Abstract
Microorganisms express multidrug resistance pumps (MDRs) that can confound antibiotic discovery. We propose the use of mutants deficient in MDRs to overcome this problem. Sensitivity to quinolones and to amphipathic cations (norfloxacin, benzalkonium chloride, cetrimide, pentamidine, etc.) was increased 5- to 30-fold in a Staphylococcus aureus mutant with a disrupted chromosomal copy of the NorA MDR. NorA was required both for increased sensitivity to drugs in the presence of an MDR inhibitor and for increased rate of cation efflux. This requirement suggests that NorA is the major MDR protecting S. aureus from the antimicrobials studied. A 15- to 60-fold increase in sensitivity to antimicrobials also was observed in wild-type cells at an alkaline pH that favors accumulation of cations and weak bases. This effect was synergistic with a norA mutation, resulting in an increase up to 1,000-fold in sensitivity to antimicrobials. The usefulness of applying MDR mutants for natural product screening was demonstrated further by increased sensitivity of the norA− strain to plant alkaloid antimicrobials, which might be natural MDR substrates.
Keywords: antimicrobials, Staphylococcus aureus, NorA, plant alkaloids
Inhibition of target cell growth is a traditional approach for screening natural antimicrobials. This method has yielded all of the natural antibiotics currently in use. New antibiotics are continued to be discovered at a rate of >500/year (1), but these almost invariably belong to previously identified classes of compounds, making it likely that pathogens will be able to rapidly build up resistance to these “new” drugs. The increasing threat of drug-resistant pathogens is causing a renewed interest in the discovery of novel antibiotics. A current trend is to identify essential genes/proteins that are not targets for known antibiotics (e.g., the bacterial cell division protein FtsZ) and use the purified proteins to search for potent binding ligands from natural sources or combinatorial libraries (2).
A different strategy is to use traditional, broad-based whole cell screening but increase the probability of finding new antibiotics by gathering producing organisms from untapped environments, such as the ocean (1). A general problem facing both in vitro and whole-cell screening is that the ambient concentration of potential drugs is prone to be low. The problem with screening using whole cells is exacerbated further by numerous multidrug pumps found in all bacteria and yeast studied so far (3–6). In Escherichia coli, for example, there are at least 10 multidrug resistance pumps (MDRs) with overlapping specificities that can protect against most antibiotics, both natural and synthetic (3, 4). During therapy, antibiotics are administered routinely at high concentrations that are sufficient to overcome the action of MDR pumps (notable exceptions exist, such as MDR-dependent drug resistance in Pseudomonas aeruginosa). However, extrusion of antibiotics present at low concentrations in the process of screening will inevitably diminish the chances of discovery. The use of MDR− mutants has the potential for improving the sensitivity of antimicrobial discovery.
Staphylococcus aureus is an important human pathogen, and many antibiotic-resistant strains have been isolated (7). The NorA MDR pump of S. aureus protects the cells from norfloxacin and a number of amphipathic cations, such as the common disinfectants benzalkonium chloride and cetrimide (8, 9). We find that a mutant of S. aureus with a knockout in the norA gene coding for the MDR pump has a substantially increased sensitivity to a large number of antimicrobials, including therapeutically significant compounds. We also find that increasing the pH of the medium that favors accumulation of basic substances in the cell acts synergistically with the norA mutation, leading to an increase in sensitivity of up to 1,000-fold. This approach may offer a significantly higher sensitivity of drug detection compared with standard in vivo screening.
MATERIALS AND METHODS
Strains and Growth Conditions.
S. aureus RN 4222 referred to as wild type served as the parent strain to generate mutant strains described in this study. Cells were cultured in tryptic soy broth medium (Difco) [except for minimal inhibitory concentration (MIC) determination, described below] with aeration at 37°C overnight.
Disruption of the norA Gene.
