Abstract
Expression of the feline immunodeficiency virus (FIV) accessory protein OrfA (or Orf2) is critical for efficient viral replication in lymphocytes, both in vitro and in vivo. OrfA has been reported to exhibit functions in common with the human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) accessory proteins Vpr and Tat, although the function of OrfA has not been fully explained. Here, we use microarray analysis to characterize how OrfA modulates the gene expression profile of T-lymphocytes. The primary IL-2-dependent T-cell line 104-C1 was transduced to express OrfA. Functional expression of OrfA was demonstrated by trans complementation of the OrfA-defective clone, FIV-34TF10. OrfA-expressing cells had a slightly reduced cell proliferation rate, but did not exhibit any significant alteration in cell cycle distribution. Reverse-transcribed RNA from cells expressing green fluorescent protein (GFP) or GFP + OrfA were hybridized to Affymetrix HU133 Plus 2.0 microarray chips representing more than 47,000 genome-wide transcripts. By using two statistical approaches, 461 (Rank Products) and 277 (ANOVA) genes were identified as modulated by OrfA expression. The functional relevance of the differentially expressed genes was explored by Ingenuity pathway analysis. The analyses revealed alterations in genes critical for RNA post-transcriptional modifications and protein ubiquitination as the two most significant functional outcomes of OrfA expression. In these two groups, several subunits of the spliceosome, cellular splicing factors and family members of the proteasome-ubiquitination system were identified. These findings provide novel information on the versatile function of OrfA during FIV infection and indicate a fine-tuning mechanism of the cellular environment by OrfA to facilitate efficient FIV replication.
Introduction
FIV is a lentivirus associated with an AIDS-like syndrome in the domestic cat (Pedersen, 1993). Like HIV, FIV can be transmitted via mucosal exposure, blood transfer, and vertically via prenatal and postnatal routes (O'Neil et al., 1995; O'Neil, e al., 1996; Obert and Hoover, 2000; Pedersen et al., 1987; Rogers and Hoover, 1998) and the primary target of infection is the CD4+ T cell. The overall genomic structure of FIV is markedly similar to HIV, although there are important distinctions (Olmsted et al., 1989; Phillips et al., 1990; Talbott et al., 1989). One such distinction is the lack of the transactivator gene, tat, and the presence of a short open reading frame termed OrfA. Translation of an approximate nine kDa protein encoded by this region occurs from a bicistronic mRNA that also encodes downstream Rev (de Parseval and Elder, 1999). The genomic location, size, and structural features of OrfA have many similarities to HIV Tat as well as to the L domain of visna virus, both of which demonstrate transactivating functions. In fact, OrfA has been shown to facilitate a net increase in translation of proteins whose expression is driven from the FIV long terminal repeats (LTRs) (de Parseval and Elder, 1999; Sparger et al., 1992; Waters et al., 1996). However, OrfA does not act via a TAR element, as is the case with HIV-1 Tat, and promotes a net increase in transcription / translation via mechanisms distinct from that of other lentiviruses (Chatterji et al., 2002; Gemeniano et al., 2003). Attempts to show direct interaction of OrfA with the FIV LTR proved negative (Chatterji et al., 2002) and the gene is dispensable for viruses adapted for propagation in adherent cell lines such as Crandell feline kidney cells (CrFK) and G355−5 cells (Phillips et al., 1990). However, OrfA is required for productive infection of the primary in vivo target cell, CD4+ T cells (Waters et al., 1996). Stable feline T-cell lines expressing OrfA can function in trans to complement an OrfA-defective FIV (this study; (Gemeniano et al., 2003)). Furthermore, cats inoculated with OrfA-mutated FIV clones had a greatly reduced plasma viremia (Pistello et al., 2002). Evidence has been presented that a 39bp deletion in OrfA gives a four-fold decrease in viral mRNA expression and a moderate decrease in Gag protein expression (Gemeniano et al., 2003). It has also been reported that OrfA may have relatedness to HIV-1 Vpr and is implicated in facilitating cell cycle arrest and virus release from the cell (Gemeniano et al., 2003; Gemeniano, et al., 2004). Overall, these findings suggest that OrfA may be a multi-functional protein, which would certainly be in keeping with the need for versatility, given the relatively small viral genome. In the present report, we used genome array analysis to study the consequence of OrfA on pleiotropic cellular gene expression in T cells. RNA was prepared from cells transduced with Mig-R1 vector expressing either green fluorescent protein (GFP) alone or both GFP and OrfA and analyzed by microarray analysis, using Affymetrix HU 133 Plus 2.0 chips. The results show many parallels with gene expression observed in HIV-infected cells (van 't Wout et al., 2003), with a down-regulated expression of factors reported to influence HIV-1 mRNA splicing. Furthermore, expression of genes encoding ubiquitin-conjugating enzymes and proteasome subunits were identified as down-regulated in the OrfA-expressing T cells.
Results
Generation and characterization of OrfA-expressing T-cells
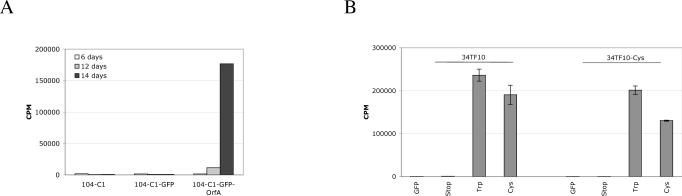
In order to better understand the function of OrfA during FIV infection, we generated stable cell lines expressing GFP +/− FIV-PPR OrfA (Chatterji et al., 2002). These clones (termed 104-C1-GFP and 104-C1-GFP-OrfA) were established in the primary IL-2-dependent T-cell line, 104-C1. Flow cytometry analysis confirmed the GFP expression in 104-C1-GFP and 104-C1-GFP-OrfA to be >95% (data not shown). To assay for expression of functional OrfA, the cells were infected with FIV-34TF10, a strain deficient in functional OrfA protein (Phillips et al., 1990; Talbott et al., 1989). The presence of OrfA complemented FIV-34TF10 in trans and rescued viral replication in 104-C1-GFP-OrfA cells (Fig. 1A). In cells lacking OrfA (104-C1 and 104-C1-GFP), only background levels of reverse transcriptase activity was detected in the culture supernatant. In addition, gene expression of OrfA in 104-C1-GFP-OrfA cells, but not in 104-C1-GFP cells, was confirmed by Q-PCR (data not shown). The OrfA mutation at residue 44 in FIV-34TF10 changes the native Tryptophan (Trp) residue to a stop codon. Repair of this mutation restores wild-type infectivity on PBMCs and T cell lines (Waters et al., 1996). Consistent with this finding, transduced 104-C1 cells expressing truncated OrfA-Stop failed to trans-complement FIV-34TF10 (Fig. 1B). An additional third base mutation was generated by alteration of the codon at residue 44 from Trp to Cys and the mutant OrfA-Cys was engineered into 104-C1 cells and FIV-34TF10. Interestingly, 104-C1-GFP-OrfA Cys was able to trans-complement the OrfA defective FIV (Fig. 1B, left). However, placing this mutation in the context of FIV-34TF10 (termed 34TF10-Cys) did not generate wild type levels of viral replication (Fig. 1B, right) until a back-mutation to Trp appeared at approximately 16 days of culture (data not shown). These findings indicate that Cys at residue 44 is a less stable residue than Trp but that the latter residue is not mandatory for OrfA function. Low-level viral replication results in rapid reversion to Trp in the context of the virus as opposed to the genetically static state of the OrfA plasmid.
Figure 1.
Confirmation of functional OrfA expression by FIV-34TF10 infection of transfected 104-C1 cells and reverse transcriptase activity assay. (A) Expression of OrfA in 104-C1 cells (104-C1-GFP-OrfA) trans complements the OrfA-defective clone FIV-34TF10 and rescues viral replication. (B) 104-C1 cells were transfected to express GFP alone or GFP together with the following OrfA variants, stop: truncated OrfA due to a stop codon at position 44; Trp: OrfA with Trp at position 44; Cys: OrfA with a Cys at position 44. The transfected cells were infected with 34TF10 or 34TF10 with Cys at position 44 (34TF10-Cys) and analyzed for viral replication by RT activity assay at 14 days post-infection. Cells expressing OrfA with a Trp or Cys at residue 44 can rescue proliferation of 34TF10. However, 34TF10-Cys has a very low proliferation rate in 104-C1 cells unless OrfA-Trp or OrfA-Cys is expressed by the cell.
