Abstract
We identified a set of cytokinin-insensitive mutants by using a screen based on the ethylene-mediated triple response observed after treatment with low levels of cytokinins. One group of these mutants disrupts ACS5, a member of the Arabidopsis gene family that encodes 1-aminocyclopropane-1-carboxylate synthase, the first enzyme in ethylene biosynthesis. The ACS5 isoform is mainly responsible for the sustained rise in ethylene biosynthesis observed in response to low levels of cytokinin and appears to be regulated primarily by a posttranscriptional mechanism. Furthermore, the dominant ethylene-overproducing mutant eto2 was found to be the result of an alteration of the carboxy terminus of ACS5, suggesting that this domain acts as a negative regulator of ACS5 function.
Keywords: hormone biosynthesis, plant hormones
Plant development is modulated by interactions among multiple hormones. An illustration of such interactions is the induction of biosynthesis of one hormone by another. Cytokinins, N6-substituted adenine derivatives, are one of the many factors that modulate the biosynthesis of the gaseous hormone ethylene (1-3). We are characterizing the induction of ethylene biosynthesis by cytokinin to understand how cytokinins and ethylene interact to regulate plant development.
Cytokinins were first identified as factors that acted synergistically with auxin to promote cell division in vitro and acted antagonistically to auxin in shoot and root initiation from callus cultures (4–6). Cytokinins have been implicated in many aspects of plant growth and development, including cell division, shoot initiation and growth, senescence, and photomorphogenesis (7–9). In contrast to the wealth of knowledge concerning the physiological effects of cytokinins, the molecular mechanisms underlying cytokinin perception and action remain largely unknown.
Ethylene is involved in a variety of plant growth and developmental processes, including senescence, leaf abscission, epinasty, and fruit ripening (2, 3). Recent studies have begun to shed light on the molecular mechanisms underlying ethylene signaling, including the identification of the genes encoding the ethylene receptor and several downstream signaling elements (10-14). The ethylene biosynthetic pathway has been worked out in great detail, and genes encoding the biosynthetic enzymes have been isolated from many plant species (1, 15, 16). The conversion of S-adenosyl methionine to 1-aminocyclopropane-1-carboxylate (ACC) by ACC synthase is the first committed step in ethylene biosynthesis and generally is considered to be the rate-limiting step. This enzyme is encoded by a divergent gene family in all plant species analyzed so far. The expression of ACC synthase has been best studied in tomato, in which the gene family consists of at least nine members (17-21). The emerging picture is that different subsets of ACC synthase genes are expressed in response to various developmental, environmental, and hormonal factors. In Arabidopsis, five ACC synthase genes have been identified (ACS1–ACS5), three of which are expressed and encode active ACC synthase enzymes (22-24). The final step of ethylene biosynthesis is the conversion of ACC to ethylene by the enzyme ACC oxidase, which also is encoded by a multigene family in many plant species and whose members also are differentially regulated (25-30).
Ethylene biosynthesis is regulated by other hormones in many plant tissues, the best-studied example of which is auxin (1, 15). The steady-state level of mRNA from specific members of the ACS gene family become elevated in response to exogenous auxin in various plant species (19, 31, 32). In Arabidopsis, auxin specifically induces expression of ACS4, and four putative auxin response elements have been identified in the 5′ regulatory sequences of this gene (32). Cytokinins also induce ethylene biosynthesis in many plant species, including Arabidopsis (33), but the mechanism for this induction is unknown. In at least one tissue, mung bean hypocotyls, the cytokinin benzyladenine (BA) has been shown to increase ACC synthase activity (34). To further understand how cytokinins regulate ethylene biosynthesis, we isolated a collection of Arabidopsis mutants that did not respond to cytokinin by increased ethylene biosynthesis. Here, we present the analysis of one such complementation group, cin5 (cytokinin insensitive), which disrupts a member of the ACS gene family. Furthermore, we demonstrate that perturbation of the carboxy terminus of this same ACS5 isoform results in ethylene overproduction.
MATERIALS AND METHODS
Plant Material.
Columbia (Col) and Wassilewskija (WS) were the two A. thaliana ecotypes used in this study. The T-DNA mutagenized population (35) was obtained from the Arabidopsis Biological Resource Center (Ohio State University). Plants were grown in Metro-Mix 200 (Grace, Boca Raton, FL) under continuous illumination with fluorescent light at 23°C. Ethyl methanesulfonate (EMS)-mutagenized seeds were obtained by treating hydrated seeds (soaked overnight in distilled water) with 0.4% EMS for 7 h. The seeds were washed thoroughly with water and then sown in flats (52 × 26 cm) at a density of 3,000 seeds per flat. Etiolated seedlings were grown as follows: seeds were surfaced sterilized for 8 min in 5% sodium hypochlorite (Clorox)/0.01% Tween 20 and rinsed five times with sterile water. The sterile seeds then were resuspended in 0.8% low-melting-point agarose and plated on 0.8% agar containing Murashige and Skoog salts (MS; GIBCO) supplemented with 2% sucrose. Seeds were incubated at 4°C for 4 days and then moved to 23°C in the dark for 3 days.
Ethylene Measurements.
