Abstract
Lack of crystal structure data of folate binding proteins has left so many questions unanswered (for example, important residues in active site, binding domain, important amino acid residues involved in interactions between ligand and receptor). With sequence alignment and PROSITE motif identification, we attempted to answer evolutionarily significant residues that are of functional importance for ligand binding and that form catalytic sites. We have analyzed 46 different FRs and FBP sequences of various organisms obtained from Genbank. Multiple sequence alignment identified 44 highly conserved identical amino acid residues with 10 cysteine residues and 12 motifs including ECSPNLGPW (which might help in the structural stability of FR).
Keywords: folate receptors (FR), folate binding proteins (FBP), multiple sequence alignment, consensus sequence, conserved motifs, evolutionary trace (ET)
Background
Folate is the major source of one carbon moiety during DNA biosynthesis in various organisms. Internalization of the folic acid into cell is mediated by folate receptors (FRs) or folate binding proteins (FBPs) or by reduced folate carriers (RFCs). [1,2] The FRs are clustered on the cell surface and associated with uncoated membrane invaginations known as caveolae. [3,4] Folate binds to the externally oriented receptor and is followed by internalization. After binding and internalization, at low pH folate receptor disassociates from folate and is transported back to the cell surface through potocytosis. [2,5,6] Based on the tissue expression and affinity of binding towards folic acid and its various analogues into three major types, FR1 (FR-α), FR2 (FR-β), FR3 (FR-γ). [7] FR1 (FR-α) and FR2 (FR-β) are membrane bound GPI anchored and expressed in adult epithelial cells and placental (fetal) tissues respectively [8,9,10], FR3 (FR-γ) is a secretory protein as it lacks signal for GPI attachment [11] and is developmentally highly regulated with a restricted spatial (tissue specific) or temporal (time specific) expression pattern. [12] The presence of fourth rare type is also reported as FR4 (FR-δ). [10] FR1 and FR2 differ not only on tissue expression but also in binding affinities and stereo specificities to various folate analogues. [10] It is been reported that, Folate receptors and Riboflavin binding proteins share more than ~30% sequence similarity. [13] In both cases, sequences exhibited 16 conserved cysteine residues that form eight bridges important for ligand binding. The N-terminal and centre portion of the sequences show very high similarity rather than C-terminal. [14] Here, we attempted to explore the important residues that may play an important role in the binding of folic acid and various analogues. This may help us to find mechanism of binding and interactions of the folate receptor subtypes with folic acid and its analogues under divergent physiologic condition. This study will provide information about the differential efficacy of the compounds such as, Methotrexate (MTX) in relation to developing new strategy in relation to cancer therapy.
Multiple sequence alignments are often used to find out conserved sequence regions in a group of sequences. Here, we have used ClustalW multiple sequence alignment tool which produces biologically meaningful alignments of divergent sequences. Through multiple sequence alignment we can also find out motifs which are short conserved sequence among set of sequences. The detection of conserved residues will be useful in identifying the functionally important residue even in the absence of structural information.
Methodology
Dataset
We retrieved 140 folate related sequences by keyword search at GenBank [15]. We manually curated the dataset to remove redundant sequences. Thus, a refined dataset of 46 non-redundant sequences of different folate receptors and folate binding proteins were created. To study evolutionarily trend among conserved functional residues in these sequences (about 250 residues in length), we grouped them based on sequence source (12 different organisms). (See: supplementary material).
Multiple sequence alignment
The 46 sequences are subjected to multiple sequence alignment as a first step to assess sequence conservation of single residue or motifs (residue stretch). Multiple sequence alignment was done using CLUSTAL-W. [16] Different parameters were tested and manual editing was performed wherever required to get significant alignment.
Motif Identification
The conserved motifs identified by multiple sequence alignment are submitted to PROSITE web server to scan against existing signatures and identify motifs unique to folate receptor [17].
Discussion
Conserved and variable amino acids
Multiple alignments of all FBP sequences are presented in Figure 2. A list of conserved amino acids is presented in figure 1 with the percentage of conservation and the major substitutions present at the position. Figure 1 shows that L217, C221, M222, H227, K228, P2449, E247, L250, C307, P309, W310, C315, C323, S328, F341, H345, C346, H370, F371, Q373, C376, E379, C380, S381, P382, N383, L384, G385, P386, W387, E409, R437, P442, L443, C444, E464, D465, C466, W470, C473, T478, C479, W486 and W490 residues are absolutely conserved in all the sequence of folate receptors. Although 44 residues showed 100% conservation, an additional 6 residues (K225, D248, T327, G347, I388, K480, and G489) showed more than 95% conservation (Figure 1).
