Abstract
The oil palm is a tropical oil bearing tree. Recently EST-derived SNPs and SSRs are a free by-product of the currently expanding EST (Expressed Sequence Tag) data bases. The development of high-throughput methods for the detection of SNPs (Single Nucleotide Polymorphism) and small indels (insertion / deletion) has led to a revolution in their use as molecular markers. Available (5452) Oil palm EST sequences were mined from dbEST of NCBI. CAP3 program was used to assemble EST sequences into contigs. Candidate SNPs and Indel polymorphisms were detected using the perl script auto_snip version 1.0 which has used 576 ESTs for detecting SNPs and Indel sites. We found 1180 SNP sites and 137 indel polymorphisms with frequency 1.36 SNPs / 100 bp. Among the six tissues from which the EST libraries had been generated, mesocarp had high frequency of 2.91 SNPs and indels per 100 bp whereas the zygotic embryos had lowest frequency of 0.15 per 100 bp. We also used the Shannon index to analyze the proportion of ten possible types of SNP/indels. ESTs from tissues of normal apex showed highest values of Shannon index (0.60) whereas abnormal apex had least value (0.02). The present report deals the use of Shannon index for comparing SNP/ indel frequencies mined from ESTlibraries and also confirm that the frequency of SNP occurrence in oil palm to use them as markers for genetic studies.
Keywords: Elaeis guineensis, in silico, molecular markers, Shannon index
Background
The oil palm is a tropical palm tree important as oilseed next only to soybean. It belongs to the species Elaeis guineensis from tropical western Africa. It is allogamous and propagated via seeds. A related species is known from South America, E. oleifera (HBK). Oil palm has a large diploid genome of 3400 MB distributed in 32 chromosomes. Oil is extracted from the mesocarp both pulp and kernel of the fruit. Oil palm is a large tree and produces thousands of fruits, in compact bunches whose weight varies between 10 and 40 kilograms.
ESTs are small pieces of DNA sequence (usually 200 to 500 nucleotides long) that are generated by sequencing either one or both ends of gene transcripts. These are bits of DNA that represent genes expressed in certain cells, tissues, or organs from different organisms these “tags” are used to fish a gene from chromosomal DNA by matching base pairs. dbEST [1] a division of GenBank that contains sequence data and other information on “single-pass” cDNA sequences, or Expressed Sequence Tags, from many organisms.
EST resources are useful in genetic studies and are helpful in understanding the tissue and species-specific gene expression. Molecular Marker types such as SSRs (Simple Sequence Repeat) and SNPs can be searched in these EST databases and employed for designing locus-specific primers. In the past, development of these markers was expensive, but now EST-derived SNPs and SSRs are a free by-product of the currently expanding EST databases. These SNPs and SSRs are obviously limited to those species with large number of ESTs. The usefulness of these EST-derived SNPs and SSRs also lies in their expected transferability, since they are based on the conserved coding region of the genome. ESTs provide valuable but incomplete information. However, because they represent expressed genomic regions, ESTs are thought to identify the parts of the genome with the most biological significance.
The development of high-throughput methods for the detection of single nucleotide polymorphisms (SNPs) and small indels (insertion / deletion) has led to a revolution in their use as molecular markers. EST derived molecular markers especially SNP and SSR are highly useful in developing linkage maps and markers assisted breeding programs. These markers are also transferable to related genera. Molecular marker techniques are advantageous as they directly reflect variations in the DNA sequences and therefore of independence of environment. Recently EST resources of oil palm are being developed at IRD, Montpellier France [2] and MPOB Kuala Lumpur Malaysia (http:// palmoils.mpob.gov.my/palmgenes.html).
SNPs are genetic markers, which are bi-allelic in nature, highly abundant and less prone to mutations than SSRs. [3] SNPs are increasingly becoming the marker of choice in genetic analysis and are used routinely as markers in agricultural breeding programs. Unlike random amplified polymorphic DNAs and RFLPs, SNPs are direct markers because sequence information provides the exact nature of the allelic variants. EST sequence data may provide the richest sources of biologically useful SNPs due to the relatively high redundancy of gene sequences, the diversity of genotypes represented within databases, and the fact that each SNP would be associated with an expressed gene. [4] Candidate SNPs can be grouped according to nucleotide substitution as either transition (C / T or G / A) or transversion (C / G, A / T, C / A or T / G). Indel sites can classified to four groups based on the nucleotide involved (A/T/C/G). Thus there are ten kinds of SNP/indel (two types of transition and four types of transversion and four groups of indels) are possible in the SNP/indel sites in EST libraries.
Oil palm representing important taxonomic group, Arecales of monocotylednous plants, hence these SNPs could be useful in other economic palms. Objective of the present study is to examine the EST libraries of oil palm and look for SNP/indel sites. The study also attempts to use the Shannon index to compare the frequencies in each of the ten possible types of SNP/indel sites.
Methodology
EST sequences were mined from dbEST(dbEST release 081007). Which contains 5452 sequences from seven tissues; mesocarp tissue, abnormal apex, normal apex, male inflorescence, female inflorescence, immature zygotic embryo and lambda zap II of oil palm The GenBank accession numbers of the sequences used in the study are BM402088, BM402089 (E. oleifera), CN599371 to CN601781, ES273633 - ES414798, EL563704 - EL930621 (Elaeis guineensis Jacq.) ESTs were separated tissue wise, most of the ESTs belonged to mesocarp tissue and lambda zap libraries contains very less ESTs compared to other. These EST sequences were used to make contigs to minimize the sequencing errors and avoid redundant sequences using cap3. [5] A perl script Auto_snip version 1.0 [6] was used to detect SNPs. Auto_snip also clusters and make contigs using the FASTA format sequences by acting as a wrapper for the clustering using cap3. Some authors use package d2cluster [7] for the purpose. ACE formatted output has given as input of Auto_snip program and an HTML format output file was generated to allow the user to browse through the SNP results. Output html files of auto_snip as a list of SNP sites and that of primer3 as list of primers are provided as zip files in supplementary information.