Seedlings (about 15 per vial) were grown in 22-ml gas chromatography vials containing 3 ml of MS medium in the dark at 23°C. Appropriate supplements as indicated were either included in the media or added just before capping. Added compounds were in a volume of 200 μl (controls were 200 μl of water). The vials were flushed with hydrocarbon-free air and then capped for the indicated times. The accumulated ethylene was measured by using a gas chromatograph (Perkin–Elmer) fitted with a PoraPLOT U column, a cryofocusing attachment, and a flame ionization detector. A sample of headspace from each sample was injected onto the column at −50°C via an autosampler, and the column was then warmed to 30°C. The ethylene peaks were quantified by using Turbochrome 4 software (Perkin–Elmer) based on comparison to a 1 μl/liter ethylene standard. Ethylene production was normalized for the number of seedlings in each vial and the time between capping and sampling. All observations were from at least three replicates, and each experiment was repeated at least once with comparable results. The measurements of the basal level of ethylene from WS and cin5 are from seven independent experiments with at least three replicates each.
Hypocotyl Measurements.
Sterilized seeds were plated on MS medium and grown for 3 days at 23°C in the dark. The length of the hypocotyl was measured under a dissecting microscope, and the data presented as the mean ± the SD. At least 80 seedlings were measured in each case.
Mapping.
cin5-3 (ecotype Col) was backcrossed to the WS ecotype to generate a mapping population. F2 seeds were plated on MS medium supplemented with 0.5 μM BA, and cin5 mutant seedlings (tall) were transferred to MS medium without BA and allowed to grow for about 1 week. DNA was extracted (36), and minisatellite markers polymorphic between ecotypes were amplified by using PCR (37). The products were separated by gel electrophoresis in 4% agarose and visualized by ethidium bromide staining.
Cloning of CIN5.
Plant DNA flanking the left border of the T-DNA insertion in cin5-1 was isolated by plasmid rescue as follows: 5 μg of genomic DNA from a plant homozygous for the cin5-1 allele was digested with SaII restriction enzyme. The reaction mixture was extracted once with an equal volume of phenol/CHCl3/isoamyl alcohol (25:24:1), once with CHCl3/isoamyl alcohol, and then ethanol precipitated. The DNA was ligated in a 500-μl reaction according to the manufacturer’s instructions (Boehringer Mannheim). The ligation mixture was precipitated with ethanol, transformed into XL1 blue, and plated on Luria–Bertani agar containing 100 μg/ml of ampicillin (LB Amp). Colonies (300) were patched onto LB Amp plates, and colony lifts were prepared by using Hybond N+ (Amersham) as described (38). The filters were hybridized to a left border probe (obtained from the Arabidopsis Biological Resource Center) as described (38). Sixty-nine positive colonies were obtained, the most common class of which displayed a restriction pattern expected for a direct repeat of T-DNA. The second most common class consisted of six clones, and one of these (pTK2) was used to probe a Southern blot containing wild-type (ecotype WS) and cin5-1 genomic DNA cleaved with various restriction enzymes. This analysis showed that the mobilities of the hybridizing bands were distinct in the cin5-1 allele, indicating that pTK2 corresponded to the DNA flanking the site of the T-DNA insertion. DNA sequence analysis showed that pTK2 contained a fusion of T-DNA and the ACS5 gene.
The various cin5 alleles were sequenced as follows: PCR using oligonucleotide primers (JV9: ATCGGATCCCAACAGACCCATTGCTTCTC and JV10: CGAGGATCCATAGCCACTGCAAATCCACA) flanking the ACS5 coding region was used to amplify ACS5 from wild type and the various cin5 alleles. The products were sequenced directly after removal of excess primers by ultrafiltration (Ultrafree-MC, Millipore). At least two independent PCR products were sequenced for each allele. DNA was sequenced by using gene-specific primers (JV1: GGCTATAAACGACGTGGACTCG; JV4: CATGAGAGTCTCATTCGCAG; JV11: ACGACTCAAGTCCAGACAGA; JV16: GCATGGATCCTAATTTCATCGTTCATCAGGTAC) by automated sequencing.
Northern Blot Analysis.
Seedlings were grown (2,000 per 100-mm plate) on MS agar for 3 days as described above except sterile filter paper was placed on the top of the agar. For cytokinin-treatments, 10 ml of 5 μM BA (in liquid MS) were added, and the seedlings were harvested at the indicated times. Total RNA was prepared by extraction with phenol/chloroform, and poly(A)+-RNA was isolated by oligo(dT)-cellulose affinity chromatography (38). Northern blot analysis was performed as described (38), and signals quantitated by using a PhosphorImager (Molecular Dynamics). The β-tubulin fragment was obtained by amplifying the gene from Arabidopsis genomic DNA using oligonucleotide primers (JVTF: GAGATTCTTCACATCCAGGG; and JVTR: CATCTCGTCCATTCCCTCAC).
Transformation of Arabidopsis with Wild-Type and eto2 ACS5.