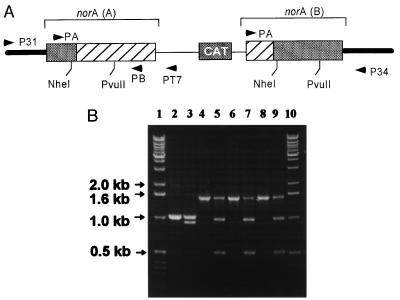
The chloramphenicol acetyltransferase gene was isolated by digesting plasmid pLI50 (10) with Sau3A (New England Biolabs). The chloramphenicol acetyltransferase gene was cloned into the BamHI site of plasmid pSP72 (Promega) to yield plasmid PCH22. Chromosomal DNA from S. aureus RN4220 was isolated according to the protocol of Qiagen (Chatsworth, CA). A 1,050-bp part of the norA gene lacking a 80-bp N terminus and a 20-bp C terminus was PCR-amplified from chromosomal DNA by using primer A: 5′-GCTCTATGTTGC-TTTTCAATT and primer B: 5′-CTGTTTATTTAAAAGATTTGGG. This fragment was cloned into the EcoRI/EcoRV site of PCH22 to yield plasmid PCH23. S. aureus were transformed by PCH23 by using electroporation (11). Cells were plated on trypticase soy agar with chloramphenicol. A transformant colony was picked after 24 h of incubation at 37°C, regrown, and plated on CAM medium, and a large colony was picked for further work. Primers P31 (5′-AACGTCATCACATGCACCAA), PT7 (5′-TAATA-CGACTCACTATAGGG), and P34 (5′-AATTAGGTATGTGGATTGCAA-3′) (Fig. 1) were used for PCR amplification to examine the disruption of the norA gene.
Figure 1.
Disruption of the chromosomal norA gene by homologous recombination. (A) The shaded bars represent the N-terminal and C-terminal portions of norA originating from the chromosome, and the hatched boxes represent portions of the norA sequence originating from integration of plasmid PCH23 lacking N- and C-termini. The resultant norA(A) lacks a C terminus and the resultant norA(B) lacks an N terminus. Primer P31 corresponds to a sequence 450 bp upstream the norA gene. Primer PT7 corresponds to the T7 promoter sequence of plasmid PCH23. (B) Using primers P31 and PT7, a 1.5-kb fragment was expected to be amplified if norA was disrupted in a manner shown in A. Lanes: 1 and 10, DNA markers; 2, PCR product amplified with primers A and B from chromosomal DNA of wild-type cells (positive control, expected size, 1 kb); 3, DNA from lane 2 digested incompletely by NheI; 4, 6, and 8, PCR products using P31 and PT7 primers and chromosomal DNA from KLE 820, KLE 821, and KLE 822, three independent putative norA disruption clones, respectively. There was no product with primers P31 and PT7 using wild-type chromosomal DNA (not shown). Lanes 5, 7, and 9, PCR products of lanes 4, 6, and 8 digested incompletely with NheI. The structure of the construct was verified further by obtaining an expected 1.4-Kb PCR product with primers PA and P34. Primers P31 and P34 produced an expected 1.7-Kb PCR product from the wild type but not from KLE 820 (not shown).
Efflux of Ethidium Bromide (EtBr).
EtBr has high fluorescence when bound to DNA, making it possible to measure efflux from the cell. Cells from an overnight culture (1 ml, OD600 = 4) were pelleted and washed twice with 20 mM Hepes/NaOH (pH 7.0) buffer. Cells then were resuspended in 1 ml of Hepes buffer containing 10 μM carbonylcyanide m-chlorophenylhydrazone and 10 μg of ethidium bromide followed by incubation at 37°C for 30 min (12). The cells were centrifuged, washed, and resuspended at 0.13 OD600 in ice-cold Hepes buffer, and fluorescence was measured with a Perkin–Elmer LS-5B luminescence spectrometer at 530-nm excitation with 600-nm emission wavelengths.
Determination of MICs.
