, M., Scholkopf, B., Weigel, D., and Lohmann, J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37 501–506. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Uhlenhaut, N.H., Godard, F., Demar, M., Bressan, R., Weigel, D., and Lohmann, J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130 6001–6012. [DOI] [PubMed] [Google Scholar]
- Sherman, F., Fink, G.R., and Hicks, J. (1983). Methods in Yeast Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence, J., Gali, R.R., Dittmar, G., Sherman, F., Karin, M., and Finley, D. (2000). Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102 67–76. [DOI] [PubMed] [Google Scholar]
- Stelter, P., and Ulrich, H.D. (2003). Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425 188–191. [DOI] [PubMed] [Google Scholar]
- Sturn, A., Quackenbush, J., and Trajanoski, Z. (2002). Genesis: Cluster analysis of microarray data. Bioinformatics 18 207–208. [DOI] [PubMed] [Google Scholar]
- Suzuki, G., Yanagawa, Y., Kwok, S.F., Matsui, M., and Deng, X.W. (2002). Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 16 554–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, S., Sakamoto, A., Sato, S., Kato, T., Tabata, S., and Tanaka, A. (2005). Roles of Arabidopsis AtREV1 and AtREV7 in translesion synthesis. Plant Physiol. 138 870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui, C., Raguraj, A., and Pickart, C.M. (2005). Ubiquitin binding site of the ubiquitin E2 variant (UEV) protein Mms2 is required for DNA damage tolerance in the yeast RAD6 pathway. J. Biol. Chem. 280 19829–19835. [DOI] [PubMed] [Google Scholar]
- Tuteja, N., Singh, M.B., Misra, M.K., Bhalla, P.L., and Tuteja, R. (2001). Molecular mechanisms of DNA damage and repair: Progress in plants. Crit. Rev. Biochem. Mol. Biol. 36 337–397. [DOI] [PubMed] [Google Scholar]
- Vicient, C.M., and Delseny, M. (1999). Isolation of total RNA from Arabidopsis thaliana seeds. Anal. Biochem. 268 412–413. [DOI] [PubMed] [Google Scholar]
- Vierstra, R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8 135–142. [DOI] [PubMed] [Google Scholar]
- Villalobo, E., Morin, L., Moch, C., Lescasse, R., Hanna, M., Xiao, W., and Baroin-Tourancheau, A. (2002). A homologue of CROC-1 in a ciliated protist (Sterkiella histriomuscorum) testifies to the ancient origin of the ubiquitin-conjugating enzyme variant family. Mol. Biol. Evol. 19 39–48. [DOI] [PubMed] [Google Scholar]
- Wang, C., Deng, L., Hong, M., Akkaraju, G.R., Inoue, J., and Chen, Z.J. (2001). TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412 346–351. [DOI] [PubMed] [Google Scholar]
- Wang, H., Datla, R., Georges, F., Loewen, M., and Cutler, A.J. (1995). Promoters from kin1 and cor6.6, two homologous Arabidopsis thaliana genes: Transcriptional regulation and gene expression induced by low temperature, ABA, osmoticum and dehydration. Plant Mol. Biol. 28 605–617. [DOI] [PubMed] [Google Scholar]
- Wen, R., Newton, L., Li, G., Wang, H., and Xiao, W. (2006). Arabidopsis thaliana UBC13: Implication of error-free DNA damage tolerance and Lys-63-linked polyubiquitylation in plants. Plant Mol. Biol. 61 241–253. [DOI] [PubMed] [Google Scholar]
- Xiao, W., Chow, B.L., Fontanie, T., Ma, L., Bacchetti, S., Hryciw, T., and Broomfield, S. (1999). Genetic interactions between error-prone and error-free postreplication repair pathways in Saccharomyces cerevisiae. Mutat. Res. 435 1–11. [DOI] [PubMed] [Google Scholar]
- Xiao, W., Lin, S.L., Broomfield, S., Chow, B.L., and Wei, Y.F. (1998). The products of the yeast MMS2 and two human homologs (hMMS2 and CROC-1) define a structurally and functionally conserved Ubc-like protein family. Nucleic Acids Res. 26 3908–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, W., and Samson, L. (1993). In vivo evidence for endogenous DNA alkylation damage as a source of spontaneous mutation in eukaryotic cells. Proc. Natl. Acad. Sci. USA 90 2117–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, M., et al. (2006. a). Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat. Immunol. 7 962–970. [DOI] [PubMed] [Google Scholar]
- Yamamoto, M., Sato, S., Saitoh, T., Sakurai, H., Uematsu, S., Kawai, T., Ishii, K.J., Takeuchi, O., and Akira, S. (2006. b). Cutting edge: Pivotal function of Ubc13 in thymocyte TCR signaling. J. Immunol. 177 7520–7524. [DOI] [PubMed] [Google Scholar]
- Zhou, H., Wertz, I., O'Rourke, K., Ultsch, M., Seshagiri, S., Eby, M., Xiao, W., and Dixit, V.M. (2004). Bcl10 activates the NF-κB pathway through ubiquitination of NEMO. Nature 427 167–171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.