The ACS5 gene was amplified from wild-type (ecotype Col) and eto2 genomic DNA using oligonucleotide primers (ACS5ka: AAAAGGATCCGTACTTCAAAAAAGTGGATATACG and ACS5kb:. AAAAGAGCTACAATACACACAAACGTTTC) with TaKaRa polymerase as described by the manufacturer (Panvera, Madison, WI). The resultant 4.15-kilobase PCR product was composed of the ACS5 coding region, 2.1 kilobases of the 5′ flanking region, and the ACS5 3′ untranslated region. The PCR products were cleaved with SacI and BamHI and ligated into the plant transformation vector pBI101 (39). The resultant plasmids then were transformed into wild-type WS and the cin5-2 mutant by vacuum infiltration (40). Transformants were selected on MS medium containing 50 μg/ml of kanamycin.
RESULTS
Isolation of Mutants Defective in Cytokinin-Induced Ethylene Biosynthesis.
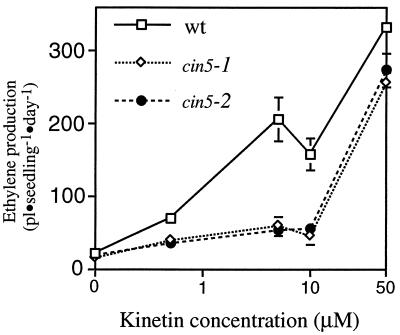
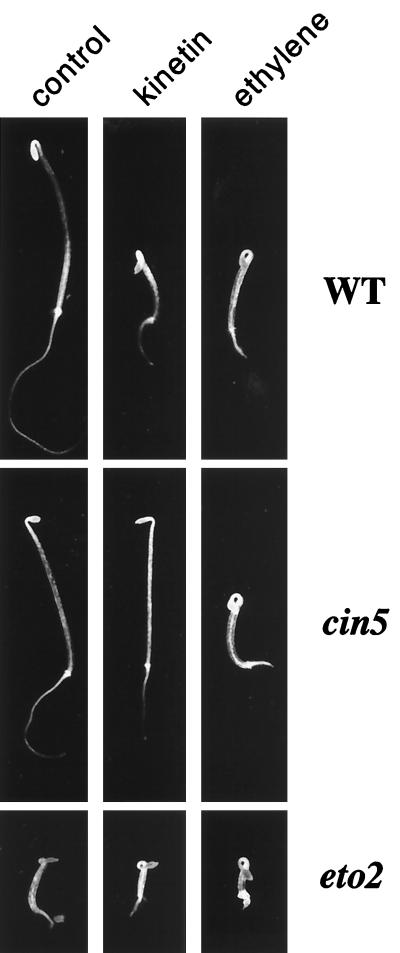
Measurements of ethylene production by etiolated Arabidopsis seedlings grown in the presence of increasing concentrations of the cytokinin kinetin show a biphasic dose-response curve with a reproducible inflection at approximately 10 μM (Fig. 1). This finding indicates that there may be two at least partially independent signaling pathways mediating cytokinin induction of ethylene biosynthesis, a possibility supported by the identification of mutations that affect only the low-dose pathway (Fig. 1 and see below). The ethylene produced in response to low doses of cytokinin (0.5-10 μM) is sufficient to induce a triple response in etiolated Arabidopsis seedlings (Fig. 2). The triple response of etiolated dicotyledonous seedlings to ethylene was first described in peas by Neljubow (41) and consists in Arabidopsis of a shortening and radial swelling of the hypocotyl, inhibition of root elongation, and exaggerated curvature of the apical hook (Fig. 2). This triple response is not observed in the presence of cytokinin if inhibitors of ethylene action or biosynthesis are also present or when ethylene-insensitive mutants are grown on media supplemented with cytokinin (not shown), indicating that cytokinin exerts its effects on seedling morphology primarily by increasing ethylene biosynthesis, as has been previously observed (33).
Figure 1.
Kinetin dose response of wild-type and cin5 mutant seedlings. Approximately 15 seeds were sown in a 22-ml gas chromatography vial containing 3 ml of MS medium supplemented with the indicated concentration of kinetin, incubated at 4°C for 4 days, flushed with hydrocarbon-free air, and sealed. Accumulated ethylene was measured after 72 h of incubation in the dark at 23°C. The means ± the SD based on three replicates are plotted. Very similar dose-response curves were obtained with the cytokinin benzyladenine (not shown). Note that the cin5 mutation only disrupts ethylene biosynthesis at lower concentrations of kinetin.
Figure 2.
Phenotype of cin5 and eto2 mutants. Wild-type (ecotype WS), cin5-2, or eto2 seedlings grown in the dark for 3 days on MS medium supplemented with no hormones, 0.5 μM kinetin, or in the presence of 10 μl/liter ethylene as indicated. Representative seedlings were picked and photographed.
We used this seedling response to screen approximately one million mutagenized Arabidopsis seedlings. After separating the ethylene-insensitive mutants and performing complementation tests, we were left with five complementation groups called cin1-cin5 (cytokinin insensitive). These cin mutants did not display the triple response in the presence of low concentrations of cytokinin but were indistinguishable from wild-type seedlings in their response to ethylene (Fig. 2). One complementation group, cin5, was comprised of three independent alleles and is the focus of this paper.