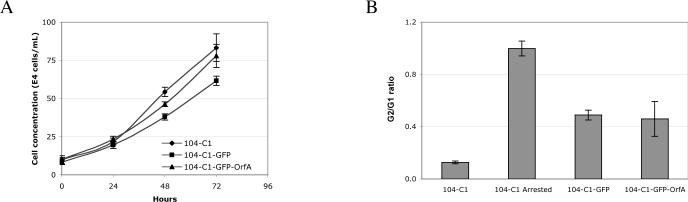
Further analyses of the stable 104-C1 clones revealed a small reduction in proliferation rate of the OrfA-expressing cells and in cells expressing only GFP (Fig. 2A). FIV-PPR OrfA has been reported to induce cell cycle arrest in the G2 phase in transiently transfected primate COS-7 cells (Gemeniano et al., 2004). Therefore, cell cycle analysis of the 104-C1 clones was performed by staining ethanol-fixed cells in a propidium-RNase solution and analyzed for cell fluorescence by flow cytometry. No change in cell cycle distribution was detected due to PPR OrfA expression in the feline T cell line 104-C1 relative to 104-C1-GFP cells (Fig. 2B) or in feline CrFK cells (data not shown).
Figure 2.
Proliferation rate and cell cycle distribution. (A) OrfA expression does not significantly change cell proliferation rate. Cells were seeded at 105/ml and counted after 24, 48 and 72 hours using a hemocytometer. (B) 104-C1-GFP and 104-C1-GFP-OrfA cells exhibit similar cell cycle distribution. Cellular DNA was stained with propidium iodide, cell fluorescence was measured by flow cytometry and discrimination of cells in G0/G1, S and G2/M phases was performed using FlowJo software. 104-C1 arrested cells were exposed to aphidicolin and Hoechst 33342 in two subsequent overnight incubations.
Microarray analysis of OrfA expressing cells
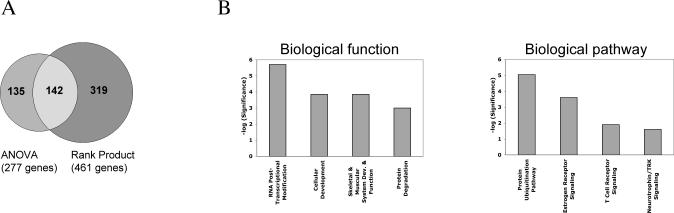
Total RNA from 104-C1-GFP and 104-C1-GFP-OrfA cells was isolated and subjected to reverse transcription followed by in vitro transcription to generate biotinylated anti-sense RNA probes. The labeled cRNA probes were hybridized to Affymetrix HU133 Plus 2.0 microarray chips representing more than 47,000 genome-wide transcripts. Differentially regulated genes were identified by ANOVA analysis and Rank Products method. The list of regulated genes was annotated using Affymetrix NetAffx software. Changes in gene expression levels ranged from 0.2- to 5.6-fold with a general trend toward down-regulated expression levels. Among the genes regulated, the percentage of genes down-regulated in OrfA+ cells were 74% and 57% for genes identified by ANOVA statistical analysis and Rank Products method, respectively. By using these two statistical analyses a total of 596 genes were identified of which 24% were present in both assays i.e. the overlapping data set (Fig. 3A). The functional relevance of the differentially expressed genes was explored by Ingenuity Pathway Analysis and Ingenuity Canonical Pathway Analysis software. Analyses of all data sets i.e. ANOVA, Rank Products or genes overlapping both statistical analyses, revealed expression changes in genes critical for “RNA post-transcriptional modification” and “protein ubiquitination” as the two most statistically significant functional outcomes of OrfA expression. The data presented in Fig. 3B is a classification of the genes in the overlapping data set based on their involvement in different biological processes. Forty percent of the genes in these two groups were found present in both statistical analyses, see Table 1-3.
Figure 3.
Analysis of microarray data. (A) Rank Products method and ANOVA analysis were both performed on the microarray data set. Indicated is the number of genes identified by each method and in the overlapping data set. (B) Exploration of the functional relevance of the genes in the overlapping array data set was done with Ingenuity pathway analysis (left) and Ingenuity canonical pathway analysis (right). Data are presented as the probability that each indicated biological function / pathway assigned to the data set is due to chance alone.
Table 1.
Post transcriptional modification genes identified by Rank Product (RP) or ANOVA statistical analysis - RNA splicing genes
| Gene | RP | Anova | Fold change | Protein name |
|---|---|---|---|---|
| CSTF3 | x | x | 0.46 | Cleavage stimulation factor, 3′ pre-RNA, subunit 3 |
| CUGBP1 | x | x | 0.55 | CUG triplet repeat, RNA binding protein 1 |
| FUSIP1 | x | 0.62 | FUS interacting protein (serine/arginine-rich) 1 | |
| hnRNP A | x | 0.86 | Heterogeneous nuclear ribonucleoprotein A1 (A) | |
| hnRNP A2B1 | x | x | 0.64 | Heterogeneous nuclear ribonucleoprotein A2/B1 |
| hnRNP H | x | 0.71 | Heterogeneous nuclear ribonucleoprotein H1 (H) | |
| hnRNP H3 | x | x | 0.72 | Heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| hnRNP Q | x | x | 0.63 | Heterogeneous nuclear ribonucleoprotein Q, SYNCRIP |
| MBNL1 | x | 0.54 | Muscleblind-like (Drosophila) | |
| NCBP1 | x | x | 0.67 | Nuclear cap binding protein subunit 1 |
| PLRG1 | x | 0.82 | Pleiotropic regulator 1 (PRL1 homolog, Arabidopsis) | |
| PPIG | x | 0.64 | Peptidylprolyl isomerase G (cyclophilin G) | |
| PTBP2 | x | 0.85 | Polypyrimidine tract binding protein 2 | |
| RP9 | x | 0.64 | Retinitis pigmentosa 9 (autosomal dominant) | |
| SFRS1 | x | 0.66 | Splicing factor, arginine/serine-rich 1 (splicing factor 2), ASF/SF2 | |
| SFRS3 | x | x | 0.67 | Splicing factor, arginine/serine-rich 3, SRP20 |
| SFRS7 | x | x | 0.56 | Splicing factor, arginine/serine-rich 7, 9G8 |
| SFRS15 | x | 0.66 | Splicing factor, arginine/serine-rich 15 | |
| SNRPA | x | x | 0.62 | Small nuclear ribonucleoprotein polypeptide A |
| SNRPA1 | x | x | 0.52 | Small nuclear ribonucleoprotein polypeptide A′ |
| SNRPB | x | x | 0.7 | Small nuclear ribonucleoprotein polypeptides B and B1 |
| TRA2A | x | 1.24 | Transformer-2 alpha | |
| U2AF35 | x | 0.67 | U2 small nuclear RNA auxiliary factor 1, U2AF1 | |
| UAP56 | x | x | 0.69 | U2AF65-associated protein 1, 56-kDa, BAT1 |
| WBP11 | x | 0.55 | WW domain binding protein 11 |
Table 3.
