Abstract
New evidence suggests a role for the plant growth hormone auxin in pathogenesis and disease resistance. Bacterial infection induces the accumulation of indole-3-acetic acid (IAA), the major type of auxin, in rice (Oryza sativa). IAA induces the expression of expansins, proteins that loosen the cell wall. Loosening the cell wall is key for plant growth but may also make the plant vulnerable to biotic intruders. Here, we report that rice GH3-8, an auxin-responsive gene functioning in auxin-dependent development, activates disease resistance in a salicylic acid signaling– and jasmonic acid signaling–independent pathway. GH3-8 encodes an IAA–amino synthetase that prevents free IAA accumulation. Overexpression of GH3-8 results in enhanced disease resistance to the rice pathogen Xanthomonas oryzae pv oryzae. This resistance is independent of jasmonic acid and salicylic acid signaling. Overexpression of GH3-8 also causes abnormal plant morphology and retarded growth and development. Both enhanced resistance and abnormal development may be caused by inhibition of the expression of expansins via suppressed auxin signaling.
INTRODUCTION
Plants respond to pathogen infection through two types of immune responses: basal and isolate-specific disease resistance (Jones and Dangl, 2006). The basal defense response is activated by virulent pathogens through the interaction of host pattern-recognition receptors and pathogen-associated molecular patterns. The isolate-specific or gene-for-gene defense response is triggered by host resistance (R) proteins recognizing isolate-specific pathogen effectors. The two types of immune responses are closely associated (Abramovitch et al., 2006; Jones and Dangl, 2006; Shen et al., 2007) and regulated by two classes of genes, the R genes and defense-responsive or defense-related genes. Plant pathogen recognition activates the signal transduction network composed of products from the two classes of genes. In addition to uncovering an increasing number of R and defense-responsive genes, studies have revealed multiple signal transduction pathways, mitogen-activated protein kinase signaling, gene-for-gene resistance, salicylic acid (SA)–dependent resistance, jasmonic acid (JA)/ethylene–dependent resistance, and induced systemic resistance, with each pathway containing multiple branches (Glazebrook, 2001; Asai et al., 2002; Hammond-Kosack and Parker, 2003; Bartsch et al., 2006). A network composed of synergistic or antagonistic crosstalk among these signal transduction pathways has been delineated based mainly on studies of dicots (Glazebrook, 2001; Hammond-Kosack and Parker, 2003; Durrant and Dong, 2004; Pieterse and Van Loon, 2004), although this network still needs to be refined to elucidate the molecular mechanism of pathogen-induced defense responses. In addition, although the accumulated information suggests that monocots may have similar fundamental modes of pathogen recognition and defense signaling as dicots, the defense responses to pathogen infection are not necessarily the same. For example, SA plays an important role in signaling of systemic acquired resistance in dicots, but rice (Oryza sativa) maintains a high endogenous level of SA without activating defense responses (Silverman et al., 1995). Characterization of more R genes and defense-responsive genes will help to explore the signal transduction network in monocotyledonous species.
Recent studies have reported that auxin promotes disease susceptibility, and repression of auxin receptors by microRNA is part of an induced immune response in Arabidopsis thaliana (Navarro et al., 2006); auxin-resistant Arabidopsis by mutation of genes functioning in auxin signaling shows enhanced disease resistance (Park et al., 2007; Wang et al., 2007; Zhang et al., 2007). Thus, auxin signaling, which is generally recognized to be involved in plant growth and development (Woodward and Bartel, 2005), is likely involved in the complicated network of plant–pathogen interactions. However, understanding of the interaction of plant defense systems and auxin signaling is in its infancy (Jones and Dangl, 2006).
In the regulation of plant development and growth, auxin can rapidly and transiently induce the expression of three groups of genes: the SMALL AUXIN-UP RNA (SAUR) family, the GH3 family, and the AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) family (Woodward and Bartel, 2005). The functions of the SAUR family are less understood compared with the other two families. The first GH3 gene was identified as a rapid auxin-responsive gene in soybean (Glycine max) (Hagen and Guilfoyle, 1985). However, not all GH3 genes are auxin-responsive (Woodward and Bartel, 2005). In Arabidopsis, some GH3 proteins are adenylate-forming enzymes that conjugate amino acids to the auxin indole-3-acetic acid (IAA) or to JA or SA (Staswick et al., 2002, 2005). The rice GH3 gene family consists of at least 12 members (Jain et al., 2006b). Aux/IAA proteins are transcription factors acting as repressors in auxin-regulated gene expression. The rice Aux/IAA gene family consists of at least 31 members (Jain et al., 2006a). The expression of most SAUR, GH3, and Aux/IAA genes is regulated by auxin response factors (ARFs), which can either activate or repress target gene expression (Woodward and Bartel, 2005). The rice ARF gene family consists of at least 11 members (Sato et al., 2001).
Our previous study showed that cDNA clone EI5P11, corresponding to GH3-8, a member of the rice GH3 gene family (Jain et al., 2006b), was pathogen-responsive (Wen et al., 2003). EI5P11 expression increased after inoculation with either incompatible pathogens in rice lines that carried different R genes conferring resistance to Xanthomonas oryzae pv oryzae (Xoo), the cause of bacterial blight disease, or to Magnaporthe grisea, the cause of fungal blast disease, or a compatible pathogen in a susceptible rice line. Furthermore, EI5P11 maps to the same genomic region as a quantitative trait locus for blast resistance on rice chromosome 7 (Wen et al., 2003). These results suggest that GH3-8 may be involved in both auxin signaling and defense signaling in a pathogen-nonspecific manner. To evaluate this hypothesis, we monitored GH3-8 expression and analyzed its function. It encodes an IAA–amido synthetase that maintains auxin homeostasis by conjugating excess IAA to amino acids. We demonstrated a novel pathway in which auxin signaling is involved in the regulation of the rice–Xoo interaction, such that GH3-8 protein functioning in auxin signaling regulates both disease resistance and growth and development in rice. Overexpressing GH3-8 activates rice resistance to bacterial blight, yet it also retards rice growth and development. Knockout of GH3-8 moderately compromises the resistance to Xoo. The molecular mechanism of such a dual regulation can be at least partly explained by the suppression of a group of auxin-responsive genes encoding expansins, proteins that control cell wall loosening and expansion, by preventing the accumulation of IAA.
RESULTS
Upregulation of GH3-8 Enhances Disease Resistance but Causes Abnormal Morphology
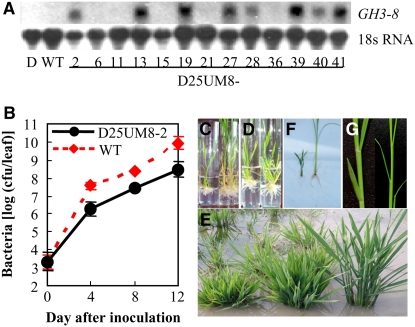
We isolated GH3-8 from resistant rice line C101LAC, which showed differential expression of GH3-8 on pathogen infection (Wen et al., 2003). GH3-8 was 2300 bp in length and had a coding region interrupted by one intron (see Supplemental Figure 1A online). The predicted protein product of GH3-8 consists of 605 amino acids and belongs to group II of the GH3 proteins based on phylogenetic analysis with Arabidopsis GH3 proteins. To determine whether GH3-8 was involved in rice defense to pathogen infection, we overexpressed the protein in a susceptible rice cultivar, then examined that cultivar's resistance to the rice pathogen Xoo. GH3-8, driven by a constitutive promoter, was transformed into the susceptible rice cv Mudanjiang 8. Sixteen of the 34 independent transformants showed significantly enhanced resistance to Xoo strain PXO61, with the lesion area ranging from 24 to 54% (lesion length/leaf length), compared with 78% in the susceptible cv Mudanjiang 8 and 15% in the resistant cv IRBB4 (see Supplemental Table 1 online). The enhanced resistance was clearly associated with overexpression of GH3-8 (Figure 1A; see Supplemental Figure 2 and Supplemental Table 1 online). A bacterial growth analysis demonstrated that the growth rate of PXO61 in resistant transgenic plants was 5.1- to 27.9-fold lower (P < 0.05) than that in the wild type at 4 to 12 d after inoculation (Figure 1B). These results suggest that GH3-8 is involved in the regulation of disease resistance.