Figure 2.
Shown in next four pages. Multiple sequence alignment of 46 members of FBPs. Conserved residues are highlighted in block (an arbitrary cut-off of 50% was used for conservation). Dashes represent gaps, introduced for the best alignment. A consensus sequence is shown at the bottom of each column, with conserved amino acid motifs underlined. For full protein names, see supplementary material
Figure 1.
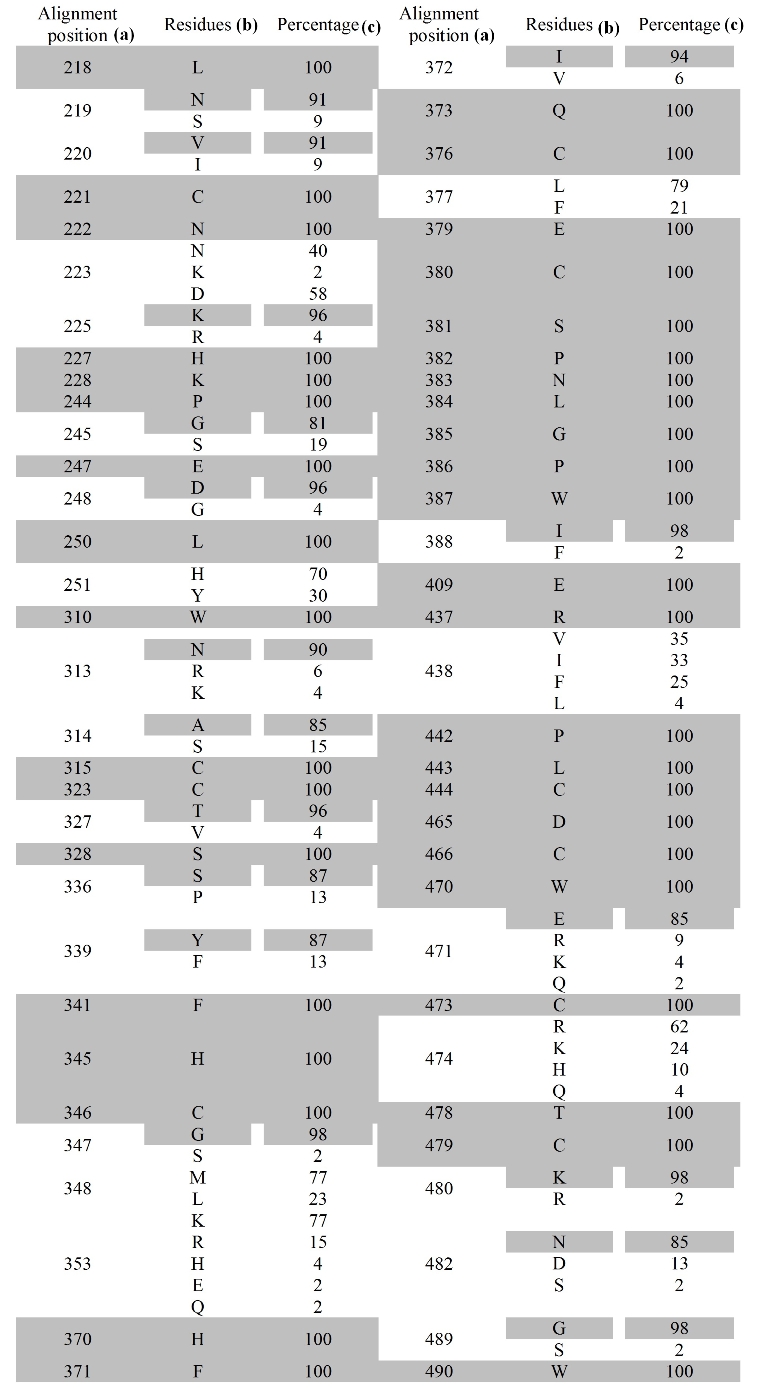
Conserved amino acids in members of FBP sequences (a) Amino acid position by alignment (b) Consensus residues given at positions are in bold. Residues over 85% are shaded (c) Percentage of 46 aligned sequences in which the listed residue appears. Percentages are rounded to the nearest number. Residues over 85% are shaded
Structural or functional domain elements are generally formed by grouping of conserved amino acids. The 44 amino acids are very well conserved, with few similar amino acid residue substitutions not altering the overall property. For example, in cases where the consensus residue is an aromatic amino acid (Trp, Tyr, Phe) an aromatic substitution occurs in 87% of cases and non aromatic amino acids occurs only 13% of cases. For example, at position 339, which has conserved Tyr, has 6 aromatic amino acid substitutions (6 Phe) which are shown in figure 1. In position 561, which has conserved Asp residue is substituted with similar amino acid Glu in 8 sequences. In some cases, aliphatic character was completely conserved, such as position 220 Val is substituted by Ile 3 times.