Protein ubiquitination pathway genes identified by Rank Product (RP) or ANOVA statistical analysis
| Gene | RP | Anova | Fold change | Protein name |
|---|---|---|---|---|
| ANAPC5 | x | 0.6 | Anaphase promoting complex subunit 5 | |
| CDC20 | x | 0.52 | CDC20 cell division cycle 20 homolog (S. cerevisiae) | |
| HSPA8 | x | 0.55 | Heat shock 70kDa protein 8 | |
| PSMA1 | x | x | 0.66 | Proteasome subunit, alpha type, 1 |
| PSMA2 | x | 0.63 | Proteasome subunit, alpha type, 2 | |
| PSMA5 | x | x | 0.68 | Proteasome subunit, alpha type, 5 |
| PSMA7 | x | x | 0.63 | Proteasome subunit, alpha type, 7 |
| PSMB7 | x | 0.81 | Proteasome subunit, beta type, 7 | |
| PSMC6 | x | x | 0.62 | Proteasome 26S subunit, ATPase, 6 |
| PSMD1 | x | 0.56 | Proteasome 26S subunit, non-ATPase, 1 | |
| PSMD8 | x | 0.75 | Proteasome 26S subunit, non-ATPase, 8 | |
| PSME2 | x | x | 0.63 | Proteasome activator subunit 2 (PA28 beta) |
| PSME3 | x | 0.69 | Proteasome activator subunit 3 (PA28 gamma) | |
| SKP1A | x | 0.67 | S-phase kinase-associated protein 1A (p19A) | |
| TAP2 | x | x | 1.23 | Transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) |
| TCEB1 | x | x | 0.61 | Transcription elongation factor B (SIII) |
| UBE2C | x | x | 0.43 | Ubiquitin-conjugating enzyme E2C |
| UBE2D2 | x | 0.72 | Ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | |
| UBE2L3 | x | 0.67 | Ubiquitin-conjugating enzyme E2L 3 | |
| UBE2N | x | x | 0.6 | Ubiquitin-conjugating enzyme E2N (UBC13 homolog, yeast) |
| UBE2V1 | x | 0.6 | Ubiquitin-conjugating enzyme E2 variant 1 | |
| UBE3A | x | 0.78 | Ubiquitin protein ligase E3A (HPV E6-associated protein) | |
| USP1 | x | x | 0.61 | Ubiquitin specific peptidase 1 |
| USP6 | x | 1.35 | Ubiquitin specific peptidase 6 (Tre-2 oncogene) | |
| USP24 | x | 1.12 | Ubiquitin specific peptidase 24 |
Gene expression of spliceosome subunits and cellular splicing factors are modulated by OrfA
In the RNA post-transcriptional modification group, the predominant portion (64%) of the identified genes are involved in the RNA splicing process (Table 1). Genes for RNA splicing proteins that were differentially regulated in OrfA-expressing cells include heterogeneous nuclear ribonucleoprotein (hnRNP) family members, serine/arginine-rich (SR) splicing factors and subunits of the spliceosome machinery including small nuclear ribonucleoprotein polypeptides (SNRPs; see Table 1). Retroviruses utilize alternative splicing to generate expression of several gene products from one primary polycistronic transcript (Purcell and Martin, 1993). This splicing process makes use of suboptimal splice sites and cis-acting elements in the viral genome. The cis-acting elements act mainly by binding hnRNP family members and SR splicing factors. In OrfA-expressing cells, the following hnRNPs exhibited down-regulated expression; hnRNP A, hnRNP A2B1, hnRNP H, hnRNP H3, and hnRNP Q (Table 1). Furthermore, the array data analysis revealed an OrfA-initiated down-regulation of four SR splicing factors, namely, SFRS1 (ASF/SF2), SFRS3 (SRp20), SFRS7 (9G8) and SFRS15 (Table 1). Most interestingly, the majority of the hnRNP and SR family members identified here, have also been reported to influence HIV-1 mRNA splicing (Caputi et al., 1999; Domsic et al., 2003; Jacquenet et al., 2001; Ropers et al., 2004; Stoltzfus and Madsen, 2006; Tange et al., 2001; Zahler et al., 2004).
The actual splicing reaction is performed by the spliceosome, which consists of the U1, U2, U4, U5 and U6 small nuclear ribonucleoproteins (snRNPs) (Graveley, 2000). The expression of the following SNRP genes were found modulated by OrfA (Table 1); SNRPA – a subunit specific of U1 snRNP (Sillekens et al., 1987); SNRPA1 – a component specific of U2 snRNP (Sillekens et al., 1989); and SNRPB – a subunit shared by all snRNPs (Luhrmann et al., 1990).
U2 auxiliary factor (U2AF) is a heterodimer of U2AF35 and U2AF65 proteins and has a role in the first step of spliceosome assembly by binding the 3' splice site (Graveley, 2000). Our array data display a down-regulation of U2AF35 in OrfA-expressing cells. UAP56 (U2AF65-associated protein), an essential splicing factor required for U2 snRNP binding to pre-mRNA branch site (Fleckner et al., 1997), is also down-regulated by OrfA (Table 1).
Among the other genes identified as coding for RNA post-transcriptional modification proteins (Table 2) were some of specific relevance for retroviral replication. The mammalian mRNA capping enzyme RNA guanylyltransferase and 5'-phosphatase (RNGTT or Mce1) has been reported to both interact with and to be regulated by HIV-1 Tat (Chiu et al., 2001). CDK7 (Cyclin-dependent kinase 7) has been implicated in general HIV-1 transcription (Kim et al., 2006) as well as Tat-mediated transcription (Nekhai et al., 2002). KHDRBS1 (KH domain containing, RNA binding, signal transduction associated 1 or SAM68) plays a role in Rev-mediated RNA export (Modem et al., 2005) and HIV-1 RNA polyadenylation (McLaren et al., 2004). According to the Ingenuity data base, the following genes identified as regulated in the array can be classified as being involved in the RNA polyadenylation process; CPSF6, GRSF1, NUDT21, PABPN1, PAPOLG, and SNRPA (Table 1 and 2).
Table 2.
Post transcriptional modification genes identified by Rank Product (RP) or ANOVA statistical analysis - without genes involved in RNA splicing
| Gene | RP | Anova | Fold change | Protein name |
|---|---|---|---|---|
| CDC2 | x | x | 0.57 | Cell division cycle 2 |
| CDK7 | x | 0.55 | Cyclin-dependent kinase 7 | |
| CPSF6 | x | 0.68 | Cleavage and polyadenylation specific factor 6 | |
| DDX17 | x | 1.24 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 | |
| GRSF1 | x | 0.65 | G-rich RNA sequence binding factor 1 | |
| hnRNP D | x | x | 0.56 | Heterogeneous nuclear ribonucleoprotein D |
| hnRNP DL | x | x | 0.47 | Heterogeneous nuclear ribonucleoprotein D-like |
| KHDRBS1 | x | 0.8 | KH domain containing, RNA binding, signal transduction associated 1 | |
| NUDT21 | x | 0.65 | Nudix (nucleoside diphosphate linked moiety X)-type motif 21 | |
| PABPN1 | x | x | 0.65 | Poly(A) binding protein, nuclear 1 |
| PAPOLG | x | 0.83 | Poly(A) polymerase gamma | |
| PCBP1 | x | 0.71 | Poly(rC) binding protein 1 | |
| RBPMS | x | 1.47 | RNA binding protein with multiple splicing | |
| RNGTT | x | x | 0.6 | RNA guanylyltransferase and 5′-phosphatase |
Modulation of RNA expression for genes in the ubiquitin-proteasome pathway
In addition to genes in the RNA post-transcriptional modification group, several genes involved in the process of protein ubiquitination and degradation were differentially regulated in the OrfA expressing T-cells. Protein degradation through the ubiquitin-proteasome pathway plays an important role in immune and inflammatory responses. There are two distinct steps in this pathway; conjugation of ubiquitin molecules to the target protein and degradation of the ubiquitinated protein by the 26S proteasome (Glickman and Ciechanover, 2002). Expression of five different E2 ubiquitin-conjugating enzymes and one E3 ubiquitin-protein ligase were down-regulated by OrfA expression (Table 3). Furthermore, three different ubiquitin-specific peptidases (USPs) were also regulated in OrfA-expressing cells (Table 3). USPs belongs to the family of de-ubiquitinating enzymes that can release ubiquitin from degraded proteins or de-ubiquitinylate proteins prior to their association with the proteasome (Glickman and Ciechanover, 2002).
The peptide cleavage of ubiquitinated proteins takes place in the 26S proteasome, which is a multicatalytic proteinase complex consisting of a 20S catalytic core and two 19S regulatory complexes responsible for recognition, binding and unfolding of ubiquitinated proteins (Kloetzel and Ossendorp, 2004). Eight different subunits of the proteasome were down-regulated in OrfA-expressing cells (Table 3). These are four 20S core alpha subunits (PSMA1, 2, 5 and 7); one 20S core beta subunit (PSMB7); and three 19S subunits (PSMC6, PSMD1 and PSMD8). Proteasome inhibitors have been reported to enhance HIV-1 infectivity 4−26 fold (Wei et al., 2005), thus suggesting a cellular defense system with proteasome-mediated degradation of viral particles or proteins. Furthermore, HIV-1 Tat and Nef can impair proteasome activity (Apcher et al., 2003; Qi and Aiken, 2007), as a mechanism to protect viral particles from degradation (Qi and Aiken, 2007).