MICs were determined by serial two-fold dilution of antimicrobials in Mueller–Hinton broth containing chloramphenicol (34 μg/ml). Most antimicrobials were from Sigma, and ciprofloxacin was from Bayer, Elkhart, CT. Around 105 cells were inoculated in 1 ml of medium for the test. MICs were determined as a concentration of an antimicrobial that completely prevented cell growth during an 18-h incubation at 37°C with aeration. The IC50 was determined by growing cells in microtiter plate wells and reading the OD after overnight growth at 650 nm. For experiments performed at pH 9.0, the pH of the growth medium was adjusted with NaOH. MIC was determined in samples that showed no bacterial growth, and in these samples there were no detectable changes in pH by the end of the experiment. MIC determinations in the strain carrying a norA gene disrupted with a chloramphenicol resistance cassette were performed in the presence of chloramphenicol to inhibit proliferation of possible cells that have lost the disrupted norA construct.
RESULTS
Sensitivity of the norA− Mutant to Antimicrobial Agents.
Disruption of the chromosomal copy of the norA gene was performed by replacement with a norA sequence interrupted by a chloramphenicol acetyltransferase cassette (see Materials and Methods). The disrupted chromosomal sequence was analyzed by PCR by using a primer immediately outside of the norA gene and a primer specific for the disrupted sequence. A PCR product of the predicted size (1.5 kb) and composition was obtained from the norA− but not from the wild-type strain, confirming that the chromosomal copy of norA was disrupted (Fig. 1).
Table 1 shows the susceptibility of the norA mutant toward a range of antimicrobials. Using either MIC or IC50 to calculate the sensitivity of the norA− strain relative to the wild type gave similar results. Increased resistance to norfloxacin and amphipathic cations of strains overproducing NorA has been described (9), and a recent study reported increased sensitivity of norA− to quinolones (13). Our data show that amphipathic cations are generally better substrates of the pump than are quinolones. Many other tested antimicrobials do not appear to be substrates of the pump (Table 1).
Table 1.
NorA dependence of antimicrobial resistance as measured by MIC
| RN 4222 (WT) | KLE 820 (norA−) | Sensitivity* | |||
|---|---|---|---|---|---|
| Drugs | −R | +R | −R | +R | |
| EtBr | 5 | 1.25 | 0.33 | 0.33 | 16 |
| TPP | 15.6 | 7.8 | 1.9 | 1.9 | 8 |
| Acriflavine | 20 | 10 | 2.5 | 2.5 | 8 |
| Norfloxacin | 1.25 | 0.67 | 0.33 | 0.33 | 4 |
| Ciprofloxacin | 0.67 | 0.33 | 0.16 | 0.16 | 4 |
| Kanamycin | 0.33 | ND | 0.33 | ND | 1 |
| Erythromycin | 0.195 | ND | 0.195 | ND | 1 |
| Gentamycin | 0.62 | ND | 0.62 | ND | 1 |
| Tetracycline | 0.097 | ND | 0.097 | ND | 1 |
| Cephalothin | 0.15 | ND | 0.15 | ND | 1 |
| Lincomycin | 0.6 | ND | 0.6 | ND | 1 |
| Vancomycin | 1.5 | ND | 1.5 | ND | 1 |
| Clindamycin | 0.047 | ND | 0.047 | ND | 1 |
MIC in micrograms per milliliter. TPP, tetraphenylphosphonium bromide; ND, not determined; −R, no reserpine; +R, 20 μg/ml reserpine.
Sensitivity of norA− compared with the wild type in the absence of reserpine.
Testing for Additional MDRs in S.
aureus. We wished to know whether additional MDR pumps took part in the quinolone and amphipathic cation resistance. Identifying and disrupting these additional MDRs would further improve the sensitivity of the strain and would be useful for drug discovery. A simple way to check the presence of MDR activity is to use an MDR inhibitor. Reserpine is a broad range MDR inhibitor that acts on diverse MDRs from NorA (major facilitator family of translocases powered by the proton motive force) to human P-glycoprotein (belonging to the ABC ATP binding cassette family of ATP-dependent translocases) (14). Addition of reserpine increased the sensitivity of the wild type to drugs in accordance with prior observations (8) but had virtually no effect on the norA mutant (Table 1). The mutant strain was consistently more sensitive than the wild type treated with reserpine, suggesting that inhibition by reserpine was incomplete. It appears that NorA is the principal S. aureus MDR pump, responsible for the resistance to the chemically diverse compounds we tested.