Characterization of CIN5.
cin5 segregates as single-gene, recessive mutation (e.g. cin5-1: 388 wild-type seedlings: 138 mutant seedlings in an F2 from a backcross to wild type; χ2 = 0.43, P > 0.4), suggesting that it is a loss-of-function allele. This hypothesis was confirmed by molecular analysis (see below). cin5 mutations have no detectable effect on the morphology of adult plants and do not significantly affect ethylene production in adult tissues (Table 1). The cin5 mutation was mapped to the bottom of chromosome 5, 32 map units from the microsatellite marker nga129 (37), close to the position of the eto2 mutation (11) as well as the ACC synthase 5 (ACS5) gene (24). Subsequent molecular analysis showed that these are, in fact, all a single gene (see below).
Table 1.
Ethylene production by various wild-type and mutant tissues
| Tissue | Ethylene
production, pl·mg−1·h−1
|
||
|---|---|---|---|
| Wild type (WS) | cin5-1 | eto2-1 | |
| Seedlings | |||
| Etiolated | 1.8 ± 3.5 | 1.2 ± 2.0 | 33.2 ± 3.2 |
| Light-grown | 11.6 ± 0.7 | 11.2 ± 1.2 | 15.7 ± 0.4 |
| Adult tissues | |||
| Flowers | 39.0 ± 5.8 | 44.0 ± 4.3 | 47.5 ± 8.0 |
| Siliques | 19.7 ± 2.4 | 19.8 ± 5.0 | 13.4 ± 3.0 |
| Leaves | 1.6 ± 0.4 | 1.4 ± 0.2 | 2.0 ± 0.2 |
The mean of at least three independent replicates ± SD. Seedlings were grown or excised tissue placed in vials containing 3 ml of MS agar. The vials were flushed with hydrocarbon-free air, sealed, and incubated at 23°C (3 days for seedlings, 7 h for tissue), and the accumulated ethylene was measured.
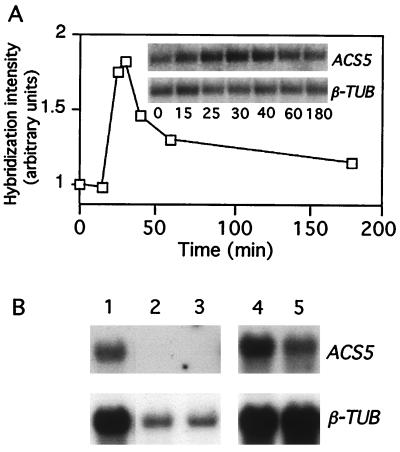
To learn more about the function of CIN5, the gene corresponding to the cin5 mutations was cloned by T-DNA tagging. A T-DNA population (35) was screened for mutants that lacked a triple response in the presence of cytokinin, and a single line segregating for a mutation (cin5-1) that failed to complement cin5-2 was identified. Analysis of the segregation of the kanamycin resistance marker carried on the T-DNA in this line suggested that it contained insertions at two unlinked loci (F2 of a backcross segregated 363 kanamycin resistant/22 kanamycin sensitive, which fits a 15:1 ratio, χ2 = 0.18 P > 0.05). One of the two T-DNA insertions was found to be closely linked to the cin5-1 mutation, and the DNA flanking the site of this insertion was isolated by plasmid rescue (see Materials and Methods). Southern blot analysis using this rescued DNA as a probe showed DNA polymorphisms between genomic DNA from wild-type and cin5-1 plants, confirming that the DNA flanking the T-DNA insertion had been cloned (not shown). The cloned DNA was sequenced, and a database search showed that the T-DNA had inserted 608 bp upstream of the start of transcription of the ACS5 gene (42). This position suggests that the T-DNA insertion disrupts expression of ACS5 mRNA, which is consistent with the finding that no ACS5 transcript is detected in RNA isolated from cin5-1 seedlings (Fig. 3B).
Figure 3.
Expression of ACS5 mRNA in wild-type and mutant lines. (A) Time course of ACS5 mRNA accumulation. Intact 3-day-old etiolated seedlings were incubated in 5 μM benzyladenine for the times indicated; total RNA was isolated and analyzed by Northern blotting. The blot was hybridized with an ACS5 probe, stripped, and rehybridized with a β-tubulin probe as a loading control. Hybridization intensities were quantified with a PhosphorImager (Molecular Dynamics). The level of ACS5 expression was normalized to that of β-tubulin and plotted. (Inset) The original images of the blot. (B) Northern analysis of RNA from variously treated seedlings. Poly(A)+ RNA was prepared from 3-day-old etiolated seedlings grown as follows: lane 1, acs5-2 plants grown on MS medium; lane 2, acs5-1 plants grown on MS medium or lane 3, MS medium plus 5 μM benzyladenine; lane 4, wild-type (Col) grown on MS medium; and lane 5, eto2 grown on MS medium. Note that lanes 4 and 5 are from an independent experiment.