Interferon-gamma (IFN-γ) can change the 26S proteasome into an immunoproteasome responsible for generation of peptides for MHC class-I presentation. This is achieved by an IFN-γ induced up-regulation of both specific proteasome subunits and the immunoproteasome regulator, PA28/11S. Interestingly, in our array analysis two PA28 subunits i.e. PSME2 and PSME3 are down-regulated in OrfA expressing cells (Table 3). Similarly, HIV-1 Tat can modulate the expression of subunits specific for the immunoproteasome (Gavioli et al., 2004; Remoli et al., 2006) and interferes with the formation of the immunoproteasome, which leads to reduced antigen presentation (Huang et al., 2002; Seeger et al., 1997).
Another interesting gene identified as being regulated in OrfA expressing cells is HMGB1 (High mobility group B1) that belongs to the HMG protein family of abundant non-histone components of chromatin. HMGB1, which was down-regulated by OrfA (data not shown), is released by activated macrophages/monocytes and can stimulate production of chemokines and induce a down-regulation of viral replication in primary HIV-1 infected monocyte-derived macrophages (Nowak et al., 2006; Wang et al., 2006). HMGB1 has also been reported to repress HIV-1 LTR-mediated transcription in monocytes and epithelial cells (Naghavi et al., 2003).
Validation of microarray data by Quantitative Real-Time PCR (Q-PCR)
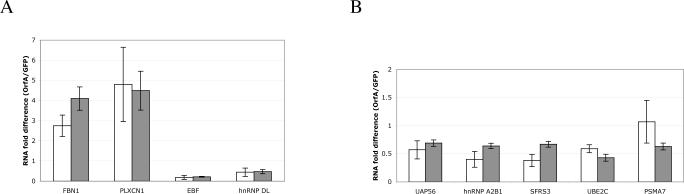
Q-PCR is the most appropriate tool for confirmation of gene expression results obtained from microarray analysis. As an independent measure of the influence of OrfA on gene expression in 104-C1 T-cells, we performed Q-PCR using primer sets corresponding to a sampling of genes identified as OrfA-responsive in the microarray analysis (Fig. 4). Primers were designed for four highly OrfA-responsive genes and the mRNA levels were analyzed in 104-C1-GFP and 104-C1-GFP-OrfA cells (Fig. 4A). In addition, mRNA levels of five regulated genes of the splice factor and proteasome families shown in Tables 1 and 3 were also analyzed (Fig. 4B). Of the nine genes analyzed by Q-PCR, eight yielded trends virtually identical to results noted in the microarray analysis (Fig. 4). One gene, PSMA7, which showed a slight down-turn in expression by microarray, was essentially unchanged in the Q-PCR analysis. Thus, the two types of gene expression analysis showed high agreement, lending support to the relevance of the array data.
Figure 4.
Q-PCR validation of microarray data. Fold difference of RNA levels of genes identified in the microarray analysis (grey bars) were analyzed in 104-C1-GFP and 104-C1-GFPOrfA cells by Q-PCR (white bars). (A) RNA fold differences for four genes identified as highly regulated by OrfA. (B) RNA fold differences for five regulated genes of the splice factor and proteasome families.
Discussion
In order to better understand the function of OrfA during FIV infection and replication, we generated stable cell lines expressing either GFP alone or both GFP and OrfA. As reported previously (Chatterji et al., 2002), expression levels for OrfA in FIV-infected cells is extremely low and is essentially undetectable by either radioimmune precipitation or Western blotting techniques. Likewise, net expression of OrfA in the stable cell line used in the present study is low, although in vitro transcription/translation readily demonstrated OrfA expression from the vector construct used to create the cell line (Chatterji et al., 2002), data not shown. The implication from these findings is that the protein has a limited half-life in both FIV-infected cells and in the stable 104-C1-GFP-OrfA cell line. Important to the virological relevance of the present study, OrfA in the context of the stable cell line was able to rescue OrfA-defective FIV-34TF10 in trans (Fig. 1A) as well as enhance FIV LTR-driven transcription/translation (data not shown). We previously noted cell toxicity with other OrfA-expressing cell lines and surmise that higher levels of expression result in cell toxicity. Thus, we feel that the cell line used here is the most appropriate for reaching conclusions regarding the mandatory role of OrfA in FIV replication in T cells. Further analyses of the stable cell lines revealed a minor down-regulation in the proliferation rate of the OrfA expressing cells (Fig. 2A) but there were no changes in cell cycle distribution due to OrfA expression in the primary T cell line 104-C1, compared to 104-C1-GFP (Fig. 2B), or in CrFK cells (data not shown). These cell cycle observations in FIV relevant feline cells diverge from what previously has been reported when expressing OrfA in primate COS-7 cells and, thus, may be connected to interspecies differences or due to relative expression levels of OrfA in the two studies.
The impact of OrfA on the pleiotropic gene expression pattern in feline T-cells was characterized by microarray analysis using RNA from the 104-C1-GFP and 104-C1-GFP-OrfA cells. Since no feline chips are available, we compare the relative reactivity of feline RNA to both murine and human chips and settled on use of the human HU133Plus 2.0 chip for these studies. Although it is unlikely that we could expect complete cross-species hybridization with this chip, we felt it appropriate to use the available technology with the knowledge that positive findings could be independently verified and pursued.
By using ANOVA analysis and Rank Products method, approximately 600 genes were identified as differentially expressed in the stable clones. The changes in expression levels are subtle for the predominant portion of the genes. This is consistent with the idea of a virus-induced modification of the cellular environment, without fully overtaking or killing the host cell, at least in the short term. Although many changes in expression level were small, families of genes were regulated in the same direction and these changes may pave the way for a downstream functional outcome relevant to virus replication. The functional relevance of the gene expression changes was explored by Ingenuity Pathway Analysis. For all data sets (ANOVA, Rank Products or genes present in both assays), “RNA post-transcriptional modification” and “protein ubiquitination” were revealed as the two most important functional outcomes of OrfA expression (Fig. 3B).
HIV-1, FIV and other retroviruses utilize alternative splicing to generate expression of several gene products from one primary polycistronic transcript while at the same time controlling the level of splicing to allow generation of full-length genomic RNA for packaging in new viral particles. The alternative splicing process is dependent on suboptimal splice sites and cis-acting elements in the viral genome. The cis-acting elements include exonic splicing silencers (ESS) and intron splicing silencers (ISS) that are regulated by hnRNP A/B family members or by hnRNP H. Another group of cis-acting elements are exonic splicing enhancers (ESE) that function by binding members of the SR protein family (Graveley, 2000). For a review of ESS, ISS and ESE function in HIV-1 RNA splicing see Kammler et al. (Kammler et al., 2006). Among the splicing genes found down-regulated by OrfA are hnRNP A, hnRNP A2B1 and hnRNP H (Table 1). Expression of these three genes have previously been described down-regulated, at a similar level, after HIV-1 infection of a human CD4+ T cell line (van 't Wout et al., 2003) and hnRNP A is also down-regulated in astrocytes expressing HIV-1 Tat (Pocernich et al., 2005). Furthermore, hnRNP A/B proteins and hnRNP H have been described as splicing inhibitors of HIV-1 RNA (Caputi et al., 1999; Jacquenet et al., 2001; Stoltzfus and Madsen, 2006). hnRNP H3 and hnRNP Q (SYNCRIP) are two other hnRNP splicing proteins identified as down-regulated in OrfA expressing cells (Table 1). Of these proteins, hnRNP H3 is known to bind HIV Env RNA (Caputi et al., 1999).