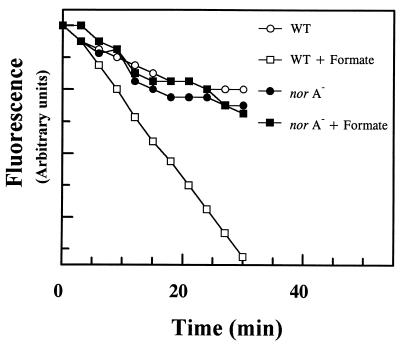
To examine this more directly, efflux of EtBr from wild-type and norA− cells was measured (Fig. 2). Cells were deenergized with an uncoupler, loaded with EtBr, washed, and diluted into a buffer medium. The rate of EtBr efflux was significantly greater in the wild type in the presence of a respiratory substrate formate compared with the norA− mutant. Addition of formate did not increase the rate of EtBr efflux from the norA cells. This result suggests that NorA is indeed the primary route for energy-dependent EtBr efflux.
Figure 2.
Efflux of EtBr from wild-type and norA− strains of S. aureus. Cells were deenergized with uncoupler and loaded with EtBr as described in Materials and Methods. The graph depicts the loss of EtBr from cells resuspended in buffer without EtBr and uncoupler. Fluorescence of EtBr is expressed in arbitrary units.
Synergy Between a norA Mutation and Alkaline pH.
It seemed possible to further increase the sensitivity toward amphipathic cations by increasing the pH of the medium. Amphipathic cations, including the ones we tested, come in two major groups—strong cations like quaternary ammonium compounds and imino-substances and weak bases such as pentamidine or chlorhexidine (Fig. 3). The cytoplasmic pH of the bacterial cell is maintained near 7.5 when the external pH is in the range 5–9 (15). At alkaline pH, an electrogenic Na+/nH+-antiporter leads to the acidification (relative to the external medium) of the cytoplasm to maintain the pH homeostasis (15). Hence, at pH 9, the pH gradient across the membrane is inverted (Fig. 4). When the pH gradient is inverted, there is a concomitant compensatory increase in the membrane potential (15). The elevated membrane potential will lead to an increased accumulation of such permeant cations as tetraphenylphosphonium and quaternary ammonium compounds. The inverted pH gradient also will act to increase the accumulation of weak bases. The permeant form is the unprotonated species (16), and with a typical pK of 9 for an amino group, this means that increasing pH from 7 to 9 will increase the intracellular concentration of the permeant species 50-fold.
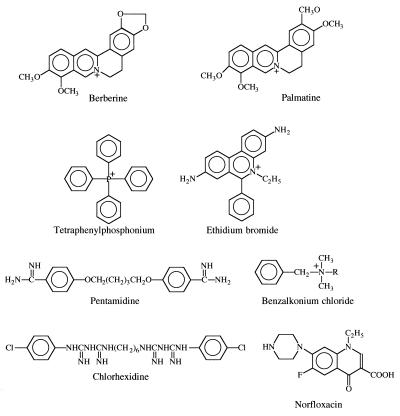
Figure 3.
Structural formulas of NorA substrates. Strong amphipathic cations are indicated by their positive charge. Berberine and palmatine are plant antimicrobial alkaloids. Tetraphenyl phosphonium has been used as a probe to measure the membrane potential. EtBr is used as an antiprotozoal (veterinary). Benzalkonium chloride is an antiseptic and disinfectant. Weak bases: Pentamidine is a systemic antiprotozoan, and chlorhexidine is an antiseptic. Norfloxacin is a quinolone DNA topoisomerase inhibitor and a systemic antibiotic.
Figure 4.