Figure 1.
Expression of the GH3-8 Gene and Phenotypes of GH3-8–Overexpressing Plants (Mudanjiang 8 Is the Wild Type).
(A) GH3-8 expression in C101LAC (donor of GH3-8; D), wild-type, and T0 transgenic plants (D25UM8) detected by RNA gel blot analysis.
(B) Growth of PXO61 in leaves of GH3-8–overexpressing (D25UM8-2) and wild-type plants. The bacterial population was determined from three leaves at each time point by counting colony-forming units (cfu) (Sun et al., 2004). 0 day, 2 h after bacterial inoculation. Each point represents a mean ± sd.
(C) Transgenic plants grown on rooting medium in the absence of auxin. Left, GH3-8–overexpressing plant; right, negative transgenic plant.
(D) GH3-8–overexpressing plants grown on rooting medium supplemented with 0.3 mg/L 2,4,5-T for 10 d.
(E) Adult plants grown in the field. From left to right are the most stunted T0 plant (D25UM8-2), a moderate dwarf T0 plant (D25UM8-28), and the wild type (Mudanjiang 8).
(F) and (G) T1 plants from transgenic plant D25UM8-27 that overexpressed GH3-8. Left, abnormal T1 plant; right, normal T1 plant.
Overexpression of GH3-8 also affected plant development. All of the transgenic plants that showed enhanced resistance and overexpression of GH3-8 had abnormal morphology from tissue culture to field growth. During tissue culture, these plants were short and had fewer short roots than normal tissue culture plants; thus, they showed poor survival on the rooting culture medium (Figure 1C). To promote rooting, these plants were transferred to a fresh rooting culture medium supplemented with 2,4,5-trichlorophenoxyacetic acid (2,4,5-T), an analog of IAA. On this medium, the plants produced more and longer roots and survived tissue culture (Figure 1D). After transplantation to the field, all of the GH3-8–overexpressing plants showed the phenotype of dwarf and tufted shape (Figure 1E). The most stunted plants remained in the vegetative phase and did not reproduce. Although most of the moderately dwarf plants did enter the reproductive stage, they showed greatly reduced fertility, with only one of them (D25UM8-27) setting a few seeds that had a very low germination rate. The T1 progeny of D25UM8-27 that showed a high level of GH3-8 transcripts produced shorter roots and fewer adventitious roots than the negative segregates (Figure 1F). Similar abnormal morphology was also observed in auxin-deficient Arabidopsis (Nakazawa et al., 2001; Takase et al., 2004). GH3-8–overexpressing plants also showed abnormal leaf formation. Normal rice leaves at the vegetative growth stage form in a distichous alternate phyllotaxic manner, and successive leaves develop on opposite sides of the shoot apical meristem, with 180° of divergence. However, the successive leaves of GH3-8–overexpressing plants showed <180° of divergence from the previous leaves (Figure 1G), which was similar to the IAA3-modified transgenic rice that is auxin-insensitive (Nakamura et al., 2006). Treatment of these dwarf plants grown either in soil or in hydroponic conditions with 0.1 to 20 mg/L 2,4,5-T did not promote the growth of these plants. These results suggest that tightly regulated low expression of GH3-8 is crucial for the normal growth and development of rice.
To examine the effects of reduced GH3-8 expression, RNA interference (RNAi) was used to suppress GH3-8 expression in the moderately resistant cv Minghui 63. We obtained 11 positive transgenic plants. None of the plants showed a significant difference (P > 0.05) in response to PXO61 infection compared with the wild type after partial suppression of GH3-8 (see Supplemental Table 2 online). Since GH3-8 is a member of a multigene family, functional redundancy among the family members may mask the effect of partial GH3-8 underexpression. This hypothesis was further evaluated by characterization of the mutant 03Z11EV19 with T-DNA inserted in the second exon of GH3-8 (see Supplemental Figure 1C online), identified from the Rice Mutant Database (Zhang et al., 2006). The 03Z11EV19 mutant had the genetic background of Zhonghua 11, which was moderately susceptible to Xoo strain PXO61. Some of the 03Z11EV19 plants showed slightly increased susceptibility to PXO61, and this increased susceptibility was associated with the insertion of T-DNA and the lack of GH3-8 expression (see Supplemental Figure 3 online), suggesting that the increased susceptibility was due to the loss of GH3-8 expression. The average lesion area of the homozygote GH3-8–knockout plants after PXO61 infection was 46 ± 4.8%, compared with 32 ± 9.5% for wild-type plants, indicating that GH3-8 loss of function has only a small effect on rice disease resistance. This result further supports the hypothesis of functional redundancy among GH3 family members in rice.
GH3-8 Is an IAA–Amido Synthetase and Modulates Auxin Homeostasis
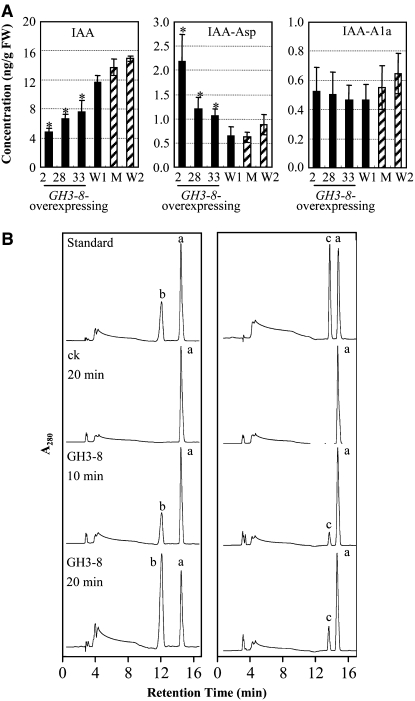
To evaluate whether the phenotype of GH3-8–overexpressing plants was associated with the level of endogenous IAA, we quantified the free IAA in GH3-8–overexpressing and knockout plants. The concentration of free IAA in the leaves of GH3-8–overexpressing plants was 1.5- to 2.4-fold lower than that of wild-type plants, suggesting that the phenotype change of these transgenic plants is very likely the result of reduced endogenous IAA (Figure 2A). GH3-8 shows 55 to 73% amino acid sequence identity and 72 to 84% sequence similarity to the Arabidopsis group II GH3 proteins GH3.2, GH3.3, GH3.4, GH3.5, GH3.6, and GH3.17, which are IAA–amido synthetases functioning to maintain auxin homeostasis by conjugating excess IAA to amino acids (see Supplemental Figure 4 online) (Staswick et al., 2005). The high degree of sequence similarity of GH3-8 with Arabidopsis IAA–amido synthetases and the correlation between GH3-8 overexpression and suppressed free IAA accumulation strongly suggest that GH3-8 encodes an IAA–amido synthetase.
Figure 2.
Function of GH3-8.
(A) Quantification of free IAA and IAA–amino acid conjugates (IAA-Asp and IAA-Ala) in the leaves of GH3-8–overexpressing plants (D25UM8-2, -28, and -33) and a GH3-8–knockout plant (03Z11EV19; M) at the booting stage. FW, fresh weight; W1, wild-type Mudanjiang 8 for GH3-8–overexpressing plants; W2, wild-type Zhonghua 11 for the GH3-8–knockout plant. Bars represent means (three replicates) ± sd. Asterisks indicate that a significant difference (P < 0.05) was detected between GH3-8–overexpressing and W1 plants.
(B) HPLC analysis of amino acid conjugates of IAA synthesized by recombinant GH3-8 protein in different time courses. Standard IAA (peak a), IAA-Asp (peak b), and IAA-Ala (peak c) were bought from Sigma-Aldrich. ck, proteins from E. coli transferred with the null vector PET28a.