Earlier, Ratnam., et al, in 1999 reported a model structure for FR-α using the crystal structure of Chicken riboflavin binding protein and have mentioned the ligand binding sites Ala49, Val104, and Glu166 as important residues. [13] Position 49 specifically has small hydrophobic amino acids Ala/Leu. At position 49, Ala is highly conserved in all FR-α and FR-δ sequences, in case of FR-β it is substituted by similar residue Leu and in case of binding proteins and FR-γ it is Ala or Leu. Position 104 contains neutral, nonpolar amino acids with larger side chains- Phe, Val and Ile. Sequences of binding proteins, FR-α, FR-γ and FR-δ have either Val or Ile, where as in FR-β it is substituted by Phe. Position 166 contains neutral, nonpolar amino acid Gly or polar amino acids of various charges (positive or negative) aspartic acid, lysine, Gln, Glu and serine indicating less conservation in this position. Glu at 166 is conserved in 75% of FR-α sequence with remaining sequences having Gly or Lys as a substitution. FR-β has Gly highly conserved except for Equus caballus sequence having Glu. FR-δ has Lys conserved except for Canis familiaris sequence where it is substituted by Lys. No such conservation can be concluded in FBP and FR-γ. These substitutions are clearly shown in supplementary material. Present studies clearly indicate about the residues in binding pocket which lead to the specific affinity of subtypes of receptors towards various folate analogues.
We also compared the amino acid sequence of chicken RfBP with those of folate binding proteins and folate receptors. The Ligand binding site residue, Tyr-75 of RfBP is highly conserved in FBPs with few exceptions (His in four sequences and Cys in one sequence). Another ligand binding site, Trp-156 of RfBP is conserved in most of the FBP sequences except for one substitution Phe residue (Data not showed).
Conserved protein motifs
A number of conserved amino acid motifs are shown at the bottom of Figure 1. Using PROSITE database we found out that 4 motifs are unique to folate binding proteins and the rest are observed in other protein sequences as well. The motif [LM]-L-[NS]-[VI]-C-M-x(2)-[KR]-[HRY]-H-K (positions 217-228), C-x(3)-[TV]-S-x-[EAH]-[ALD]-[HT]-x-[DEA]-x-[SP]-x-[LS]-[YF]-x-F-[NST]-x(2)-H-C-[GS]-x-[ML]-x(3)-[CR] (positions 323-353), H-F-[IV]-Q-[DAN]-x-C-[LF]-[YHC]-E-C-S-P-N-L-G-P-W-[IF] (positions 370-388), E-D-C-x(2)-[WR]-W-x-[DA]-C-x(2)-[SY]-x-T-C-[KR]-x-[NDS] (positions 464-482) are uniquely present in folate receptors and folate binding proteins only. The other motifs such as, P-[GS]-[PQ]-E-[DG]-x-L-[HY] (positions 244-251), P-W-x(2)-[NRK]-[AS]-C (positions 309-315), [QRD]-x-[VAE]-x-[QSP]-x-[WGR]-x-E (positions 400-409), R-[FVI]-x(3)-P-L-C (positions 437-444), W-x(2)-[GS]-W-x-[WC] (positions 486-492), [YNHVF]-[FA]-P-[TGS]-[PGS]-[AKDTEV] (positions 523-528), [WC]-[STNDL]-[HNRVF]-[STD]-[YFN]-[KNE] positions 535-540, [YEA]-[SRGQ]-[RK]-[GNT]-[SQ]-G-[RQK]-[CLG]-[ILK]-[QD]--[MKH]-[WP]-F-[DE]-[PSAL]-[ATIVEF]-[QLEH]-[GSD]-N-P-N-[EV]-[EADV]-V-[AV]-[RKL]-[FLYH]-[YF]-[AL] (positions 548-576) (where x is any amino acid). This motif E-C-S-P-N-L-G-P-W (position 379-387) is 100% conserved in all the folate receptor sequences.
Conserved cysteines residues
Cysteine residues form disulfide bridges that help to keep the molecule intact and to maintain the confirmation of elements of the active site. Monaco, in 1997, comparing amino acid sequences of chicken RfBP, bovine milk folate-binding protein and human folate-binding protein, identified 16 conserved cysteine residues that formed intra molecular disulfide bonds. [14] All analyzed 46 sequences in these substitution 16 cysteine residues. Out of 16, 10 cysteines are 100% aligned in our alignment positions 221, 315, 323, 346, 376, 380, 444, 466, 473, and 479. Remaining 6 cysteine residues are not fully conserved in our alignment (positions 307, 353, 499, 517, 531, and 555).