A model of increased accumulation of amphipathic cations. (A) Wild type, pH 7. Strong amphipathic cations are accumulated in the cell driven by the membrane potential (inside negative). Weak bases are extruded from the cell along the pH gradient. Both species are extruded by NorA that acts as a drug/proton antiporter. (B) norA mutant, pH 9. The pH gradient inverts, leading to a compensatory increase in the membrane potential and increased accumulation of strong permeant cations. Inversion of the pH gradient also inverts the direction of the flux of weak bases that become accumulated in the cell.
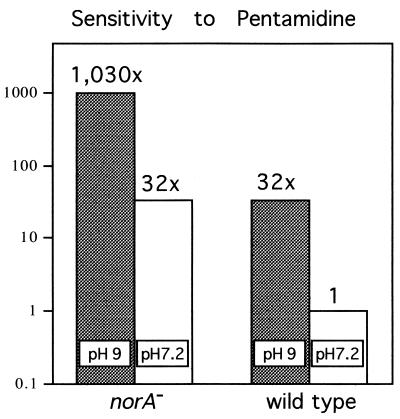
At pH 9, S. aureus grew at ≈70% of the growth rate at pH 7.2 (not shown). The sensitivity to all amphipathic cations increased significantly at pH 9 (Table 2). For example, sensitivity to pentamidine increased ≈30-fold at pH 9 in the wild type. The NorA pump is electrogenic (17) and continues to protect the cell at alkaline pH. A norA mutation also lead to a 30-fold increase in the sensitivity to pentamidine (Fig. 5). When combined, pH 9 and the norA mutation provided a strong synergistic effect, increasing sensitivity by an impressive 1,000-fold. This effect decreased the MIC for pentamidine to 10 ng/ml. Puromycin is a weak base and apparently not a substrate of NorA. Puromycin potency increased 8-fold at higher pH, but a combination of high pH and a norA mutation did not further increase its antimicrobial action. Analogously, sensitivity to norfloxacin [that is predominantly neutral or anionic (16)] only was affected by a norA mutation but not by increase in pH.
Table 2.
Synergistic effect of high pH and a norA mutation
| Drugs | Sensitivity
|
||||
|---|---|---|---|---|---|
| RN4222 (WT)
|
KLE 820 (norA−)
|
norA− at pH 9 vs. WT at pH 7.2 | |||
| pH 7.2 | pH 9 | pH 7.2 | pH 9 | ||
| Pentamidine | 10 | 0.3125 | 0.3125 | 0.0097 | 1,030 |
| EtBr | 5 | 0.078 | 0.3125 | 0.01 | 500 |
| Berberine | 60 | 3.75 | 7.5 | 0.23 | 260 |
| Palmatine | 200 | 12.5 | 50 | 0.78 | 256 |
| TPP | 15.6 | 0.48 | 1.98 | 0.12 | 130 |
| Benzalkonium chloride | 0.67 | 0.08 | 0.16 | 0.0104 | 64 |
| Chlorhexidine | 0.5 | 0.06 | 0.125 | 0.0156 | 32 |
| Puromycin | 20 | 2.5 | 20 | 2.5 | 8 |
| Norfloxacin | 1.25 | 1.25 | 0.33 | 0.33 | 4 |
MIC in micrograms per milliliter. TPP, tetraphenylphosphonium bromide.
Figure 5.
Synergistic action of high pH and pentamidine. Data are taken from Table 2. MIC of the wild type at pH 7.2 is divided by the MIC of the other samples to produce an index of sensitivity. pH 9 and a norA mutation equally contribute to increased sensitivity in this case, and a combination of both produces a synergistic effect.
Among the substances we tested for antimicrobial activity were two plant alkaloids, berberine and palmatine, which are strong amphipathic cations (Fig. 3). Both appeared to be substrates of the NorA pump. It is possible that these alkaloids are natural MDR substrates, and it seems that artificial amphipathic cations were found to be MDR substrates because of their resemblance to these natural substrates. Sensitivity to both alkaloids was increased further at pH 9. These observations validate the idea of using MDR mutants and high pH media for discovery of natural antimicrobials.