To further examine this hypothesis, we quantified IAA–amino acid conjugates in the same samples used for the quantification of free IAA. The concentration of IAA-Asp in the leaves of GH3-8–overexpressing plants was 1.7- to 3.4-fold higher than that in wild-type plants, but the concentration of IAA-Ala showed no significant difference between GH3-8–overexpressing and wild-type plants (Figure 2A). We then tested the enzyme activity of recombinant GH3-8 in the reaction mixture containing IAA and Asp or Ala. Quantification analysis showed that the reactions yielded new products compared with controls (Figure 2B, ck). The new products had the same retention times as IAA-Asp and IAA-Ala standards. There was more IAA-Asp than IAA-Ala generated (Figure 2B). These results suggest that GH3-8 is an IAA-amido synthetase that is more capable of catalyzing the synthesis of IAA-Asp than IAA-Ala.
The levels of endogenous IAA, IAA-Asp, and IAA-Ala in the leaves of GH3-8–knockout mutants showed no significant difference from those in the wild type (Figure 2A). This result also supports the hypothesis of functional redundancy among GH3 family proteins in rice. Thus, only GH3-8–overexpressing plants were examined in the following analyses.
Bacterial Infection Induces Local Accumulation of IAA
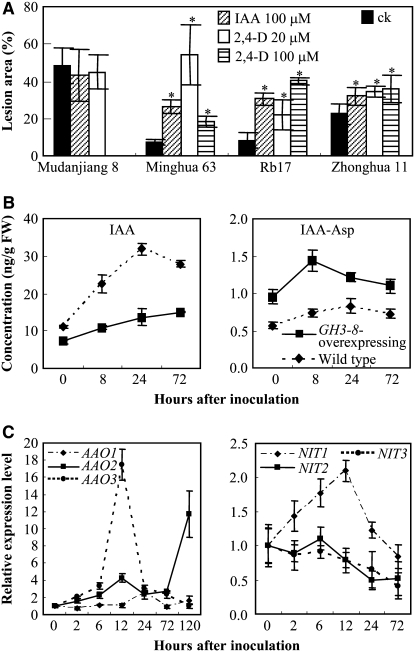
To test whether auxin influences rice resistance to Xoo, we treated the resistant rice lines Minghui 63 and Rb17, carrying bacterial blight resistance gene Xa3/Xa26, and the susceptible rice lines Mudanjiang 8 and Zhonghua 11, with IAA and 2,4-D (an analog of IAA), before inoculation with Xoo strain PXO61. Rb17 is a transgenic line carrying a single copy of Xa3/Xa26 with the genetic background of Mudanjiang 8 (Sun et al., 2004). Treating resistant rice lines with either IAA or 2,4-D significantly promoted disease symptoms; the lesion area of these treated rice lines increased 2.4- to 7.3-fold after PXO61 infection compared with that of control plants (Figure 3A). IAA or 2,4-D treatment further increased the susceptibility of moderately susceptible Zhonghua 11 and showed no influence on the susceptibility of highly susceptible Mudanjiang 8 to PXO61. The influence of auxin on the response of resistant rice lines to Xoo infection is associated with the growth rate of Xoo in plants. A bacterial growth analysis showed that the growth rate of PXO61 in resistant Minghui 63 and GH3-8–overexpressing plants increased (P < 0.01) 9- to 16-fold and 4- to 8-fold, respectively, after treating with 2,4-D at 8 to 12 d after bacterial inoculation, but treating susceptible Mudanjiang 8 with 2,4-D showed no obvious influence on the growth rate of PXO61 (see Supplemental Figure 5 online).
Figure 3.
Effect of IAA on the Development of Disease and Expression Patterns of Auxin Synthesis–Related Genes.
(A) Exogenous application of IAA or 2,4-D increased lesion area in resistant rice lines Minghui 63 and Rb17 after PXO61 inoculation. Mudanjiang 8 was not treated with 100 μM 2,4-D. Bars represent means (five replicates) ± sd. Asterisks indicate that a significant difference (P < 0.05) was detected between IAA- or 2,4-D–treated plants and untreated (ck) plants.
(B) Bacterial infection induced the accumulation of free IAA and IAA-Asp in both GH3-8–overexpressing (D25UM-28) and wild-type (Mudanjiang 8) plants. Approximately 6-cm-long leaf fragments right next to the inoculation site were used for the analysis. Each point represents a mean (three replicates) ± sd. 0 h, immediately after inoculation with PXO61. FW, fresh weight.
(C) Expression patterns of IAA synthesis–related genes on PXO61 infection in Mudanjiang 8. The expression level of each gene was calculated relative to that in the plants immediately after inoculation (0 h). Each point represents a mean (three replicates) ± sd.
To examine whether Xoo could induce IAA accumulation in infected rice, we quantified the content of free IAA after PXO61 inoculation (Figure 3B). Bacterial infection significantly (P < 0.01) increased the content of free IAA in the infected leaves of both wild-type (2.9-fold) and GH3-8–overexpressing (2.1-fold) plants, although the induction was more dramatic in the wild-type plants (Figure 3B). Furthermore, the IAA concentration was significantly higher (P < 0.01) in the wild-type plants than in the transgenic plants. Free IAA concentration in wild-type plants was 1.6-fold higher than that in GH3-8–overexpressing plants immediately after bacterial inoculation (0 h), and the difference reached 2.3-fold at 24 h after inoculation. Accompanying the accumulation of free IAA, the concentration of IAA-Asp maximally increased ∼1.5-fold in both GH3-8–overexpressing and wild-type plants (Figure 3B). However, the IAA-Asp concentration was 1.5- to 2-fold higher (P < 0.01) in GH3-8–overexpressing plants than in the wild type. These results suggest that Xoo infection induces the local accumulation of free IAA and GH3-8 suppresses the free IAA accumulation by increasing the synthesis of IAA-Asp.
To determine whether the pathogen-induced accumulation of IAA is synthesized endogenously in rice, we examined the expression of indole-3-acetaldehyde oxidase (AAO) and nitrilase (NIT) functioning in two Trp-dependent IAA biosynthesis (indole-3-pyruvic acid and indole-3-acetaldoxime) pathways (Woodward and Bartel, 2005). We searched the amino acid sequences of Arabidopsis NIT1 (protein databases accession number P32961) and AAO1 (Q7G193), which are known to be involved in IAA production (Bartling et al., 1992; Seo et al., 1998), against the rice whole genomic, EST, and full-length cDNA sequences using the TBLASTN program (Altschul et al., 1997). The search identified three putative rice AAO genes, AAO1 (GenBank accession number AK072847), AAO2 (AK103597), and AAO3 (AK065990), and three putative rice NIT genes, NIT1 (AK104033), NIT2 (AK058965), and NIT3 (AK069786). The encoding products of AAO1, AAO2, AAO3, NIT1, NIT2, and NIT3 showed 72, 69, 76, 78, 78, and 42% amino acid sequence similarity to their Arabidopsis homologs, respectively. Gene expression assays showed that bacterial infection significantly induced (P < 0.01) the expression of AAO1 (2.6-fold), AAO2 (11.7-fold), AAO3 (17.5-fold), and NIT1 (2.1-fold) at 12 to 120 h after infection (Figure 3C). These results suggest that Xoo infection induces the endogenous synthesis of IAA.
Upregulation of GH3-8 and Disease Resistance Suppress Auxin Signaling
To examine whether GH3-8 is involved in auxin signaling, the expression of auxin signaling–related genes, Aux/IAA and ARF families, was analyzed by quantitative reverse transcription–PCR (qRT-PCR) in GH3-8–overexpressing plants. The expression of IAA1, IAA4, IAA9, IAA14, IAA20, IAA24, ARF6a, ARF6b, and ARF8 was rapidly induced by IAA in wild-type plants (Figure 4A). IAA treatment also induced the expression of IAA1, IAA9, IAA14, and IAA20 in GH3-8–overexpressing plants, but the transcript levels were markedly lower. In addition, IAA did not markedly influence the expression of IAA4, IAA24, ARF6a, ARF6b, and ARF8 in GH3-8–overexpressing plants. Expression analysis of other genes that are not associated with auxin signaling suggested that the differential expression of these auxin signaling–related genes between transgenic and wild-type plants was not due to other effects, such as somatic mutation caused by tissue culture in GH3-8–overexpressing plants (see Supplemental Figure 6 online). These results suggest that the GH3-8 protein influences auxin signaling by restraining the accumulation of free IAA.
Figure 4.
Expression of Auxin Signaling–Related Genes Was Influenced by IAA and Pathogen Infection.