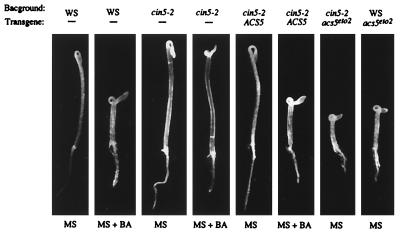
To verify that the cin5 phenotypes were caused by mutations in ACS5, we transformed cin5-2 with a 4.15-kilobase genomic clone containing the wild-type ACS5 gene. In five independent transformants, ACS5 was able to complement the seedling phenotype of cin5-2 (Fig. 4). To further confirm that cin5 disrupts ACS5, two ethyl methanesulfonate-induced cin5 alleles were sequenced (Fig. 5). cin5-2 was found to have a G to A transition that is predicted to result in a Ser to Asn substitution at amino acid 269. The cin5-3 allele has a C to T transition that is predicted to result in a Ser to Phe substitution at amino acid 201. Both of these amino acid residues are highly conserved in other ACC synthases and the Ser at position 269 corresponds to a predicted active site residue of aminotransferases (43). This analysis suggests that these mutations disrupt the catalytic activity of ACS5.
Figure 4.
Seedling phenotypes of Arabidopsis transformants containing a wild-type or eto2 ACS5 transgene. The transgene contained in each line is indicated above the seedling pictures, as is the background into which the transgene was transformed; − indicates that the line was not transformed. Seedlings were plated on either MS medium or MS medium supplemented with 5 μM BA as indicated below each picture. The seedlings were grown for 3 days in the dark, and representative seedlings were picked and photographed. Note that transformation with the eto2 version of ACS5 results in a constitutive triple-response phenotype.
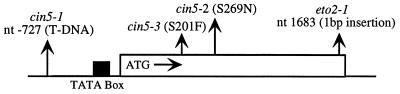
Figure 5.
Sequence alterations in the cin5 and eto2 alleles of the ACS5 gene. The putative TATA box is indicated (from ref. 42). Both indicated Ser residues are highly conserved in the aminotransferase superfamily to which ACC synthase enzymes belong. The insertion in eto2 is a C (on the coding strand) at the indicated position that is predicted to disrupt the carboxy-terminal 12 amino acids.
Role of ACS5 in Ethylene Biosynthesis.
The loss-of-function cin5 mutations allowed us to characterize the role of ACS5 in ethylene biosynthesis under various conditions. The basal level of ethylene biosynthesis is lower in etiolated cin5 mutant seedlings (13.1 ± 2.1 pl⋅seedling−1⋅day−1 for wild-type seedlings and 7.5 ± 1.4 pl⋅seedling−1⋅day−1 for cin5-1 mutant seedlings); accordingly, the hypocotyls of etiolated cin5 mutant seedlings are slightly longer than are those of wild-type seedlings (X = 5.2 ± 1.0 mm for wild-type WS etiolated seedlings and X = 8.2 ± 1.4 mm for cin5-1 etiolated seedling; P < 0.05 with Student’s t test). At low cytokinin concentrations, cin5 seedlings produce much less ethylene than do wild-type seedlings, but there is no significant difference at higher concentrations (Fig. 1). This finding suggests that ACS5 is responsible for almost all of the ethylene produced in response to low levels (<10 μM) of cytokinin, but does not play a significant role in ethylene biosynthesis at higher cytokinin concentrations. The residual response of cin5 mutants to low doses of cytokinin could be caused by a slight leakiness of our cin5 alleles, partial induction of the high dose pathway, or a minor response of other ACS isoforms.
Wild-type, 2-day-old etiolated Arabidopsis seedlings also produce high levels of ethylene in response to CuSO4, AgCl, and auxin (Table 2). Auxin has been found to increase the steady-state level of ACS4 mRNA in Arabidopsis (32). Silver ions most likely increase ethylene production by blocking negative feedback regulation (44). cin5 mutants produce wild-type levels of ethylene in response to the synthetic auxin 2,4-dichlorophenoxyacetic acid and AgCl (Table 2). cin5-1 and cin5-2, which are in the WS ecotype of Arabidopsis, produce slightly less ethylene than do wild-type seedlings in response to CuSO4 (Table 2 and not shown). However, acs5-3 (Col ecotype) produces wild-type levels of ethylene in response to CuSO4 (not shown), indicating that these two ecotypes regulate ethylene in response to CuSO4 slightly differently. An additional difference between these ecotypes is that Col seedlings produce only half as much ethylene as do WS seedlings in response to cytokinin (not shown).
Table 2.
Ethylene production by etiolated seedlings
| Treatment | Ethylene production,
pl·seedling−1·day−1
|
|
|---|---|---|
| Wild type (WS) | cin5-1 | |
| Control | 18 ± 7 | 12 ± 2 |
| H2O | 21 ± 6 | 14 ± 8 |
| AgCl (17 μg/ml) | 46 ± 8 | 48 ± 7 |
| 2,4-D (50 μM) | 94 ± 16 | 86 ± 22 |
| CuSO4 (20 mM) | 114 ± 18 | 68 ± 10 |
Seeds were grown on MS agar for 48 h as described in Materials and Methods, and 200 μl of the indicated solution was added just before the vials were sealed. The control samples had nothing added to the vials. The vials then were incubated for an additional 24 h, and the accumulated ethylene was measured. For the silver ion treatment, seeds were sown directly on medum containing AgCl. Numbers represent the mean ± the SD based on three replicates. 2,4-D, 2,4-dichlorophenoxyacetic acid.