We have used Shannon index for working out the indexing the distribution of SNP/Indels into ten possible categories. Frequency of each of the ten types of SNP/indel sites was scored. From this value, proportion (Pi) of occurrence of each type (nature of transition / transversion / indel) to the total SNP/indels in each tissue library was worked out. Shannon index estimates (1949) have been worked out using the formula (1) under supplementary material. We divided the summation value by 0.5N*In0.5 to normalize the index for easy comparison among different contigs where N is the total number of EST sequences used in analysis.
Results and discussion
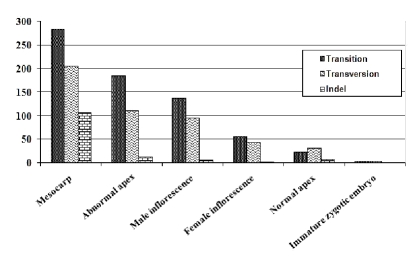
We found a total of 1180 SNP sites and 137 indel polymorphisms in 576 ESTs analyzed with frequency 1.36 SNPs / 100 bp. Results of the tissue wise SNP and indel discovery are listed in table 1 (supplementary material) and figure 1. Lambda Zap II tissue represents only two ESTs (BM402088, BM402089) and it contained no SNP/Indel hence we have eliminated Lambda Zap II library for further analysis.
Figure 1.
Frequency of SNPs and indel polymorphisms in EST libraries of different tissues of oil palm
In oil palm candidate SNPs could be detected at frequency of 1.36 SNPs/100bp. When comparing human DNA from two individuals, SNPs are found on average every 1 to 2 kb [8] and one SNP per 48 or 130 bp in 3' untranslated regions and coding regions respectively in maize. [9]
Among the six tissues from which the EST libraries had been generated, mesocarp had high frequency of 2.91 SNPs and indels per 100 bp (Table 1 under supplementary material) whereas the zygotic embryos had lowest frequency of 0.15 per 100 bp. Mesocarp tissue of oil palm had undergone selection pressure by human and nature. Billotte et al 2005 [10] have linked an AFLP marker to the shell thickness locus in the oil palm genome a single gene governs the shell thickness, in oil palm, which in dominant homozygous state offers thick-shelled fruits. It is botanically known as dura. Recessive form of the gene gives the fruits with thin of no shell also called as pisifera. Pisiferas are female sterile lines and the embryo gets aborted. Heterozygous forms (tenera) are intermediate in shell thickness and are commercially important with high oil yield.
There was a relative increase in the proportion of transition (690) over transversion (490) in oil palm ESTs except in normal apex libraries (Figure 1). C / T transition was found to be high in oil palm (Table 1 under supplementary material). High frequency of the C to T mutation is usually seen due to methylation. [11]
We also used the Shannon information index to analyze the proportion of ten possible types of SNP/indels. ESTs from tissues of normal apex showed highest values of indices (0.60) whereas abnormal apex had the least value (0.02). Our study on higher number and Shannon index of SNP/indel sites in apex tissue than other tissues also gives the additional information about in genomic variation in genes expressed specifically in apex tissue. Ratio of transition to transversion (Ts/Tv) was very useful to compare the genotypes of hepatitis virus C [12] and also differences among the mitochondrial genomes [13] of animals. Our study gives a method, which compares the ten possible types of SNP/ indels in a single index.
Conclusion
Potential SNP sites from the study could also prove useful to detect polymorphism in oil palm germplasm and also linkage mapping. The present data confirm that the frequency of SNP occurrence in oil palm is sufficient to make them appropriate markers for any kind of genetic studies. The study also highlights the use of Shannon index to analyze and compare the frequencies of SNP/indel sites in EST libraries.
Supplementary material
Acknowledgments
We are grateful to Department of Biotechnology Government of India for the financial support.
References
- 1.Boguski MS, et al. Nature Genetics. 1993;4:332. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 2.Jouannic S, et al. FEBS. 2005;579:2709. doi: 10.1016/j.febslet.2005.03.093. [DOI] [PubMed] [Google Scholar]
- 3.Giordano M, et al. Genomics. 1999;56:247. doi: 10.1006/geno.1998.5715. [DOI] [PubMed] [Google Scholar]
- 4.Picoult-Newberg L. Genome Res. 1999;9:167. [PMC free article] [PubMed] [Google Scholar]
- 5.Huang X, Madan A. Genome Research. 1999;9:868. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker G, et al. Bioinformatics. 2003;19:421. doi: 10.1093/bioinformatics/btf881. [DOI] [PubMed] [Google Scholar]
- 7.Burke J, et al. Genome Research. 1999;9:1135. doi: 10.1101/gr.9.11.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deutsch S, et al. Genome Research. 2001;11:300. doi: 10.1101/gr.164901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tenaillon M, et al. P Natl Acad Sci. 2001;98:9161. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Billotte N, et al. Theor Appl Genet. 2005;110:754. doi: 10.1007/s00122-004-1901-8. [DOI] [PubMed] [Google Scholar]
- 11.Coulondre C, et al. Nature. 1978;274:775. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka T, et al. FEBS Lett. 1993;315:201. doi: 10.1016/0014-5793(93)81163-t. [DOI] [PubMed] [Google Scholar]
- 13.Belle EM, et al. Gene. 2005;355:58. doi: 10.1016/j.gene.2005.05.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.