Mudanjiang 8 is the wild type. Each point represents a mean (three replicates) ± sd.
(A) Expression of Aux/IAA and ARF gene families was influenced after IAA treatment (10 μM) in GH3-8–overexpressing (D25UM8-2) and wild-type plants. The expression level of each gene was calculated relative to that in untreated (ck) wild-type plants.
(B) Both resistant and susceptible reactions influenced the expression of GH3-8 as well as IAA and ARF genes. Plants were inoculated with Xoo strain PXO61. 0 h (control), immediately after inoculation. The expression level of each gene was calculated relative to that in control wild-type plants.
(C) Both resistant and susceptible reactions influenced the accumulation of free IAA and IAA-Asp. 0 h (control), immediately after inoculation. Each point represents a mean (three replicates) ± sd. FW, fresh weight.
To further examine the role of auxin signaling in disease resistance, we examined the expression of GH3-8 and auxin signaling–related genes in compatible (susceptible) and incompatible (resistant) host–pathogen interactions in susceptible wild-type Mudanjiang 8 and resistant transgenic lines Rb49 and D49O, carrying bacterial blight resistance gene Xa3/Xa26 (Sun et al., 2004) and Xa21, respectively. GH3-8 expression was induced at 2 h after inoculation of Xoo strain PXO61, and this inoculation resulted in approximately threefold and twofold increases of GH3-8 transcripts in Rb49 and D49O, respectively (Figure 4B). Pathogen infection first suppressed and then induced (maximum, 3.9-fold) GH3-8 expression in the wild type. Accompanying the upregulation of GH3-8 induced by PXO61, the expression of IAA9 and ARF1 was first induced and then suppressed in both resistant lines and the wild type. In addition, the transcript levels of IAA9, ARF1, and ARF8 were markedly lower in resistant lines than in the wild type after pathogen infection, although the expression of ARF8 was only tentatively repressed in resistant lines and induced in the wild type on pathogen infection.
PXO61 infection significantly induced (P < 0.01) the accumulation of free IAA in both resistant transgenic lines Rb49 (2.1-fold) and D49O (1.9-fold) and the susceptible wild type (2.5-fold). But the IAA level in the wild type was significantly higher than that in Rb49 and D49O (Figure 4C). The IAA-Asp level was also increased in both resistant (1.6- to 1.9-fold) and susceptible (1.6-fold) plants at 8 to 24 h after pathogen infection. However, the IAA-Asp levels in Rb49 and D49O were 1.6- to 2.4-fold higher than that in the wild type at 8 to 24 h after pathogen infection. These results suggest that suppressing IAA signaling by GH3-8 may be important in R gene–mediated Xoo resistance.
Activation of Disease Resistance by GH3-8 Upregulation Is Independent of SA and JA
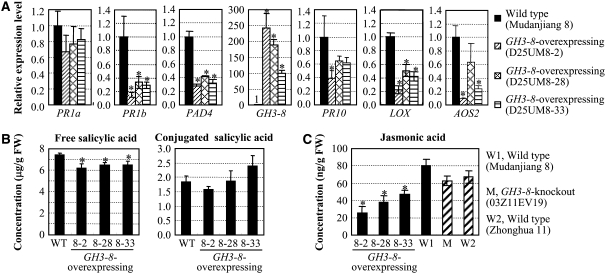
Our previous study revealed that activation of the SA-dependent pathway in the genetic background of Mudanjiang 8 can enhance rice resistance to both Xoo and M. grisea (Qiu et al., 2007). To examine whether this pathway is also involved in GH3-8–mediated resistance in Mudanjiang 8, we examined the expression of defense-responsive genes, pathogenesis-related (PR) genes PR1a (for acidic PR protein1) and PR1b (for basic PR protein1), and SA synthesis–related gene PAD4 (for phytoalexin-deficient4), which are known to function in SA signaling during disease resistance in both Arabidopsis and rice (Durrant and Dong, 2004; Qiu et al., 2007). The qRT-PCR assays showed that the transcript levels of PR1b and PAD4 in GH3-8–overexpressing plants were 2.1- to 3.7-fold lower than those in wild-type plants (Figure 5A). PR1a also tended to be suppressed in GH3-8–overexpressing plants. Accompanying the suppression of these defense-responsive genes, the free SA levels in GH3-8–overexpressing plants were also comparatively lower than those of wild-type plants (Figure 5B).
Figure 5.
Overexpression of GH3-8 Suppresses the Accumulation of SA and JA and the Expression of Defense-Responsive Genes Functioning in SA- and JA-Dependent Pathways.
Bars represent means (three replicates) ± sd. Asterisks indicate that a significant difference (P < 0.05) was detected between GH3-8–overexpressing plants and wild-type plants.
(A) Expression patterns of defense-responsive genes.
(B) SA and conjugated SA levels in rice leaves. FW, fresh weight.
(C) JA levels in rice leaves.
Lipoxygenase (LOX) and allene oxide synthase (AOS) are important enzymes in JA biosynthesis (Zhao et al., 2005). The expression of the PR10 (ribonuclease) gene was suppressed accompanying the suppression of LOX and AOS2 in rice var Mudanjiang 8 during disease resistance, suggesting that it functions in a JA-dependent pathway (Qiu et al., 2007). Overexpression of GH3-8 significantly suppressed the accumulation of endogenous JA; the JA level in GH3-8–overexpressing plants was 1.7- to 3.1-fold lower than that in wild-type plants (Figure 5C). The transcript levels of PR10, LOX, and AOS2 in GH3-8–overexpressing plants were also significantly lower than those in the wild type (Figure 5A). These results suggest that the disease resistance conferred by the upregulation of GH3-8 does not require the activation of the SA- and JA-dependent pathways; it may be a basal resistance.
Activation of GH3-8 and Disease Resistance Accompanies the Suppression of Expansin Genes
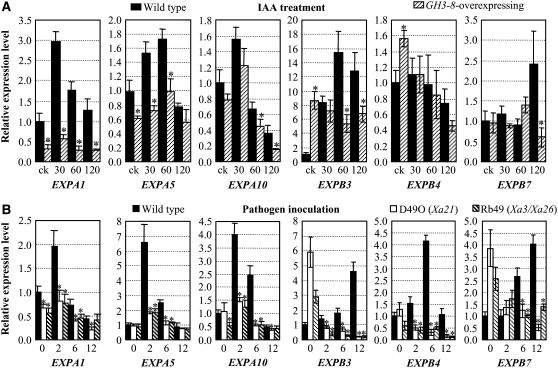
Expansins, a multiprotein family, are wall-loosening proteins of plant cell walls and are known to function in auxin-regulated growth (McQueen-Mason et al., 1992; Hager, 2003). However, loosening cell walls can make the cells vulnerable to invaders. To evaluate whether GH3-8 influences the function of expansins, we analyzed the expression of three rice α-expansin genes, EXPA1, EXPA, and EXPA10, and three rice β-expansin genes, EXPB3, EXPB4, and EXPB7, named according to Kende et al. (2004), in leaf tissue. IAA induced the expression of EXPA1, EXPA5, EXPB3, and EXPB7 and first induced and then suppressed EXPA10 in wild-type plants (Figure 6A). EXPA1, EXPA5, EXPA10, and EXPB7 displayed similar expression patterns in GH3-8–overexpressing plants as well, but the maximum expression levels were 6.2-, 2.1-, 2.4-, and 4.0-fold lower than those in the wild type, respectively (Figure 6A). The EXPB3 transcript level was significantly higher in GH3-8–overexpressing plants than in wild-type plants without IAA treatment but was significantly lower in GH3-8–overexpressing plants after IAA treatment. IAA treatment appeared to suppress EXPB4 expression in both GH3-8–overexpressing and wild-type plants, but EXPB4 expression was significantly higher in GH3-8–overexpressing plants than in wild-type plants without IAA treatment. These results suggest that GH3-8 inhibited some expansin production, which was induced by IAA.
Figure 6.
IAA and Bacteria Influence the Expression of Expansin Genes.
Bars represent means (three replicates) ± sd. Asterisks indicate that a significant difference (P < 0.05) was detected between GH3-8–overexpressing plants or R-gene–carrying transgenic plants and wild-type (Mudanjiang 8) plants.