SR splicing factors have several functions in splicing of precursor mRNA and are required both for selection of alternative splice sites and for removal of constitutively spliced introns. In the array data set, the OrfA expressing T-cells displayed a down-regulation of four SR splicing factors, namely, SFRS1 (ASF/SF2), SFRS3 (SRp20), SFRS7 (9G8) and SFRS15. Analogous to our study, three of these SR splicing factors (ASF/SF2, SFRS3, and 9G8) have been reported down-regulated to the same extent in earlier studies using HIV-1 infected T-cells (Ryo et al., 2000; van 't Wout et al., 2003). ASF/SF2 and 9G8 have been identified as factors that influence the splicing pattern of HIV-1 pre-mRNA, (Caputi et al., 1999; Domsic et al., 2003; Ropers et al., 2004; Tange et al., 2001; Zahler et al., 2004) and an altered expression of these genes/proteins have a prominent effect on HIV-1 protein expression and virion production (Jacquenet et al., 2001; Ropers et al., 2004; Wang et al., 1998). In our array data set, three SNRP subunits of the spliceosome were also identified as modulated by the presence of OrfA (Table 1).
The HIV accessory gene product Vpr has been reported to influence splicing of viral RNA and has a differential effect on the level of spliced viral RNA in integrase-defective HIV (Poon et al., 2007). Vpr has also been shown to interact with the spliceosomal protein SAP145 and thereby mediate inhibition of cellular precursor mRNA splicing and induction of G2 arrest (Hashizume et al., 2007; Terada and Yasuda, 2006). These findings are consistent with parallels drawn by Gemeniano et al. between HIV-1 Vpr and FIV OrfA (Gemeniano et al., 2004).
Ubiquitin-proteasome degradation of proteins plays an important role in a variety of basic cellular processes as well as in immune and inflammatory responses. Proteins are directed to degradation by conjugation of ubiquitin molecules. The degradation of the ubiquitinated protein is accomplished by the 26S proteasome (Glickman and Ciechanover, 2002). Three types of enzymes are needed for the ubiquitination process i.e. E1 ubiquitin-activating enzyme; E2 ubiquitin-conjugating enzyme; and E3 ubiquitin-protein ligase. The role of E1 is to activate ubiquitin molecules, which are then transferred to the protein substrate by E2. Finally, conjugation of ubiquitin to the protein is performed by E3 by binding of both E2 and the protein substrate (Glickman and Ciechanover, 2002). The ubiquitin system has a hierarchical structure where E1 interacts with all E2s, each type of E2 can interact with many different E3s and each E3 targets several substrates. In our array data set, several E2 ubiquitin-conjugating enzymes and one ubiquitin-protein ligase exhibited a down-regulated gene expression induced by OrfA (Table 3). Thus, one way OrfA may facilitate viral replication is by limiting ubiquitination/degradation of viral proteins in the host cell.
The immunosuppressive activity of HIV-1 Tat (Viscidi et al., 1989) can partly be explained by alteration of immunoproteasome activity. Tat inhibits the activity of 20S and competes with PA28/11S for binding to 20S. Therefore, formation of the immunoproteasome is blocked and presentation of antigenic peptides is reduced (Huang et al., 2002; Seeger et al., 1997).
Additionally, Tat can modulate the expression of specific subunits of the immunoproteasome and thereby change the peptide antigen profile of cells expressing or being exposed to Tat (Gavioli et al., 2004; Remoli et al., 2006). In this study, we identified a down-regulation by OrfA of two PA28 subunits (PSME2 and PSME3). This finding is consistent with an additional role of OrfA in deregulation of antigen presentation during FIV infection.
Furthermore, eight genes encoding general proteasome subunits are down-regulated in OrfA expressing cells (Table 3). Similarly, HIV-1 Tat has also been shown to alter the activity of the proteasome. Among the proteasome subunits that interact with Tat is PSMA7 (Apcher et al., 2003), a protein also identified in our microarray analysis as down-regulated by OrfA. Another HIV-1 accessory factor, Nef, has recently been implicated in protection of HIV-1 particles from proteasomal degradation (Qi and Aiken, 2007), and HIV Vpr-mediated G2 arrest has been reported to be dependent on interaction of Vpr with ubiquitin-ligase components (Wen et al., 2007). However, the action of Vpr here most likely promotes proteasomal degradation of a cellular factor (Wen et al., 2007). Additionally, recent studies suggest that one phase of rhesus TRIM5α restriction of HIV-1 infection is accomplished by blockage of late reverse transcriptase product formation via proteasome activity (Wu et al., 2006).
The down-regulation of ubiquitin conjugating enzymes and proteasome subunits by OrfA imply a defense system initiated by FIV to avoid proteasome-mediated degradation of either cellular proteins facilitating efficient FIV replication or of FIV viral proteins / particles. Thus, the down-regulation of components in the ubiquitin-proteasome pathway may help explain the finding that OrfA is required for efficient FIV particle formation (Gemeniano et al., 2003).
The present study suggests that OrfA mediates a fine-tuning of the cellular splicing and protein degradation machinery and thus gives insight into the mechanism that makes OrfA critical for efficient FIV replication in lymphocytes.
Material and Methods
Cell lines and virus
The primary IL-2-dependent T cell line 104-C1 was isolated from feline PBMCs by limiting dilution (Lerner et al., 1998) and generously provided by Dr. Chris Grant (Custom Monoclonals Int., W. Sacramento, CA). 104-C1 cells were maintained in RPMI 1640 (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA), 2 mM l-glutamine (Gibco), 1 mM sodium pyruvate (Gibco), 10 mM HEPES buffer (Gibco), 1 × minimal essential medium-vitamins (Cellgro Mediatech, Herndon, VA), 1 × nonessential amino acids (Gibco), 1 × β-mercaptoethanol (Gibco), 2.5 μg/ml concanavalin A (Sigma, St. Louis, MO), 50 U/ml of human recombinant IL-2 (gift from Hoffman-La Roche) and 50 μg/ml gentamycin (Gemini Bioproducts, West Sacramento, CA). Construction of 104-C1 cells expressing GFP (104-C1-GFP) or co-expressing GFP and FIV-PPR (Phillips et al., 1990) OrfA (104-C1-GFP-OrfA) has been described previously (Chatterji et al., 2002). Briefly, GP2−293 packaging cells (Clontech, Palo Alto, CA) were transiently transfected with pVSV-G and Mig R1 (Pear et al., 1998) or Mig R1-OrfA to produce a retroviral stock, which was used to transduce 104-C1 cells by spinoculation. GFP-expressing cells were sorted with a FACS Vantage SE I (Becton Dickinson, Franklin Lakes, NJ) at the TSRI Flow Cytometry Core Facility. GFP+ cells were scaled up and sorted a second time followed by confirmation of GFP expression by flow cytometric analysis. OrfA expression in 104-C1-GFP-OrfA was confirmed by 35S-labeling followed by immunoprecipitation and has been described previously (Chatterji et al., 2002).
FIV-34TF10, a cell line-adapted FIV strain derived from the Petaluma isolate (Talbott et al., 1989), was used as an OrfA-defective strain for trans-complementation assays.
Micro-reverse transcriptase (RT) activity assay
Micro-RT activity assay was performed as previously described (de Parseval and Elder, 2001; de Parseval et al., 1997). Briefly, primary and transfected 104-C1 cells were infected with FIV-34TF10, which lacks functional OrfA expression, or with mutants of 34TF10 in which the stop codon in OrfA was replaced with the codon for Cysteine (Cys), rendering the gene functional. Virus production was monitored by analysis of RT activity. Fifty μl cell-free supernatant was incubated 10 minutes at room temperature with 10 μl of lysis buffer (0.75 M KCl, 20 mM dithiothreitol, 0.5% Triton X-100). Forty μl of a mixture containing 125 mM Tris-HCl (pH 8.1), 12.5 mM MgCl2, 1.25 μg poly(rA)-poly(dT)12−18 (Amersham Biosciences, Piscataway, NJ) and 1.25 μCi of [3H]dTTP (DuPont, Boston, MA) was added to the sample followed by 2 hours incubation at 37°C. RT activity was measured as previously described (de Parseval et al., 1997).
Proliferation assay
Growth rates of control and transfected cells were measured in order to assess the influence of recombinant protein expression. Control 104-C1, 104-C1-GFP and 104-C1-GFP-OrfA cells were seeded at 105 cells per well, in triplicate. Cells were then counted after 24, 48, and 72 hours using a hemocytometer.