Ethylene biosynthesis also increases sharply during flower and fruit development in many plant species (1-3, 15), including in Arabidopsis (Table 1). Light-grown Arabidopsis seedlings also produce more ethylene than do comparable etiolated seedlings. Flowers, siliques, and leaves from cin5 mutant plants all produced wild-type levels of ethylene (Table 1), indicating that ACS5 does not play a major role in the developmental regulation of ethylene biosynthesis in these tissues. Three-day-old light-grown eto2 seedlings produce slightly more ethylene than do wild type, but not nearly the many-fold excess observed in etiolated seedlings.
Regulation of ACS5 mRNA Levels by Cytokinin.
Liang et al. (42) have found that ACS5 transcription is induced by Li+ and cycloheximide. To determine whether ACS5 gene expression is regulated by cytokinin, we examined ACS5 mRNA levels at various times after exposure to cytokinin (Fig. 3A). The steady-state level of ACS5 mRNA displayed a rapid, transient and modest (less than 2-fold) increase in response to exogenous cytokinin. Seedlings pretreated with the protein synthesis inhibitor cycloheximide displayed a similar response (not shown), indicating that protein synthesis is not required for this induction. Because the induction of ACS5 by cytokinin is transient and modest, increased ACS5 mRNA levels alone most likely do not account for the rise in ethylene production observed in response to cytokinin (the mRNA level increases less than 2-fold for 1 h, but ethylene biosynthesis increases more than 8-fold over 4 days), especially because ACC synthase proteins generally have a short half-life (45-47). Taken together, these observations indicate that increases in ACS5 mRNA most likely do not contribute significantly to the sustained rise in ethylene biosynthesis observed in response to cytokinin. We conclude that posttranscriptional modulation of ACS5 very likely plays the major role in the regulation of cytokinin-mediated ethylene biosynthesis.
The eto2 Mutation Disrupts ACS5.
Additional evidence that posttranscriptional modifications play a role in the regulation of ACS5 was provided by the analysis of the eto2 mutation. eto2 was identified as a mutant that displays a triple response in the absence of exogenous ethylene (Fig. 2) and is inherited as a single gene, dominant mutation (11). Three-day-old etiolated eto2 seedlings produce approximately 20-fold more ethylene than do wild-type seedlings (Table 1). Because eto2 and cin5 are closely linked, we sequenced the ACS5 gene from an eto2 mutant and found a single base-pair insertion 35 bp upstream from the stop codon. This frame-shift mutation is predicted to change the 12 terminal amino acids of ACS5 from RVSYTDRVPDER to PGFMDRSCT. To confirm that ethylene overproduction in the eto2 mutant is the result of this disruption of ACS5, an eto2 version of ACS5 was transformed into wild-type Arabidopsis as well as the cin5-2 mutant. Five of seven and 5/5 independent transformants with the eto2 version of ACS5 into wild type and cin5-2, respectively, displayed a constitutive triple response phenotype (Fig. 4). In contrast, 0/7 independent transformants of cin5-2 with the wild-type ACS5 displayed a constitutive triple response phenotype. Measurements of ethylene production from etiolated seedlings confirm that the WS∷ACS5eto2 lines overproduce ethylene: two independent WS∷ACS5eto2 lines produce 6-fold and 4.5-fold more ethylene than do even homozygous eto2 mutants. These data confirm that the alteration of the ACS5 carboxy-terminus results in an Eto phenotype.
In 3-day-old etiolated eto2 seedlings, the steady-state level of ACS5 mRNA is slightly lower than in wild-type seedlings (Fig. 3B). This result indicates that the increased ethylene biosynthesis observed in eto2 is not caused by an increase in ACS5 mRNA levels, but rather to an alteration in the carboxy domain of the enzyme. The decrease in the steady-state level of ACS5 mRNA in eto2 mutant seedlings may be caused by negative feedback regulation by the elevated ethylene levels in these seedlings.
Adult tissues from eto2 mutants and light-grown eto2 seedlings produce a level of ethylene that is indistinguishable from or only slightly higher than that produced by wild-type tissues (Table 1). This finding is consistent with the analysis of the loss-of-function acs5 mutations that indicates that this gene does not contribute in a significant manner to ethylene production in light-grown seedlings or various adult tissues. This may be because of a very low level of ACS5 expression in these tissues, or, alternatively, the eto2 change in the carboxy-terminus of ACS5 may not increase the function of the protein in these tissues.
DISCUSSION
By using the triple response as a genetic screen, we have identified recessive and dominant mutations in ACS5 that lead to a disruption of cytokinin-induced ethylene biosynthesis and to ethylene overproduction, respectively. Our analysis indicates that cytokinin elevates ethylene biosynthesis primarily by a posttranscriptional modification of ACS5. Furthermore, ethylene overproduction in the dominant eto2 mutation was found to be caused by a perturbation of the carboxy terminus of ACS5, indicating that this domain negatively affects ACS5 function. It is plausible that cytokinin acts by relieving the negative influence of the carboxy terminus by a modification of the ACS5 protein.