(A) Expression patterns of expansin genes in GH3-8–overexpressing (D25UM8-2) or wild-type plants after IAA treatment. The plants were treated with IAA for 30, 60, or 120 min. ck, without treatment.
(B) Pathogen infection influenced the expression of expansin genes in both R gene–carrying (D49O and Rb49) and wild-type plants. Plants were inoculated with PXO61 for 0 (measured immediately after inoculation), 2, 6, or 12 h.
To further examine the role of expansin in disease resistance, we examined the expression of expansin genes in susceptible wild-type Mudanjiang 8 and resistant transgenic lines Rb49 and D49O carrying the R gene. The susceptible reaction markedly induced the expression of all six expansin genes (Figure 6B). The resistant reaction suppressed EXPA1, EXPA10, EXPB3, EXPB4, and EXPB7. Although PXO61 inoculation slightly induced EXPA5 in rice lines Rb49 and D49O, the expression level of EXPA5 was significantly lower in the resistant lines than in the susceptible wild type. In addition, the expression levels of the other five expansin genes in the resistant lines were also significantly lower than in the wild type after pathogen challenge. These results suggest that the suppression of expansin genes may be important for the resistance against Xoo.
To examine the above hypothesis, we overexpressed EXPA1, EXPA5, and EXPA10 in moderately susceptible rice var Zhonghua 11. All of the transgenic plants overexpressing EXPA5 showed significantly increased susceptibility to Xoo strain PXO61; the lesion area of these transgenic plants increased 1.8- to 2.7-fold (Figure 7). The EXPA1- and EXPA10-overexpressing plants also showed a tendency toward increased susceptibility to PXO61 infection (Figure 7). These results suggest that pathogen-induced expression of expansin genes is one of the causes of susceptibility.
Figure 7.
Overexpression of Expansin Genes (EXPA1, EXPA5, and EXPA10) Was Associated with Increased Susceptibility to Xoo Strain PXO61.
Bars represent means (three replicates) ± sd. Asterisks indicate that a significant difference (P < 0.05) was detected between transgenic plants (OVEXPA1-, OVEXPA5-, and OVEXPA10-) and wild-type Zhonghua 11 (W).
GH3-8 Expression Is Regulated by Multiple Factors
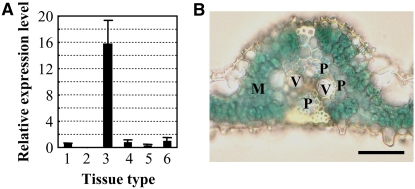
To study the regulation of GH3-8, we examined its expression after treatment with IAA, SA, and JA as well as in different plant tissues. GH3-8 expression was induced at 30 min and reached an ∼11-fold increase after treatment with IAA for 2 h (see Supplemental Figure 7 online). In addition to IAA, SA also rapidly induced the expression of GH3-8 at 5 min after treatment. Compared with the wounding control, the expression of GH3-8 was first suppressed at 5 min and then induced at 1 h after JA treatment. GH3-8 showed variable levels of expression in different tissues (Figure 8A). It had very low expression levels in leaf, pistil, root, and young panicle but an ∼16-fold higher level in stamen. No GH3-8 expression was detected in the sheath. Analysis of GH3-8 promoter (PGH3-8) and marker gene β-glucuronidase (GUS) fusion revealed that GH3-8 was preferentially expressed in the parenchyma cells surrounding the vascular vessels and mesophyll cells in leaf tissue (Figure 8B). These results indicate that the function of GH3-8 is tissue-specific, growth stage–specific, and signal molecule–regulated.
Figure 8.
Expression Pattern of GH3-8.
(A) GH3-8 had different expression levels in various tissues analyzed by qRT-PCR. Tissues were obtained from rice var Minghui 63. Column 1, leaf; column 2, sheath; column 3, stamen; column 4, pistil; column 5, root; column 6, young panicle (3- to 5-cm stage). Bars represent means (three replicates) ± sd.
(B) PGH3-8:GUS expression in transgenic rice plants. Blue indicates the expression of GUS. M, mesophyll cell; P, parenchyma cells; V, vascular elements. Bar = 30 μm.
DISCUSSION
Although slowly accumulating data indicate that auxin is a pathogen virulence factor (Sequeira and Kelman, 1962; Glickmann et al., 1998; O'Donnell et al., 2003; Navarro et al., 2006; Park et al., 2007; Wang et al., 2007; Zhang et al., 2007), it is largely unknown how this factor functions at the molecular level during pathogen infection and how the plant defense system interacts with it. Our results suggest that Xoo-induced auxin production may weaken the rice cell wall, the native defense barrier; rice in turn defends itself from auxin-facilitated bacterial infection by preventing the loosening of its cell wall by suppressing auxin signaling. Rice does this by activation of an IAA–amino synthetase that inactivates IAA, the major form of auxin, by conjugating it to amino acids.
Xoo Infection Induces Endogenous Synthesis of IAA in Rice
Auxin, the essential plant hormone regulating development and growth, has been reported to be used by some pathogens as a virulence factor for infection. Tobacco (Nicotiana tabacum) infected with the vascular pathogen Pseudomonas solanacearum exhibits symptoms associated with auxin imbalance; the IAA content increased 100-fold in infected plants (Sequeira and Kelman, 1962). Bacterial infection accompanied the accumulation of IAA in Arabidopsis (O'Donnell et al., 2003). Exogenous application of an auxin analog enhances bacterial disease symptoms (Navarro et al., 2006). However, the origin of IAA in infected tissues remains elusive. Limited information suggests that the host contributes most of the auxin during early stages of pathogenesis (Sequeira, 1965). Other studies indicate that some pathogenic bacteria, such as Pseudomonas syringae, can produce IAA (Glickmann et al., 1998). Some plant bacteria carry Trp monooxygenase (iaaM) and indole-3-acetamide hydrolase (iaaH) genes, which catalyze the biosynthesis of IAA through Trp-dependent indole-3-acetamide pathways (Woodward and Bartel, 2005). In addition, other bacteria produce IAA but do not carry genes highly similar to iaaM and iaaH (Glickmann et al., 1998).
Consistent with previous observations, our results suggest that IAA is also a virulence factor in Xoo-induced disease. Xoo infection induced the local accumulation of IAA accompanying the upregulation of IAA synthesis–related genes in rice plants. In addition, R gene–mediated Xoo resistance accompanies the suppression of auxin signaling. A search of the amino acid sequences of Pseudomonas syringae pv syringae iaaH and iaaM (Mazzola and White, 1994) against the putative encoding products of the Xoo (strains KACC10331 and MAFF311018) whole genome sequence (Lee et al., 2005; Ochiai et al., 2005) did not reveal any homologous sequence. We then searched databases and found that Xoo carries NIT proteins (National Center for Biotechnology Information protein database accession numbers AAW74756, AAW74754, and YP_200139), which may contribute to the accumulation of IAA. However, as demonstrated by the results presented in Figure 1B, the number of bacteria increased continuously in the leaf tissue during the 12 d after infection, whereas the infection-induced IAA accumulation leveled off ∼1 d after inoculation (Figure 3B). These results indicate that the contribution of IAA synthesized by the bacterial genes is trivial, meaning that endogenous synthesis by the host genes is the major source for IAA accumulation during the rice–Xoo interaction. Similar results were also observed in another study; the IAA accumulation in Arabidopsis was independent of bacterial colonization, and the magnitude and timing of host accumulation of IAA in response to different bacteria (Xanthomonas campestris pv campestris versus Pseudomonas syringae pv tomato) infection were the same (O'Donnell et al., 2003). Thus, endogenously produced auxin may amplify the bacterial infection signal, which can rapidly activate the plant's defense responses, such as the suppression of auxin signaling via the activation of GH3-8, or facilitate pathogen colonization.