Cell cycle analysis
104-C1, 104-C1-GFP and 104-C1 GFP-OrfA cells were harvested and resuspended in 0.5 ml Phosphate Buffered Saline (PBS) to achieve a single cell suspension at 2×106 cells/ml. Cells were fixed in 70% ethanol for 2 hours before being washed in PBS and suspended in Propidium Iodide (PI) / Triton X-100 staining solution containing 0.1% (v/v) Triton X-100 (Kodak, Rochester, NY), 0.2 mg/ml DNase-free RNase A (Sigma) and 0.02 mg/ml PI (Sigma) for 30 min at room temperature. Cell fluorescence was measured by flow cytometry and analyzed according to Basic protocol 1 in Current Protocols in Cytometry (Darzynkiewicz, and Juan, 1997). Discrimination of cells in G0/G1, S and G2/M phases of the cell cycle was performed using FlowJo 4.6 software. As a control, 104-C1 cells were arrested at G2/M phase by overnight incubation with 4μg/ml Aphidicolin (Fisher, Pittsburgh, PA) followed by another overnight incubation in 0.1μg/ml Hoechst 33342 (Promega, Madison, WI). Arrested cells were stained and analyzed as above.
RNA isolation
Total RNA for quantitative real time PCR and microarray assays was isolated from 2.5×106 104-C1-GFP or 104-C1-GFP-OrfA cells using TRIZOL Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol.
The GeneChip Human Genome U133 Plus 2.0 Array
As no feline microarray is yet available, labeled RNA (see below) originating from the transfected 104-C1 cells, was hybridized to the commercially available Affymetrix GeneChip oligonucleotide arrays Human Genome U133 Plus 2.0 (HU133Plus 2.0) and Mouse 430 2.0 (Affymetrix, Santa Clara, CA). As the human array gave a higher number of present transcripts in this cross-species analysis (9.7% compared to 8.3% for the mouse array) the HU133Plus 2.0 array was used throughout this study. The HU133Plus 2.0 oligonucleotide array has a total of 54,675 probe sets representing over 47,000 transcripts. A complete description and annotation for the HU133Plus 2.0 array is available at http://www.affymetrix.com/products/arrays/specific /hgu133plus.affx.
Microarray RNA processing, labeling and chip hybridization
Labeled cRNA probes were generated from total RNA samples using the MessageAmp II-Biotin Enhanced Kit (Ambion Inc., Austin, TX). Briefly, 500 ng of total RNA was reverse transcribed with a T7 oligo(dT) primer bearing a T7 promoter sequence followed by in vitro transcription of the resulting DNA with T7 RNA polymerase to generate anti-sense RNA copies of each mRNA. Biotinylated cRNA probes were hybridized to Human Genome U133 Plus 2.0 arrays using the Hybridization Oven 640 according to the manufacturer's standard protocol. Arrays were processed using the Affymetrix Fluidics Station 450 and scanned using the ScanArray 3000 with default settings and a target intensity of 250 for image scaling (Affymetrix).
Microarray data analysis
Diagnostic plots showing distribution of raw signals and RNA degradation profiles of all probe sets from 5' to 3' positions were used to eliminate outlier chips. One chip was excluded from the study due to outlying distribution and RNA degradation slope. Robust Multichip Average normalization was applied on the data followed by adjustment of the data for batch effects using an empirical Bayes framework (Johnson et al., 2007). Subsequently, both the Rank Products method (Breitling et al., 2004) and ANOVA analysis were performed using BRB Array tools (Radmacher et al., 2002), to discover differential gene expression. Differentially expressed genes were identified from the Rank Products method at 10% false positives rate and from the ANOVA analysis at a univariate p-value level of 0.001. The list of regulated genes was annotated using the Affymetrix NetAffx website (http://www.affymetrix.com/analysis/index.affx). Microarray processing, data analysis and classification of genes were performed at the TSRI DNA Array Core Facility, La Jolla, CA.
Classification and functional analyses of genes
Ingenuity Pathways Knowledge Base, a knowledge bank of biological findings for human, mouse and rat genes (Ingenuity® Systems, http://www.ingenuity.com/) was used for classification of the differentially expressed genes based on their involvement in biological processes. The functional analyses of the genes were generated through the use of Ingenuity Pathways Analysis (Ingenuity® Systems) and used to identify the biological functions that were most significant to the array data set. Ingenuity Canonical Pathways Analysis was also used to identify the pathways from the Ingenuity analysis library of canonical pathways that were most significant to the data set. Fischer's exact test was used to calculate p-values determining the probability that each biological function / canonical pathway assigned to that data set is due to chance alone.
cDNA synthesis
Residual genomic DNA was removed from the total RNA by the Turbo DNA-free kit (Ambion). 1μg of total RNA was reverse transcribed using Transcriptor First Strand cDNA synthesis kit and random hexamer primers according to manufacturer's instructions (Roche Applied Science, Indianapolis, IN)
Q-PCR
For each reaction, 1μl of a 1:10 dilution of cDNA, 11.25 μl of dH2O, 12.5μl of SYBR Green PCR master mix (Qiagen, Valencia, CA) and 500 nM of each forward and reverse primers (see below), were mixed in 25μl of total volume. Quantitative Real-Time PCR (Q-PCR) was performed using an ABI PRISM 7700 Sequence Detector (Perkin Elmer, Waltham, MA) and the conditions: 50°C, 2 min and 95°C, 10 min, followed by 45 cycles of 95°C, 15 sec, and 60°C, 1 min. The deltadelta crossing thresholds (Ct) method was used for data analysis (according to the Applied Biosystems manual). For each primer set gene expression levels were quantified relative to that of 18S rRNA. Confirmation of single amplicons, and lack of primer-dimer formation, was performed by melting-curve analysis and agarose gel electrophoresis.
Q-PCR primers
Primers for the following feline genes were used for Q-PCR quantification analyses, 18S; fibrillin 1 (FBN1); plexcin C1 (PLXCN1); early B cell factor (EBF), heterogeneous nuclear ribonucleoprotein D-like (hnRNP DL), UAP56, hnRNP A2B1, SFRS3, UBE2C, and PSMA7.
18S fwd: CGGCTACCACATCCAAAGGAA, rev: GCTGGAATTACCGCGGCT
FBN1 fwd: CCACTGGCTTCTTCTTGGTG, rev: GTCTCGCAAGTTGGGATGTT
PLXCN1 fwd: GGCTGGAAGAAGCTCAGAAA, rev: GACAGTGCCCAAAGTGGAAC
EBF fwd: TGCAGAACCCTTCCAATTTC, rev: ATCTGGCATGATCGTTCCTC
hnRNP DL fwd: GCGTTGTATGGAGCTGGATT, rev: CCTCTGCTCCCGCTACTTTA
UAP56 fwd: TGCCTTCAACTCTGACATGC, rev: CTCCTGAAGAGGGACAGACG
hnRNP A2B1 fwd: CCATGATCCTGGCTAATAGCTT, rev: GACACCAGAGCAGATGCAGA
SFRS3 fwd: AGAGGTGGTCCAGTTGGTGT, rev: CCCAAGGATTCTCACAAACA
UBE2C fwd: CTAGCAGGCTCTGGATGGAC, rev: CTGCCTTCCCTGAATCAGAC
PSMA7 fwd: CCAAAGTCGAAACCCACAAT, rev: GTTCTGGGCATGGAGAAGAA
Nucleotide sequences for the feline genes were identified by cross-species megaBLAST analysis in which the sequence for the human gene was compared with the 2× sequence assembly of the domestic cat genome from The Broad Institute and Agencourt Bioscience Corporation. Primers were designed using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi (Rozen and Skaletsky, 2000).