The isolation of loss-of-function ACS5 alleles allowed us to characterize the role played by ACS5 in the biosynthesis of ethylene under various conditions. ACS5 seems to be responsible for almost all of the ethylene produced in response to low levels of cytokinin by etiolated seedlings and is not responsible for any significant portion of the ethylene produced under the other conditions tested, with the possible exception of CuSO4 in WS ecotype plants. This finding, together with the studies of ACS message levels, suggests that different ACS isoforms respond to distinct regulatory inputs.
The analysis of eto2 is consistent with the idea that ACS5 is regulated posttranscriptionally. One model for the increased ethylene biosynthesis observed in eto2 seedlings is that the alteration of the carboxy terminus of ACS5 may create a hyperactive enzyme. The carboxy termini of ACC synthases are the least-conserved portions of the molecule (15, 16). This hypervariable carboxy-terminal domain has been shown to affect the activity and dimerization of tomato ACC synthase when expressed in Escherichia coli; truncation of the 46-52 carboxy-terminal amino acids resulted in a monomeric enzyme that had 4-fold higher specific activity (48). However, the magnitude of the increase in activity of ACS5 would have to be much greater than 4-fold to account for the 20-fold increase in ethylene biosynthesis observed in eto2 mutant seedlings. Additionally, we have found that the wild-type and eto2 versions of ACS5 have comparable specific activities when expressed in E. coli (Hui Li and J.J.K., unpublished observations). An alternative possibility is that the carboxy terminus of ACS5 is the target of a negative modification in vivo, perhaps phosphorylation (49), or that the wild-type carboxy terminus of ACS5 acts to destabilize the protein in vivo.
A simple model for the action of cytokinin in etiolated seedlings is that it increases the activity of the ACS5 protein, possibly by relieving the negative effect of the carboxy terminus. This work suggests that modifications of ACC synthase proteins, rather than simply altered transcription of these genes, play an important role in the regulation of ethylene biosynthesis. Consistent with this, Spanu et al. (49) found that perturbation of protein phosphorylation affects the regulation of tomato ACC synthase in response to elicitor treatment. Negative regulation of ACS function by the carboxy terminus may be a general feature of these enzymes; this domain may be a target of posttranslational modification in response to a variety of regulatory inputs.
We have demonstrated that the triple response can be used as a powerful screen to identify mutants disrupted in cytokinin signaling to ethylene biosynthesis. Cytokinin signaling has been recalcitrant to genetic analysis, possible because of its essential role in plant development or the complex nature of cytokinin responses. The genetic screen used here may overcome these difficulties, and analysis of the additional cin mutants may help to elucidate the mechanism of cytokinin perception.
Acknowledgments
We thank the Arabidopsis stock center at Ohio State University for supplying the T-DNA population and the Agrobacterial left border clones. This work was supported by U.S. Department of Agriculture Grants 95-37304-2294 and 97-01425 to J.J.K.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: BA, benzyladenine; ACC, 1-aminocyclopropane-1-carboxylic acid; Col, Columbia; MS, Murashige and Skoog salts; WS, Wassilewskija.
References
- 1.Yang S F, Hoffman N E. Annu Rev Plant Physiol. 1984;35:155–189. [Google Scholar]
- 2.Mattoo A K, Suttle J C. The Plant Hormone Ethylene. Boca Raton, FL: CRC; 1991. [Google Scholar]
- 3.Abeles F B, Morgan P W, Saltveit M E., Jr . Ethylene in Plant Biology. San Diego: Academic; 1992. [Google Scholar]
- 4.Miller C O, Skoog F, Saltza M H von, Strong F M. J Am Chem Soc. 1955;77:1392–1293. [Google Scholar]
- 5.Miller C O, Skoog F, Okomura F S, Saltza M H von, Strong F M. J Am Chem Soc. 1956;78:1345–1350. [Google Scholar]
- 6.Skoog F, Miller C O. Symp Soc Exp Bot. 1957;11:118–131. [PubMed] [Google Scholar]
- 7.Binns A N. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:173–196. [Google Scholar]
- 8.Brzobohaty B, Moore I, Palme K. Plant Mol Biol. 1994;26:1483–1497. doi: 10.1007/BF00016486. [DOI] [PubMed] [Google Scholar]
- 9.Mok D W S, Mok M C. Cytokinins: Chemistry, Activity, and Function. Boca Raton, FL: CRC; 1994. [Google Scholar]
- 10.Kieber J J. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:277–296. doi: 10.1146/annurev.arplant.48.1.277. [DOI] [PubMed] [Google Scholar]