GH3-8 Mediates a Basal Resistance That Does Not Require the Activation of SA- or JA-Dependent Signaling Pathways
Plants appear to have evolved different ways to suppress auxin signaling during disease resistance. Suppressing the expression of auxin receptors by microRNA is one way for Arabidopsis to prevent infection by the bacterium P. syringae; this type of resistance appears specific to virulent pathogens, but race-specific resistance does not (Navarro et al., 2006). Suppression of auxin signaling by activation of a GH3-type protein enhanced bacterial resistance by activating the expression of the PR-1 gene, whose activation is an indicator of systemic acquired resistance, in Arabidopsis (Durrant and Dong, 2004; Park et al., 2007). SA was involved in the suppression of auxin signaling during disease resistance in Arabidopsis (Wang et al., 2007; Zhang et al., 2007). Our results indicate that rice at least partly resists Xoo infection by local and pathogen-induced activation of an IAA–amido synthetase that prevents cell wall disturbance by suppressing auxin signaling.
The molecular mechanisms of auxin as a virulence factor to overcome the defense system of plants are unknown. Our results suggest that Xoo-induced auxin-stimulated local expansin production may be one of the mechanisms used by this pathogen to infect rice plants. The cell wall plays an important role in basal resistance against pathogens (Huckelhoven, 2007), but it is also a restricting factor in plant cell growth. In general, the loosening cell wall may facilitate secretion pilus penetration or pathogen entry or may provide more nutrient leakage, resulting in a suitable growth environment for pathogens (Huckelhoven, 2007). Expansins are wall-loosening proteins in acid-dependent cell extension growth activated by auxin (McQueen-Mason et al., 1992; Hager, 2003). Although only limited information indicates the involvement of expansins in pathogen invasion (Balestrini et al., 2005; Wieczorek et al., 2006), the following evidence supports expansins playing a role in the rice–Xoo interaction. First, auxin, the virulence factor associated with Xoo infection, can induce the expression of some members of the expansin families. This auxin-induced expansin expression was suppressed in GH3-8–overexpressing plants and accompanied the enhanced disease resistance. Second, virulent pathogen induced the accumulation of expansin transcripts in susceptible plants, but avirulent pathogen suppressed expansin expression in resistant plants. Last, overexpressing expansin genes increased rice susceptibility. This evidence suggests that GH3-8–mediated bacterial resistance may be at least partly due to inhibiting the expression of expansins by suppressing auxin signaling, which results in the maintenance of the physical barrier of plant cells to infection. In addition, GH3-8 is induced by multiple pathogens, including different strains of Xoo, the cause of bacterial blight, and M. grisea, the cause of fungal blast, suggesting that its function in disease resistance is not isolate-specific (Wen et al., 2003). Thus, GH3-8 regulates basal resistance.
Antagonistic interaction with defense signaling pathways may be another molecular mechanism of auxin in pathogenesis. A recent study reported that auxin may downregulate the host defense response by inhibiting the full induction of SA-mediated PR-1 expression and that inhibition of auxin signaling is part of the SA-mediated disease resistance in Arabidopsis (Wang et al., 2007). SA- and JA-dependent pathways are known to be involved in the regulation of defense against different pathogens (Thomma et al., 1998). However, the reduced accumulation of SA and JA and the suppressed expression of SA- and JA-responsive genes in GH3-8–overexpressing plants strongly suggest that GH3-8–mediated Xoo resistance does not require the activation of SA and JA signaling. This result is consistent with a previous report that showed that the basal resistance for bacterial pathogens in tomato (Solanum lycopersicum) is SA-independent (O'Donnell et al., 2001). However, suppressing auxin signaling by the activation of GH3-8 repressed JA and SA signaling (Figure 5). Both JA and SA, in addition to IAA, influenced GH3-8 expression (see Supplemental Figure 7 online). Studies have suggested that basal resistance may overlap with R gene–mediated resistance, resulting in a network of crosstalk between the defense pathways deployed (Abramovitch et al., 2006; Jones and Dangl, 2006). Our results suggest that auxin-dependent signaling interacts with JA- and SA-dependent signaling during bacterial resistance, although activation of the two known defense transduction pathways was not required for this bacterial resistance. The putative molecular mechanisms of the interaction are predicted as follows, although further studies are needed to examine these hypotheses. First, GH3-8 might conjugate amino acids to SA and JA in addition to IAA, resulting in reduced accumulation of free JA and SA in the transgenic plants. Some GH3 family members in Arabidopsis are known to function as adenylate-forming enzymes that conjugate amino acids to IAA, JA, or SA; At GH3.5 can adenylate IAA as well as SA (Staswick et al., 2002, 2005). However, overexpression of GH3-8 may cause nonphysiological adenylation of JA and SA. Second, transcription factors functioning in IAA-dependent signaling may regulate the expression of genes functioning in JA- and SA-dependent signaling. The ARF6 and ARF8 transcription factors promote JA production in Arabidopsis (Nagpal et al., 2005). This may explain the suppressed expression of ARF6a, ARF6b, and ARF8 accompanied by reduced accumulation of JA and suppressed expression of JA synthesis–related genes in GH3-8–overexpressing rice.
Because of the functional redundancy of the GH3 family in rice, GH3-8–suppressing and –knockout plants are not informative for studying the interaction of auxin signaling and defense responses. Although overexpressing a gene may have pleiotropic effects that are not directly related to the normal gene function, the following evidence suggests that GH3-8 plays a role in disease resistance in physiological conditions. First, different avirulent isolates induced GH3-8 expression in different resistant rice lines, suggesting its involvement in defense responses in an isolate-nonspecific way (Wen et al., 2003). Second, suppressing the accumulation of IAA, auxin signaling, and the expression of expansin genes accompanied the upregulation of GH3-8 in disease resistance (Figures 4B, 4C, and 6B). Thus, activation of GH3-8 is required in pathogen-induced defense responses.
Dual Roles of GH3-8 in Development and Disease Resistance Are Antagonistically Regulated
The results presented here also indicate that GH3-8 plays important roles in plant growth and development. The expressional characteristics of GH3-8 indicate that its function is restricted to vegetative and early reproductive development. Thus, constitutive overexpression of GH3-8 suppressed auxin action, which resulted in abnormal morphology similar to that reported in auxin-deficient plants (Nakazawa et al., 2001; Takase et al., 2004). Xoo is a vascular pathogen. The bacteria live in the vascular system of rice plants. GH3-8 was preferentially expressed in the cells surrounding the vascular vessels. The expressional location and bacteria-induced expression of GH3-8 further confirm its native role in bacterial resistance. Thus, as a regulator of auxin homeostasis, GH3-8 plays two roles in the rice life cycle. In the absence of pathogen invasion, it functions as a repressor of auxin-dependent developmental signaling; its function is limited to various tissues and developmental stages to maintain normal growth and development. When facing bacterial infection, GH3-8 functions as an activator of disease resistance by inducing a transient and cell-specific suppression of auxin signaling. The dual roles of GH3-8 also indicate that it is one of the points of crosstalk between development and disease resistance pathways, which may partly explain the fitness cost in disease resistance.
METHODS
Gene Isolation and Structure Analysis
EI5P11, the partial cDNA sequence of a GH3-8 allele from the rice (Oryza sativa) cv Minghui 63, was used as a query to search GenBank to identify similar sequence and design primers for PCR isolation of the GH3-8 gene from the resistant rice line C101LAC. The PCR product was cloned into the pUC19 vector, and the plasmid was named T5P11. The structure of GH3-8 was determined by sequencing cDNA of the transcript of the gene (see Supplemental Methods online).
Transformation
The overexpression construct of GH3-8 was produced by removing GH3-8 from plasmid T5P11 using the restriction enzymes NotI and ApaI and ligating it into the transformation vector pU1301 (see Supplemental Figure 1A online). To construct an RNAi vector for GH3-8, a 456-bp cDNA fragment of GH3-8 was obtained from cDNA clone EI5P11 of rice line Minghui 63 and inserted into the pDS1301 vector (see Supplemental Figure 1B online). The overexpression constructs of EXPA1, EXPA5, and EXPA10 were produced by amplifying the coding regions of the genes in Minghui 63 using gene-specific primers (see Supplemental Table 3 online) and ligating the PCR products into vector pU1301. Agrobacterium tumefaciens–mediated transformation was performed according to the protocol of Lin and Zhang (2005).