Conclusion
In our study 46 different FBP sequences were subjected to multiple sequence alignment to identify sequence homology and evolutionarily conserved residues, which found to be functionally important. Multiple sequence alignment has indicated that all 46 sequences have 44 highly conserved amino acids including 10 cysteines and 12 sequence motifs. The motifs obtained from multiple sequence alignment were compared with PROSITE database and we identified 4 motifs unique to folate binding protein sequences which further strengthened our belief that they are functionally important residues and highly conserved during evolution.
Two unique conserved motifs [LM]-L- [NS]-[VI]-C-M-x(2)-[KR]-[HRY]-H-K and C-x(3)-[TV]-S-x-[EAH]-[ALD]-[HT]-x-[DEA]-x-[SP]-x-[LS]-[YF]-x-F-[NST]-x(2)-H-C-[GS]-x-[ML]-x(3)-[CR] are predominantly helices and coils in most of the sequences. The other motif E-D-C-x(2)-[WR]-W-x-[DA]-C-x(2)-[SY]-x-T-C-[KR]-x-[NDS] form coil-helix-coil in most cases (FR-α, γ, and δ) or only coil or helix-coil in case of beta. This conserved motif ECSPNLGPW is forming a coil between a strand and a helix. Most of these conserved motifs form coils are mostly not part of any secondary structures in all 46 sequences emphasizing its probable functional importance as loop regions frequently form binding sites, active sites. The predominance of helices along with coils may be due to its transmembrane location.
Ratnam et al in 1999 have reported functionally important residues in FR-α as Ala49, Val104, and Glu166 and in FR-β as Leu49, Phe104 and Gly166. [13] Our data has shown that functionally important residues in FR-α and FR-β are highly conserved in all organisms. In FR-α Ala in 49, Val/Ile in 104 similar amino acid substitution, Glu in 166, in case of FR-β Leu in 49, Phe in 104 and Gly in 166. Our analytical data also supported the findings of similar functionally important residues in δ as Ala49 and Val104. Similar amino acid substitution Leu/Ile was observed in three cases and Lys166 with one exception of Serine. This change of amino acid from Glu to Lys (166) in alpha and delta respectively may be the reason for their differences in function even though the other two residues Ala49 and Val104 are the same. This explains the probable difference in affinities of various receptors to folate and its analogues although it needs to be validated experimentally.
Supplementary material
References
- 1.Brzezinska A, et al. Acta Biochem Pol. 2000;47:735. [PubMed] [Google Scholar]
- 2.Andeson RG, et al. Science. 1992;255:410. [Google Scholar]
- 3.Verma RS, et al. J Biol Chem. 1992;67:4119. [PubMed] [Google Scholar]
- 4.Rothberg KG, et al. J Cell Biol. 1990;111:2931. doi: 10.1083/jcb.111.6.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen F, et al. Biochem. 1995;34:5660. doi: 10.1021/bi00016a042. [DOI] [PubMed] [Google Scholar]
- 6.Kamen A, et al. J Biol Chem. 1988;263:13602. [PubMed] [Google Scholar]
- 7.Elnakat H, Ratnam M. Adv Drug Deliv Rev. 2004;56:1064. doi: 10.1016/j.addr.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Ratnam M, et al. Biochem. 1989;28:8249. doi: 10.1021/bi00446a042. [DOI] [PubMed] [Google Scholar]
- 9.Elwood PC. J Biol Chem. 1989;264:14893. [PubMed] [Google Scholar]
- 10.Wang H. Nucleic Acids Res. 1998;26:2132. doi: 10.1093/nar/26.9.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan W, Ratnam M. Biochem. 1995;34:14594. doi: 10.1021/bi00044a039. [DOI] [PubMed] [Google Scholar]
- 12.Spiegelstein O, et al. An International Journal of Genes Genomes and Evol. 2000;258:117. [Google Scholar]
- 13.Maziarz KM, et al. J Biol Chem. 1999;274:11086. doi: 10.1074/jbc.274.16.11086. [DOI] [PubMed] [Google Scholar]
- 14.Monaco LH. EMBO Journal. 1997;16:1475. doi: 10.1093/emboj/16.7.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benson A, et al. Nucleic Acids Res. 2002;30:17. [Google Scholar]
- 16.Thompson JD, et al. Nucleic Acids Res. 1994;22:4673. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. http://www.expasy.ch/PROSITE/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