- 11.Kieber J J, Rothenberg M, Roman G, Feldmann K A, Ecker J R. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 12.Chang C, Kwok S F, Bleecker A B, Meyerowitz E M. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 13.Schaller G E, Bleecker A B. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- 14.Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker J R. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- 15.Kende H. Ann Rev Plant Physiol Mol Biol. 1993;44:283–307. [Google Scholar]
- 16.Zarembinski T I, Theologis A. Plant Mol Biol. 1994;26:1579–1597. doi: 10.1007/BF00016491. [DOI] [PubMed] [Google Scholar]
- 17.Rottmann W H, Peter G F, Oeller P W, Keller J A, Shen N F, Nagy B P, Taylor L P, Campbell A D, Theologis A. J Mol Biol. 1991;222:937–961. doi: 10.1016/0022-2836(91)90587-v. [DOI] [PubMed] [Google Scholar]
- 18.Olson D C, White J A, Edelman L, Harkins R N, Kende H. Proc Natl Acad Sci USA. 1991;88:5340–5344. doi: 10.1073/pnas.88.12.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yip W K, Moore T, Yang S F. Proc Natl Acad Sci USA. 1992;89:2475–2479. doi: 10.1073/pnas.89.6.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lincoln J E, Campbell A D, Oetiker J, Rottmann W H, Oeller P W, Shen N F, Theologis A. J Biol Chem. 1993;268:19422–19430. [PubMed] [Google Scholar]
- 21.Van der Straeten D, Van Wiemeersch L, Goodman H M, Van Montagu M. Proc Natl Acad Sci USA. 1990;87:4859–4863. doi: 10.1073/pnas.87.12.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang X, Abel S, Keller J A, Shen N F, Theologis A. Proc Natl Acad Sci USA. 1992;89:11046–11050. doi: 10.1073/pnas.89.22.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van der Straeten D, Rodrigues-Pousada R A, Villarroel R, Hanley S, Goodman H M, van Montagu M. Proc Natl Acad Sci USA. 1992;89:9969–9973. doi: 10.1073/pnas.89.20.9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang X, Oono Y, Shen N F, Köhler C, Li K, Scolnik P A, Theologis A. Gene. 1995;167:17–24. doi: 10.1016/0378-1119(95)00694-x. [DOI] [PubMed] [Google Scholar]
- 25.Nadeau J A, Zhang X S, Nair H, O’Neill S D. Plant Physiol. 1993;103:31–39. doi: 10.1104/pp.103.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim W T, Yang S F. Planta. 1994;194:223–229. [PubMed] [Google Scholar]
- 27.Tang X, Wang H, Brandt A S, Woodson W R. Plant Mol Biol. 1993;23:1151–1164. doi: 10.1007/BF00042349. [DOI] [PubMed] [Google Scholar]
- 28.Barry C S, Blume B, Bouzayen M, Cooper W, Hamilton A J, Grierson D. Plant J. 1996;9:525–535. doi: 10.1046/j.1365-313x.1996.09040525.x. [DOI] [PubMed] [Google Scholar]
- 29.Tang X, Gomes A M T R, Bhatia A, Woodson W R. Plant Cell. 1994;6:1227–1239. doi: 10.1105/tpc.6.9.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lasserre E, Bouquin T, Hernandez J A, Bull J, Pech J C, Balague C. Mol Gen Genet. 1996;251:81–90. doi: 10.1007/BF02174348. [DOI] [PubMed] [Google Scholar]
- 31.Botella J R, Arteca J M, Schlagnhaufer C D, Arteca R N, Phillips A T. Plant Mol Biol. 1992;20:425–436. doi: 10.1007/BF00040602. [DOI] [PubMed] [Google Scholar]
- 32.Abel S, Nguyen M D, Chow W, Theologis A. J Biol Chem. 1995;270:19093–19099. doi: 10.1074/jbc.270.32.19093. [DOI] [PubMed] [Google Scholar]
- 33.Cary A J, Liu W, Howell S H. Plant Physiol. 1995;107:1075–1082. doi: 10.1104/pp.107.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshii H, Imaseki H. Plant Cell Physiol. 1981;22:369–379. [Google Scholar]
- 35.Feldman K A. Plant J. 1991;1:71–82. [Google Scholar]
- 36.Edwards K, Johnstone C, Thompson C. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bell C J, Ecker J R. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 38.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1996. [Google Scholar]
- 39.Jefferson R A, Kavanagh T A, Bevan M W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bechtold N, Ellis J, Pelletier G. C R Acad Sci. 1993;316:1194–1199. [Google Scholar]
- 41.Neljubow D N. Pflanzen Beih Bot Zentralbl. 1901;10:128–139. [Google Scholar]
- 42.Liang X, Shen N F, Theologis A. Plant J. 1996;10:1027–1036. doi: 10.1046/j.1365-313x.1996.10061027.x. [DOI] [PubMed] [Google Scholar]
- 43.Sung M-H, Tanizawa K, Tanaka H, Kuramitsu S, Kagamiyama H, Hirotsu K, Okamoto A, Higuchi T, Soda K. J Biol Chem. 1991;266:2567–2572. [PubMed] [Google Scholar]
- 44.Guzman P, Ecker J R. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kende H, Boller T. Planta. 1981;151:476–481. doi: 10.1007/BF00386542. [DOI] [PubMed] [Google Scholar]
- 46.Yoshii H, Imaseki H. Arch Biochem Biophys. 1982;189:280–286. [Google Scholar]
- 47.Spanu P, Felix G, Boller T. Plant Physiol. 1990;93:1482–1585. doi: 10.1104/pp.93.4.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li N, Mattoo A K. J Biol Chem. 1994;269:6908–6917. [PubMed] [Google Scholar]
- 49.Spanu P, Grosskopf D G, Felix G, Boller T. Plant Physiol. 1994;106:529–535. doi: 10.1104/pp.106.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]