Acknowledgements
We wish to thank Dr. Steven Head and the DNA Array Core Facility for valuable assistance and for supplying array chips used in the analyses, under the auspices of the SNAPS Core Grant at TSRI (from the Mental Health Institute of the National Institutes of Health, MH062261, Dr. Howard S. Fox, PI). We also thank Dr. Christoph Huber for valuable Q-PCR advice, Dr. Erika Assarsson, Dr. Ying-Chuan Lin and Dr. Aymeric de Parseval for valuable comments, Jovylyn Gatchalian for excellent technical assistance and Nancy Dorman for help with manuscript preparation. This work was supported by grant R01 AI025825 from the National Institute Allergy and Infectious Disease Institute of the National Institute of Health. Magnus Sundstrom was supported by a fellowship from the Sweden-America Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apcher GS, Heink S, Zantopf D, Kloetzel PM, Schmid HP, Mayer RJ, Kruger E. Human immunodeficiency virus-1 Tat protein interacts with distinct proteasomal alpha and beta subunits. FEBS Lett. 2003;553(1−2):200–4. doi: 10.1016/s0014-5793(03)01025-1. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573(1−3):83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Caputi M, Mayeda A, Krainer AR, Zahler AM. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. Embo J. 1999;18(14):4060–7. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji U, de Parseval A, Elder JH. Feline immunodeficiency virus OrfA is distinct from other lentivirus transactivators. J Virol. 2002;76(19):9624–34. doi: 10.1128/JVI.76.19.9624-9634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YL, Coronel E, Ho CK, Shuman S, Rana TM. HIV-1 Tat protein interacts with mammalian capping enzyme and stimulates capping of TAR RNA. J Biol Chem. 2001;276(16):12959–66. doi: 10.1074/jbc.M007901200. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Juan G. Current protocols in cytometry. John Wiley & Sons; New York: 1997. [Google Scholar]
- de Parseval A, Elder JH. Demonstration that orf2 encodes the feline immunodeficiency virus transactivating (Tat) protein and characterization of a unique gene product with partial rev activity. J Virol. 1999;73(1):608–17. doi: 10.1128/jvi.73.1.608-617.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Parseval A, Elder JH. Binding of recombinant feline immunodeficiency virus surface glycoprotein to feline cells: role of CXCR4, cell-surface heparans, and an unidentified non-CXCR4 receptor. J Virol. 2001;75(10):4528–39. doi: 10.1128/JVI.75.10.4528-4539.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Parseval A, Lerner DL, Borrow P, Willett BJ, Elder JH. Blocking of feline immunodeficiency virus infection by a monoclonal antibody to CD9 is via inhibition of virus release rather than interference with receptor binding. J Virol. 1997;71(8):5742–9. doi: 10.1128/jvi.71.8.5742-5749.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domsic JK, Wang Y, Mayeda A, Krainer AR, Stoltzfus CM. Human immunodeficiency virus type 1 hnRNP A/B-dependent exonic splicing silencer ESSV antagonizes binding of U2AF65 to viral polypyrimidine tracts. Mol Cell Biol. 2003;23(23):8762–72. doi: 10.1128/MCB.23.23.8762-8772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckner J, Zhang M, Valcarcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997;11(14):1864–72. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- Gavioli R, Gallerani E, Fortini C, Fabris M, Bottoni A, Canella A, Bonaccorsi A, Marastoni M, Micheletti F, Cafaro A, Rimessi P, Caputo A, Ensoli B. HIV-1 tat protein modulates the generation of cytotoxic T cell epitopes by modifying proteasome composition and enzymatic activity. J Immunol. 2004;173(6):3838–43. doi: 10.4049/jimmunol.173.6.3838. [DOI] [PubMed] [Google Scholar]
- Gemeniano MC, Sawai ET, Leutenegger CM, Sparger EE. Feline immunodeficiency virus ORF-A is required for virus particle formation and virus infectivity. J Virol. 2003;77(16):8819–30. doi: 10.1128/JVI.77.16.8819-8830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemeniano MC, Sawai ET, Sparger EE. Feline immunodeficiency virus Orf-A localizes to the nucleus and induces cell cycle arrest. Virology. 2004;325(2):167–74. doi: 10.1016/j.virol.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- Graveley BR. Sorting out the complexity of SR protein functions. Rna. 2000;6(9):1197–211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume C, Kuramitsu M, Zhang X, Kurosawa T, Kamata M, Aida Y. Human immunodeficiency virus type 1 Vpr interacts with spliceosomal protein SAP145 to mediate cellular pre-mRNA splicing inhibition. Microbes Infect. 2007;9(4):490–7. doi: 10.1016/j.micinf.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Huang X, Seifert U, Salzmann U, Henklein P, Preissner R, Henke W, Sijts AJ, Kloetzel PM, Dubiel W. The RTP site shared by the HIV-1 Tat protein and the 11S regulator subunit alpha is crucial for their effects on proteasome function including antigen processing. J Mol Biol. 2002;323(4):771–82. doi: 10.1016/s0022-2836(02)00998-1. [DOI] [PubMed] [Google Scholar]
- Jacquenet S, Mereau A, Bilodeau PS, Damier L, Stoltzfus CM, Branlant C. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J Biol Chem. 2001;276(44):40464–75. doi: 10.1074/jbc.M104070200. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kammler S, Otte M, Hauber I, Kjems J, Hauber J, Schaal H. The strength of the HIV-1 3' splice sites affects Rev function. Retrovirology. 2006;3:89. doi: 10.1186/1742-4690-3-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Bourgeois CF, Pearson R, Tyagi M, West MJ, Wong J, Wu SY, Chiang CM, Karn J. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. Embo J. 2006;25(15):3596–604. doi: 10.1038/sj.emboj.7601248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloetzel PM, Ossendorp F. Proteasome and peptidase function in MHC-class-I-mediated antigen presentation. Curr Opin Immunol. 2004;16(1):76–81. doi: 10.1016/j.coi.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Lerner DL, Grant CK, de Parseval A, Elder JH. FIV infection of IL-2-dependent and -independent feline lymphocyte lines: host cells range distinctions and specific cytokine upregulation. Vet Immunol Immunopathol. 1998;65(2−4):277–97. doi: 10.1016/S0165-2427(98)00162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhrmann R, Kastner B, Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim Biophys Acta. 1990;1087(3):265–92. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- McLaren M, Asai K, Cochrane A. A novel function for Sam68: enhancement of HIV-1 RNA 3' end processing. Rna. 2004;10(7):1119–29. doi: 10.1261/rna.5263904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modem S, Badri KR, Holland TC, Reddy TR. Sam68 is absolutely required for Rev function and HIV-1 production. Nucleic Acids Res. 2005;33(3):873–9. doi: 10.1093/nar/gki231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naghavi MH, Nowak P, Andersson J, Sonnerborg A, Yang H, Tracey KJ, Vahlne A. Intracellular high mobility group B1 protein (HMGB1) represses HIV-1 LTR-directed transcription in a promoter- and cell-specific manner. Virology. 2003;314(1):179–89. doi: 10.1016/s0042-6822(03)00453-7. [DOI] [PubMed] [Google Scholar]
- Nekhai S, Zhou M, Fernandez A, Lane WS, Lamb NJ, Brady J, Kumar A. HIV-1 Tat-associated RNA polymerase C-terminal domain kinase, CDK2, phosphorylates CDK7 and stimulates Tat-mediated transcription. Biochem J. 2002;364(Pt 3):649–57. doi: 10.1042/BJ20011191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak P, Barqasho B, Treutiger CJ, Harris HE, Tracey KJ, Andersson J, Sonnerborg A. HMGB1 activates replication of latent HIV-1 in a monocytic cell-line, but inhibits HIV-1 replication in primary macrophages. Cytokine. 2006;34(1−2):17–23. doi: 10.1016/j.cyto.2006.03.010. [DOI] [PubMed] [Google Scholar]
- O'Neil LL, Burkhard MJ, Diehl LJ, Hoover EA. Vertical transmission of feline immunodeficiency virus. Semin Vet Med Surg (Small Anim) 1995;10(4):266–78. [PubMed] [Google Scholar]