DISCUSSION
Disruption of the norA gene coding for a pmf-dependent MDR pump increased the sensitivity of S. aureus 5- to 30-fold to NorA substrates, primarily amphipathic cations. Reserpine, a general inhibitor of MDRs, failed to further increase sensitivity of norA− cells to these antimicrobials. The rate of efflux of EtBr from deenergized cells was stimulated by a respiratory chain substrate in wild-type, but not in norA− cells. Taken together, these observations strongly suggest that NorA is the major multidrug pump responsible for efflux of amphipathic cations from S. aureus. These substances (Fig. 3) are also good substrates for a majority of other MDR pumps from Gram-positive organisms. In Gram-negative bacteria, the known repertoire of MDR pumps includes neutral compounds [the EmrAB pump (18)] as well as amphipathic cations and anions [the AcrAB pump (19)]. Experimental data and DNA analysis from whole genome sequencing projects suggest that several different MDRs are present in a given microbial species. This number varies from a putative high of 25 in the yeast S. cerevisiae to a low of 4 in the parasite H. influenzae (4). It is noteworthy that H. influenzae has a “bare-bones” genome two times smaller than that of E. coli and does not have a complete Krebs cycle (20) yet has four different MDR pumps that seem to be important for survival. Without access to the complete S. aureus genomic sequence, it is hard to tell whether NorA is the only MDR pump. There may be additional MDRs that require specific inducers, for example. The B. subtilis MDR pump BMR (21) and the EmrAB pump of E. coli (22) are induced by their substrates. At the same time, two other B. subtilis MDRs, BLT (23) and BMRIII (24), are not expressed under normal growth conditions or in the presence of known MDR substrates.
It is interesting to note that, although amphipathic cations are “favorite” substrates of many MDR pumps, few natural antimicrobials of this type are known. It is not surprising that weak bases are rarely antimicrobial because these compounds will be excluded from the cell by the pH gradient when the external pH is <7.5. Discovered initially as weak antimicrobials, many of these substances (doxorubicin, mytomicin) are now used as antineoplastic agents. Strong amphipathic cations, on the other hand, will accumulate in the cell in accordance with the membrane potential and would be expected to be found as antibiotics more frequently. Yet, the only currently known examples come from a small group of plant alkaloids (Fig. 3). Perhaps it is precisely the ubiquity of MDR pumps among the test strains that is responsible for the underdetection of these compounds in the process of drug discovery.
We have recently tested the feasibility of an E. coli strain defective in two major MDR pumps for drug screening (S.A.S., D.D., and K.L., unpublished work). Ideally, one would want to design a test strain completely devoid of MDR pumps that would potentially improve the chances of discovering a wide variety of antimicrobials. With the availability of whole genome sequences for a number of bacterial species, this has become a realistic project. Once identified with the aid of MDR mutants, new antimicrobials could be deployed effectively either in combination with an MDR inhibitor or simply at a concentration sufficiently high to overcome the pump. Other possibilities would include chemical modification of the newly discovered drugs to increase their potency or to decrease their affinity to MDRs or both.
We took advantage of the accumulation of amphipathic cations in the cell at high pH to further increase cell sensitivity to these antimicrobials. A norA mutation and high pH acted synergistically and increased the sensitivity to pentamidine, for example, 1,000-fold. Our findings of an MDR−/high pH synergy will be useful for the discovery of amphipathic cationic antimicrobials. It seems that sensitivity of an antimicrobial screen could be increased further with the use of alkiliphilic bacteria that can grow at pH 10.5 and maintain a high inverted pH gradient (25). A somewhat similar strategy can be used in the discovery of weak acids that accumulate in the cell using the pH gradient. Weak acid antimicrobials have been used in food preservation (salicylate, benzoate) and as antineoplastic agents (mycophenolic acid, nogalamycin). Decreasing the pH of the medium and using acidophilic bacteria in combination with MDR mutations or MDR inhibitors should significantly increase the sensitivity of discovery of these compounds as well.