Pathogen Inoculation
Plants were inoculated with Philippine Xanthomonas oryzae pv oryzae (Xoo) strain PXO61 by the leaf-clipping method at the booting stage, as described previously (Sun et al., 2004). Disease was scored according to percentage lesion area (lesion length/leaf length) at 2 weeks after inoculation. Mock-inoculated (control) plants were treated under the same conditions, except that the pathogen suspension was replaced with deionized water.
For studying the effect of IAA on disease development, rice plants were grown in a greenhouse at 25°C, at 70% RH, and with a 12-h photoperiod. Plants at the six-leaf stage were sprayed with solution containing 20 or 100 μM 2,4-D or 100 μM IAA diluted in 0.02% Tween 20. The control plants were sprayed with solution containing 0.02% Tween 20. The plants were then immediately inoculated with PXO61. The bacterial inoculum was prepared as described previously by Sun et al. (2004), except that the inoculum for IAA or 2,4-D–treated plants contained 20 or 100 μM 2,4-D or 100 μM IAA.
Quantification of IAA, IAA-Ala, IAA-Asp, JA, and SA
The sample preparation protocol for IAA, IAA–amino acid conjugates, and JA quantification was provided by Katayoon Dehesh of the University of California at Davis (see Supplemental Methods online). In brief, 1 g of leaves from plants at the booting stage was used for sample preparation. D2-IAA (Sigma-Aldrich) was used as an internal standard for IAA and IAA–amino acid quantification, and 10-Dihydro-JA (Olchemim) was used as an internal standard for JA quantification. The sample was purified using a C18-SepPak cartridge (Waters). IAA, IAA–amino acids, and JA were quantified using the HPLC/electrospray ionization/tandem mass spectrometry system. The quantitative data of IAA, IAA-Ala, and IAA-Asp were obtained using the peaks of the precursor ions 176.3, 247.2, and 291.2, respectively, and the peak of the product ion 130 for all molecules. The quantitative data of D2-IAA were obtained using the peaks of the precursor ion 178.3 and the product ion 132. The quantitative data of JA and 10-Dihydro-JA were obtained using the peaks of the precursor ions 209.1 and 211.2 and the product ions 109 and 59, respectively.
To quantify free and conjugated SA, each sample was harvested from plants at the booting stage. The SA samples were extracted and quantified as described previously (Qiu et al., 2007).
Enzyme Assay
The coding region of GH3-8 was obtained by PCR amplification of cDNA from rice line C101LAC using primers OsDR2F2 and OsDR2R1 (see Supplemental Table 3 online). The product was cloned into a PET28a vector (EMD Biosciences) to generate His-GH3-8. The fusion protein construct and null PET28a vector were expressed in Escherichia coli (BL21) according to the manufacturer's protocol. The expressed proteins were purified with the B-PER 6xHis fusion protein spin purification kit (Pierce) according to the manufacturer's instructions. The reaction for IAA–amino acid conjugate formation was performed according to the procedure described previously (Staswick et al., 2005). IAA and IAA–amino acid conjugates in the reaction mixture were quantified using HPLC. The sample (20 μL) of reaction product was diluted with 80 μL of methanol. An aliquot (5 μL) of the diluted sample was injected into the HPLC system (Agilent 1100; Agilent Technologies) equipped with an Agilent C18 Zorbax ODS column (250 × 5 mm). The flow rate was 1 mL/min, and the sample was eluted with 0.1% H3PO4 (5 min), followed by a linear methanol gradient to 40% methanol in 4 min and holding at this composition for an additional 8 min. The column effluent was monitored at 280 nm. Under these conditions, the retention times of IAA-Asp, IAA-Ala, and IAA were 12.2, 13.5, and 14.6 min, respectively.
RNA Gel Blot and qRT-PCR
Aliquots (20 μg) of total RNA were used for RNA gel blot analysis. A 493-bp cDNA fragment of GH3-8, which was amplified using primers 5P11F2 and 5P11GSP2 (see Supplemental Table 3 online), was used as a hybridization probe. RT-PCR was conducted as described by Wen et al. (2003). Quantitative PCR was performed using the ABI 7500 real-time PCR system (Applied Biosystems) according to the manufacturer's instructions. Supplemental Table 4 online lists the PCR primers for all of the genes. The expression level of actin was used to standardize the RNA sample for each qRT-PCR. For each gene, qRT-PCR assays were repeated at least twice, with each repetition having three replicates; similar results were obtained in repeated experiments.
Hormone Treatment
Chemical treatments were applied as described previously (Qiu et al., 2007). Minghui 63 was cultivated in a greenhouse at 25°C, at 70% RH, and with a 12-h photoperiod for 21 d. Approximately 2-cm-long leaf segments were cut from the fully expanded leaves and floated on 30 mL of solution containing the test compound in covered sterile Petri dishes. Leaf segments floated on deionized water served as an appropriate control, which is also called wounding treatment by cut in these experiments. The concentration of working solution was 100 μM for the hormones.
GH3-8 Promoter–GUS Analysis
The promoter region of GH3-8 from rice var Zhonghua 15 (∼1.8 kb) was obtained by PCR amplification (see Supplemental Methods online). The promoter-GUS fusion was introduced to rice var Zhonghua 15 with Agrobacterium-mediated transformation.
Accession Numbers
The sequence of GH3-8 from this article can be found in the GenBank/EMBL data libraries under accession number EF103572. Additional accession numbers can be found in Supplemental Table 4 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Diagrams of the GH3-8 Gene, the Transformation Constructs of GH3-8, and the T-DNA Insertion Mutant of GH3-8.
Supplemental Figure 2. GH3-8 Expression in T0 GH3-8–Overexpressing Plants (D25UM8) and the Wild Type (Mudanjiang 8).
Supplemental Figure 3. Knockout of GH3-8 (Line 03A11EV19) Increases Rice Susceptibility to Xoo Strain PXO61.
Supplemental Figure 4. Sequence Comparison of Rice GH3-8 Protein and Arabidopsis GH3 Proteins.
Supplemental Figure 5. Effect of Auxin on the Growth Rate of Xoo and the Development of Disease.
Supplemental Figure 6. Expression of Two Tissue-Specific Expressed Genes after IAA Treatment.
Supplemental Figure 7. Effects of Different Signal Molecules on the Expression of GH3-8 Analyzed by qRT-PCR.
Supplemental Table 1. Performance of GH3-8–Overexpressing Plants (D25UM8) after Pathogen (Xoo Strain PXO61) Inoculation.
Supplemental Table 2. Performance of GH3-8–Suppressing Plants (D26RMH) after Pathogen (Xoo Strain PXO61) Inoculation.
Supplemental Table 3. PCR Primers Used for Gene Structure and Expression Analyses and Vector Construction.
Supplemental Table 4. Gene-Specific Primers for qRT-PCR.
Supplemental Methods.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Program of High Technology Development of China, the National Program on the Development of Basic Research in China, and the National Natural Science Foundation of China.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Shiping Wang (swang@mail.hzau.edu.cn).
Online version contains Web-only data.