- O'Neil LL, Burkhard MJ, Hoover EA. Frequent perinatal transmission of feline immunodeficiency virus by chronically infected cats. J Virol. 1996;70(5):2894–901. doi: 10.1128/jvi.70.5.2894-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obert LA, Hoover EA. Relationship of lymphoid lesions to disease course in mucosal feline immunodeficiency virus type C infection. Vet Pathol. 2000;37(5):386–401. doi: 10.1354/vp.37-5-386. [DOI] [PubMed] [Google Scholar]
- Olmsted RA, Hirsch VM, Purcell RH, Johnson PR. Nucleotide sequence analysis of feline immunodeficiency virus: genome organization and relationship to other lentiviruses. Proc Natl Acad Sci U S A. 1989;86(20):8088–92. doi: 10.1073/pnas.86.20.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, Baltimore D. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92(10):3780–92. [PubMed] [Google Scholar]
- Pedersen NC. Immunogenicity and efficacy of a commercial feline leukemia virus vaccine. J Vet Intern Med. 1993;7(1):34–9. doi: 10.1111/j.1939-1676.1993.tb03166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen NC, Ho EW, Brown ML, Yamamoto JK. Isolation of a T-lymphotropic virus from domestic cats with an immunodeficiency-like syndrome. Science. 1987;235(4790):790–3. doi: 10.1126/science.3643650. [DOI] [PubMed] [Google Scholar]
- Phillips TR, Talbott RL, Lamont C, Muir S, Lovelace K, Elder JH. Comparison of two host cell range variants of feline immunodeficiency virus. J Virol. 1990;64(10):4605–13. doi: 10.1128/jvi.64.10.4605-4613.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistello M, Moscardini M, Mazzetti P, Bonci F, Zaccaro L, Isola P, Freer G, Specter S, Matteucci D, Bendinelli M. Development of feline immunodeficiency virus ORF-A (tat) mutants: in vitro and in vivo characterization. Virology. 2002;298(1):84–95. doi: 10.1006/viro.2002.1442. [DOI] [PubMed] [Google Scholar]
- Pocernich CB, Boyd-Kimball D, Poon HF, Thongboonkerd V, Lynn BC, Klein JB, Calebrese V, Nath A, Butterfield DA. Proteomics analysis of human astrocytes expressing the HIV protein Tat. Brain Res Mol Brain Res. 2005;133(2):307–16. doi: 10.1016/j.molbrainres.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Poon B, Chang MA, Chen IS. Vpr is required for efficient nef expression from unintegrated human immunodeficiency virus type 1 DNA. J Virol. 2007;81(19):10515–23. doi: 10.1128/JVI.00947-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell DF, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J Virol. 1993;67(11):6365–78. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M, Aiken C. Selective restriction of Nef-defective human immunodeficiency virus type 1 by a proteasome-dependent mechanism. J Virol. 2007;81(3):1534–6. doi: 10.1128/JVI.02099-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radmacher MD, McShane LM, Simon R. A paradigm for class prediction using gene expression profiles. J Comput Biol. 2002;9(3):505–11. doi: 10.1089/106652702760138592. [DOI] [PubMed] [Google Scholar]
- Remoli AL, Marsili G, Perrotti E, Gallerani E, Ilari R, Nappi F, Cafaro A, Ensoli B, Gavioli R, Battistini A. Intracellular HIV-1 Tat protein represses constitutive LMP2 transcription increasing proteasome activity by interfering with the binding of IRF-1 to STAT1. Biochem J. 2006;396(2):371–80. doi: 10.1042/BJ20051570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AB, Hoover EA. Maternal-fetal feline immunodeficiency virus transmission: timing and tissue tropisms. J Infect Dis. 1998;178(4):960–7. doi: 10.1086/515692. [DOI] [PubMed] [Google Scholar]
- Ropers D, Ayadi L, Gattoni R, Jacquenet S, Damier L, Branlant C, Stevenin J. Differential effects of the SR proteins 9G8, SC35, ASF/SF2, and SRp40 on the utilization of the A1 to A5 splicing sites of HIV-1 RNA. J Biol Chem. 2004;279(29):29963–73. doi: 10.1074/jbc.M404452200. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Ryo A, Suzuki Y, Arai M, Kondoh N, Wakatsuki T, Hada A, Shuda M, Tanaka K, Sato C, Yamamoto M, Yamamoto N. Identification and characterization of differentially expressed mRNAs in HIV type 1-infected human T cells. AIDS Res Hum Retroviruses. 2000;16(10):995–1005. doi: 10.1089/08892220050058416. [DOI] [PubMed] [Google Scholar]
- Seeger M, Ferrell K, Frank R, Dubiel W. HIV-1 tat inhibits the 20 S proteasome and its 11 S regulator-mediated activation. J Biol Chem. 1997;272(13):8145–8. doi: 10.1074/jbc.272.13.8145. [DOI] [PubMed] [Google Scholar]
- Sillekens PT, Beijer RP, Habets WJ, van Verooij WJ. Molecular cloning of the cDNA for the human U2 snRNA-specific A' protein. Nucleic Acids Res. 1989;17(5):1893–906. doi: 10.1093/nar/17.5.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillekens PT, Habets WJ, Beijer RP, van Venrooij WJ. cDNA cloning of the human U1 snRNA-associated A protein: extensive homology between U1 and U2 snRNP-specific proteins. Embo J. 1987;6(12):3841–8. doi: 10.1002/j.1460-2075.1987.tb02721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparger EE, Shacklett BL, Renshaw-Gegg L, Barry PA, Pedersen NC, Elder JH, Luciw PA. Regulation of gene expression directed by the long terminal repeat of the feline immunodeficiency virus. Virology. 1992;187(1):165–77. doi: 10.1016/0042-6822(92)90305-9. [DOI] [PubMed] [Google Scholar]
- Stoltzfus CM, Madsen JM. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr HIV Res. 2006;4(1):43–55. doi: 10.2174/157016206775197655. [DOI] [PubMed] [Google Scholar]
- Talbott RL, Sparger EE, Lovelace KM, Fitch WM, Pedersen NC, Luciw PA, Elder JH. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc Natl Acad Sci U S A. 1989;86(15):5743–7. doi: 10.1073/pnas.86.15.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange TO, Damgaard CK, Guth S, Valcarcel J, Kjems J. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. Embo J. 2001;20(20):5748–58. doi: 10.1093/emboj/20.20.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y, Yasuda Y. Human immunodeficiency virus type 1 Vpr induces G2 checkpoint activation by interacting with the splicing factor SAP145. Mol Cell Biol. 2006;26(21):8149–58. doi: 10.1128/MCB.01170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van 't Wout AB, Lehrman GK, Mikheeva SA, O'Keeffe GC, Katze MG, Bumgarner RE, Geiss GK, Mullins JI. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4(+)-T-cell lines. J Virol. 2003;77(2):1392–402. doi: 10.1128/JVI.77.2.1392-1402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscidi RP, Mayur K, Lederman HM, Frankel AD. Inhibition of antigen-induced lymphocyte proliferation by Tat protein from HIV-1. Science. 1989;246(4937):1606–8. doi: 10.1126/science.2556795. [DOI] [PubMed] [Google Scholar]
- Wang H, Ward MF, Fan XG, Sama AE, Li W. Potential role of high mobility group box 1 in viral infectious diseases. Viral Immunol. 2006;19(1):3–9. doi: 10.1089/vim.2006.19.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xiao SH, Manley JL. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 1998;12(14):2222–33. doi: 10.1101/gad.12.14.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AK, De Parseval AP, Lerner DL, Neil JC, Thompson FJ, Elder JH. Influence of ORF2 on host cell tropism of feline immunodeficiency virus. Virology. 1996;215(1):10–6. doi: 10.1006/viro.1996.0002. [DOI] [PubMed] [Google Scholar]
- Wei BL, Denton PW, O'Neill E, Luo T, Foster JL, Garcia JV. Inhibition of lysosome and proteasome function enhances human immunodeficiency virus type 1 infection. J Virol. 2005;79(9):5705–12. doi: 10.1128/JVI.79.9.5705-5712.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Duus KM, Friedrich TD, de Noronha CM. The HIV1 Protein Vpr Acts to Promote G2 Cell Cycle Arrest by Engaging a DDB1 and Cullin4A-containing Ubiquitin Ligase Complex Using VprBP/DCAF1 as an Adaptor. J Biol Chem. 2007;282(37):27046–57. doi: 10.1074/jbc.M703955200. [DOI] [PubMed] [Google Scholar]
- Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci U S A. 2006;103(19):7465–70. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Damgaard CK, Kjems J, Caputi M. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J Biol Chem. 2004;279(11):10077–84. doi: 10.1074/jbc.M312743200. [DOI] [PubMed] [Google Scholar]