Acknowledgments
We thank Dr. John Tomashek and Dr. Frank R. Stermitz for helpful discussions and critical reading of the manuscript. S. aureus strains were provided kindly by Dr. C. Y. Lee. This study was supported by Phytera, Inc., Worcester, MA, and by National Science Foundation Grant MCB-9496291 and National Institutes of Health Grant RO1 GM54412 to K.L.
ABBREVIATIONS
- MIC
minimal inhibitory concentration
- EtBr
ethidium bromide
- MDR
multidrug resistance pump
References
- 1.Strohl W R. In: Biotechnology of Antibiotics. Strohl W R, editor. New York: Marcel Dekker; 1997. pp. 1–47. [Google Scholar]
- 2.Desnottes J F. Trends Biotechnol. 1996;14:134–140. doi: 10.1016/0167-7799(96)10015-9. [DOI] [PubMed] [Google Scholar]
- 3.Lewis K. Trends Biochem Sci. 1994;19:119–123. doi: 10.1016/0968-0004(94)90204-6. [DOI] [PubMed] [Google Scholar]
- 4.Paulsen I T, Brown M H, Skurray R A. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolhuis H, van Veen H W, Poolman B, Driessen A J M, Konings W N. FEMS Microbiol Rev. 1997;21:55–84. doi: 10.1111/j.1574-6976.1997.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 6.Lewis K, Hooper D, Ouellette M. ASM News. 1997;63:605–610. [Google Scholar]
- 7.Kluytmans J, van Belkum A, Verbrugh H. Clin Microbiol Rev. 1997;10:505–520. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neyfakh A A, Borsch C M, Kaatz G W. Antimicrob Agents Chemother. 1993;37:128–129. doi: 10.1128/aac.37.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng E Y, Trucksis M, Hooper D C. Antimicrob Agents Chemother. 1994;38:1345–1355. doi: 10.1128/aac.38.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray O, Chang S. J Bacteriol. 1981;145:422–428. doi: 10.1128/jb.145.1.422-428.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schenk S, Laddaga R A. FEMS Microbiol Lett. 1992;94:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 12.Tennent J M, Lyon B R, Midgley M, Jones I G, Purewal A S, Skurray R A. J Gen Microbiol. 1989;135:1–10. doi: 10.1099/00221287-135-1-1. [DOI] [PubMed] [Google Scholar]
- 13.Yamada H, Kurose-Hamada S, Fukuda Y, Mitsuyama J, Takahata M, Minami S, Watanabe Y, Narita H. Antimicrob Agents Chemother. 1997;41:2308–2309. doi: 10.1128/aac.41.10.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neyfakh A A, Bidnenko V, Chen L B. Proc Natl Acad Sci USA. 1991;88:4781–4785. doi: 10.1073/pnas.88.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padan E, Schuldiner S. Biochim Biophys Acta. 1994;1187:206–210. doi: 10.1016/0005-2728(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 16.Nikaido H, Thanassi D G. Antimicrob Agents Chemother. 1993;37:1393–1399. doi: 10.1128/aac.37.7.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grinius L L, Siehnel R J, Morris C N. FASEB J. 1997;11:958. [Google Scholar]
- 18.Lomovskaya O, Lewis K. Proc Natl Acad Sci USA. 1992;89:8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleischmann I D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed M, Borsch C M, Taylor S S, Vazquez-Laslop N, Neyfakh A A. J Biol Chem. 1994;269:28506–28513. [PubMed] [Google Scholar]
- 22.Lomovskaya O, Lewis K, Matin A. J Bacteriol. 1995;177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed M, Lyass L, Markham P N, Taylor S S, Vazquez-Laslop N, Neyfakh A A. J Bacteriol. 1995;177:3904–3910. doi: 10.1128/jb.177.14.3904-3910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohki R, Murata M. J Bacteriol. 1997;179:1423–1427. doi: 10.1128/jb.179.4.1423-1427.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krulwich T A, Ito M, Gilmour R, Sturr M G, Guffanti A A, Hicks D B. Biochim Biophys Acta. 1996;1275:21–26. doi: 10.1016/0005-2728(96)00044-8. [DOI] [PubMed] [Google Scholar]