References
- Abramovitch, R.B., Anderson, J.C., and Martin, G.B. (2006). Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol. 7 601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, T., Tena, G., Plotnikova, J., Willmann, M.R., Chiu, W.L., Gomez-Gomez, L., Boller, T., Ausubel, F.M., and Sheen, J. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415 977–983. [DOI] [PubMed] [Google Scholar]
- Balestrini, R., Cosgrove, D.J., and Bonfante, P. (2005). Differential location of alpha-expansin proteins during the accommodation of root cells to an arbuscular mycorrhizal fungus. Planta 220 889–899. [DOI] [PubMed] [Google Scholar]
- Bartling, D., Seedorf, M., Mithofer, A., and Weiler, E.W. (1992). Cloning and expression of an Arabidopsis nitrilase which can convert indole-3-acetonitrile to the plant hormone, indole-3-acetic acid. Eur. J. Biochem. 205 417–424. [DOI] [PubMed] [Google Scholar]
- Bartsch, M., Gobbato, E., Bednarek, P., Debey, S., Schultze, J.L., Bautor, J., and Parker, J.E. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant, W.E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42 185–209. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr. Opin. Plant Biol. 4 301–308. [DOI] [PubMed] [Google Scholar]
- Glickmann, E., Gardan, L., Jacquet, S., Hussain, S., Elasri, M., Petit, A., and Dessaux, Y. (1998). Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol. Plant Microbe Interact. 11 156–162. [DOI] [PubMed] [Google Scholar]
- Hagen, G., and Guilfoyle, T.J. (1985). Rapid induction of selective transcription by auxins. Mol. Cell. Biol. 5 1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager, A. (2003). Role of the plasma membrane H+-ATPase in auxin-induced elongation growth: Historical and new aspects. J. Plant Res. 116 483–505. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Parker, J.E. (2003). Deciphering plant-pathogen communication: Fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14 177–193. [DOI] [PubMed] [Google Scholar]
- Huckelhoven, R. (2007). Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45 101–127. [DOI] [PubMed] [Google Scholar]
- Jain, M., Kaur, N., Garg, R., Thakur, J.K., Tyagi, A.K., and Khurana, J.P. (2006. a). Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genomics 6 47–59. [DOI] [PubMed] [Google Scholar]
- Jain, M., Kaur, N., Tyagi, A.K., and Khurana, J.P. (2006. b). The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct. Integr. Genomics 6 36–46. [DOI] [PubMed] [Google Scholar]
- Jones, J.D., and Dangl, J.L. (2006). The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Kende, H., Bradford, K., Brummell, D., Cho, H.T., Cosgrove, D., Fleming, A., Gehring, C., Lee, Y., McQueen-Mason, S., Rose, J., and Voesenek, L.A. (2004). Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol. Biol. 55 311–314. [DOI] [PubMed] [Google Scholar]
- Lee, B.M., et al. (2005). The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.J., and Zhang, Q. (2005). Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep. 23 540–547. [DOI] [PubMed] [Google Scholar]
- Mazzola, M., and White, F.F. (1994). A mutation in the indole-3-acetic acid biosynthesis pathway of Pseudomonas syringae pv. syringae affects growth in Phaseolus vulgaris and syringomycin production. J. Bacteriol. 176 1374–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason, S., Durachko, D.M., and Cosgrove, D.J. (1992). Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal, P., Ellis, C.M., Weber, H., Ploense, S.E., Barkawi, L.S., Guilfoyle, T.J., Hagen, G., Alonso, J.M., Cohen, J.D., Farmer, E.E., Ecker, J.R., and Reed, J.W. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132 4107–4118. [DOI] [PubMed] [Google Scholar]
- Nakamura, A., Umemura, I., Gomi, K., Hasegawa, Y., Kitano, H., Sazuka, T., and Matsuoka, M. (2006). Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J. 46 297–306. [DOI] [PubMed] [Google Scholar]
- Nakazawa, M., Yabe, N., Ichikawa, T., Yamamoto, Y.Y., Yoshizumi, T., Hasunuma, K., and Matsui, M. (2001). DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 25 213–221. [DOI] [PubMed] [Google Scholar]
- Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., Voinnet, O., and Jones, J.D. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312 436–439. [DOI] [PubMed] [Google Scholar]
- Ochiai, H., Inoue, Y., Takeya, M., Sasaki, A., and Kaku, H. (2005). Genome sequence of Xanthomonas oryzae pv. oryzae suggests contribution of large numbers of effector genes and insertion sequences to its race diversity. Jpn. Agric. Res. Q. 39 275–287. [Google Scholar]
- O'Donnell, P.J., Jones, J.B., Antoine, F.R., Ciardi, J., and Klee, H.J. (2001). Ethylene-dependent salicylic acid regulates an expanded cell death response to a plant pathogen. Plant J. 25 315–323. [DOI] [PubMed] [Google Scholar]
- O'Donnell, P.J., Schmelz, E.A., Moussatche, P., Lund, S.T., Jones, J.B., and Klee, H.J. (2003). Susceptible to intolerance—A range of hormonal actions in a susceptible Arabidopsis pathogen response. Plant J. 33 245–257. [DOI] [PubMed] [Google Scholar]
- Park, J.E., Park, J.Y., Kim, Y.S., Staswick, P.E., Jeon, J., Yun, J., Kim, S.Y., Kim, J., Lee, Y.H., and Park, C.M. (2007). GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 282 10036–10046. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M., and Van Loon, L.C. (2004). NPR1: The spider in the web of induced resistance signaling pathways. Curr. Opin. Plant Biol. 7 456–464. [DOI] [PubMed] [Google Scholar]
- Qiu, D., Xiao, J., Ding, X., Xiong, M., Cai, M., Cao, Y., Li, X., Xu, C., and Wang, S. (2007). OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 20 492–499. [DOI] [PubMed] [Google Scholar]
- Sato, Y., Nishimura, A., Ito, M., Ashikari, M., Hirano, H.Y., and Matsuoka, M. (2001). Auxin response factor family in rice. Genes Genet. Syst. 76 373–380. [DOI] [PubMed] [Google Scholar]
- Seo, M., Akaba, S., Oritani, T., Delarue, M., Bellini, C., Caboche, M., and Koshiba, T. (1998). Higher activity of an aldehyde oxidase in the auxin-overproducing superroot1 mutant of Arabidopsis thaliana. Plant Physiol. 116 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira, L. (1965). Origin of indoleacetic acid in tobacco plants infected by Pseudomonas solanacearum. Phytopathology 55 1232–1236. [Google Scholar]
- Sequeira, L., and Kelman, A. (1962). The accumulation of growth substances in plants infected by Pseudomonas solanacearum. Phytopathology 52 439–448. [Google Scholar]
- Shen, Q.H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., Seki, H., Ulker, B., Somssich, I.E., and Schulze-Lefert, P. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315 1098–1103. [DOI] [PubMed] [Google Scholar]
- Silverman, P., Seskar, M., Kanter, D., Schweizer, P., Metraux, J.P., and Raskin, I. (1995). Salicylic acid in rice (biosynthesis, conjugation, and possible role). Plant Physiol. 108 633–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M.T., Maldonado, M.C., and Suza, W. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Tiryaki, I., and Rowe, M.L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Cao, Y., Yang, Z., Xu, C., Li, X., Wang, S., and Zhang, Q. (2004). Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encoding a LRR receptor kinase-like protein. Plant J. 37 517–527. [DOI] [PubMed] [Google Scholar]
- Takase, T., Nakazawa, M., Ishikawa, A., Kawashima, M., Ichikawa, T., Takahashi, N., Shimada, H., Manabe, K., and Matsui, M. (2004). ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 37 471–483. [DOI] [PubMed] [Google Scholar]
- Thomma, B., Eggermont, K., Penninckx, I., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D., Pajerowska-Mukhtar, K., Culler, A.H., and Dong, X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17 1784–1790. [DOI] [PubMed] [Google Scholar]
- Wen, N., Chu, Z., and Wang, S. (2003). Three types of defense-responsive genes are involved in resistance to bacterial blight and fungal blast diseases in rice. Mol. Genet. Genomics 269 331–339. [DOI] [PubMed] [Google Scholar]
- Wieczorek, K., Golecki, B., Gerdes, L., Heinen, P., Szakasits, D., Durachko, D.M., Cosgrove, D.J., Kreil, D.P., Puzio, P.S., Bohlmann, H., and Grundler, F.M. (2006). Expansins are involved in the formation of nematode-induced syncytia in roots of Arabidopsis thaliana. Plant J. 48 98–112. [DOI] [PubMed] [Google Scholar]
- Woodward, A.W., and Bartel, B. (2005). Auxin: Regulation, action, and interaction. Ann. Bot. (Lond.) 95 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Li, Q., Li, Z., Staswick, P.E., Wang, M., Zhu, Y., and He, Z. (2007). Dual regulation role of GH3.5 in salicylic acid and auxin signaling during Arabidopsis-Pseudomonas syringae interaction. Plant Physiol. 145 450–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Li, C., Wu, C., Xiong, L., Chen, G., Zhang, Q., and Wang, S. (2006). RMD: A rice mutant database for functional analysis of the rice genome. Nucleic Acids Res. 34 D745–D748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Davis, L.C., and Verpoorte, R. (2005). Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol. Adv. 23 283–333. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.