Abstract
Subunits a and c of Fo are thought to cooperatively catalyze proton translocation during ATP synthesis by the Escherichia coli F1Fo ATP synthase. Optimizing mutations in subunit a at residues A217, I221, and L224 improves the partial function of the cA24D/cD61G double mutant and, on this basis, these three residues were proposed to lie on one face of a transmembrane helix of subunit a, which then interacted with the transmembrane helix of subunit c anchoring the essential aspartyl group. To test this model, in the present work Cys residues were introduced into the second transmembrane helix of subunit c and the predicted fourth transmembrane helix of subunit a. After treating the membrane vesicles of these mutants with Cu(1,10-phenanthroline)2SO4 at 0°, 10°, or 20°C, strong a–c dimer formation was observed at all three temperatures in membranes of 7 of the 65 double mutants constructed, i.e., in the aS207C/cI55C, aN214C/cA62C, aN214C/cM65C, aI221C/cG69C, aI223C/cL72C, aL224C/cY73C, and aI225C/cY73C double mutant proteins. The pattern of cross-linking aligns the helices in a parallel fashion over a span of 19 residues with the aN214C residue lying close to the cA62C and cM65C residues in the middle of the membrane. Lesser a–c dimer formation was observed in nine other double mutants after treatment at 20°C in a pattern generally supporting that indicated by the seven landmark residues cited above. Cross-link formation was not observed between helix-1 of subunit c and helix-4 of subunit a in 19 additional combinations of doubly Cys-substituted proteins. These results provide direct chemical evidence that helix-2 of subunit c and helix-4 of subunit a pack close enough to each other in the membrane to interact during function. The proximity of helices supports the possibility of an interaction between Arg210 in helix-4 of subunit a and Asp61 in helix-2 of subunit c during proton translocation, as has been suggested previously.
During oxidative phosphorylation, F1Fo–ATP synthases couple H+ transport to the synthesis of ATP from ADP and Pi using the energy of a transmembrane H+ electrochemical gradient. These enzymes are found embedded in the inner membranes of mitochondria and bacteria and in the thylakoid membrane of chloroplasts, and consist of two distinct sectors termed F1 and Fo (1–4). The F1 sector of the enzyme contains the catalytic sites for ATP synthesis and extends from the surface of the membrane via a narrow stalk. It is easily removed from the membrane as a water-soluble complex which has ATPase activity. The Fo sector extends through the membrane and, on removal of F1, mediates passive H+ translocation. When F1 is bound to Fo in the membrane, the complex acts as a reversible, H+-transporting ATP synthase or ATPase. In Escherichia coli, F1 has five subunits in an α3β3γδɛ ratio, and Fo consists of three different types of subunits with an a1b2c9–12 stoichiometry (4).
The atomic resolution structure of the majority of the F1 part of mitochondrial F1Fo–ATPase gave new insights into the mechanism of cooperative ATP synthesis (5), and provided a structural framework for novel experiments and interpretation (6). For example, the hypothesis of subunit γ rotation during catalysis is now independently supported by several different types of experiments that relied upon the new structural information (7–9). Intermolecular cross-linking experiments between subunits of F1 (10–13) and between F1 subunits ɛ and γ and subunit c of Fo (12, 14, 15) provide information on a possible pathway of conformational changes from the site of proton translocation to the site of ATP synthesis.
The function of the three subunits of the E. coli Fo complex are incompletely understood, but all are necessary for the reconstitution of proton translocation function (16). Subunit b has a large polar cytoplasmic region and one transmembrane domain which is anchored in the membrane (4). The cytoplasmic domain is essential for F1 binding (17). Subunit c is a protein of 79 amino acid residues which folds in a hairpin-like structure with two membrane spanning α-helices linked by a polar loop at the F1-binding side of the membrane (3, 4). The carboxyl group of Asp61, which is centered in the second transmembrane helix, is the site of H+ binding and release during proton transport and the site of reaction with dicyclohexylcarbodiimide. Subunit a is a very hydrophobic protein of 271 amino acid residues, the membrane topology of which has been unclear. We now favor a five-transmembrane helix model, similar to that proposed by Hatch et al. (18), based on the side-dependent accessibility of Cys introduced into extramembrane loops (19). Subunits a and c of Fo are thought to function together in ATPase-coupled H+ transport by F1Fo (2–4). Three suppressor mutations which optimize the function of the A24D/D61G double mutant of subunit c cluster on a single face of the fourth of five transmembrane helices, i.e., at residues Ala217, Ile221, and Leu224, and the putative helix-4 was suggested as a possible site of interaction with subunit c (20). Arg210 also lies on the fourth transmembrane helix and is thought to play a key role in ATPase-coupled H+ transport (18, 21), perhaps by interacting with Asp61 of subunit c (2–4). In this paper, we report the first direct physical evidence for the interaction of these two functionally important transmembrane helices, i.e., the fourth helix of subunit a and the C-terminal helix of subunit c. The neighboring residues were revealed by the introduction of Cys into both helices and disulfide bridge formation between the two subunits.
EXPERIMENTAL PROCEDURES
Materials and General Methods.
Strain MM180 (pyrE41, entA403, argH1, rpsL109, supE44) and strain MJM63 (pyrE41, entA403, argH1, rpsL109, supE44, ΔuncE334, ilv∷Tn10) are described elsewhere (22, 23). DNA polymerase, DNA ligase, and restriction enzymes were purchased from New England Biolabs or Promega Corp. DNA fragments were separated on an agarose gel and extracted with the GeneClean Kit (Bio101 Labs). Double-stranded DNAs were sequenced with the dideoxynucleotide termination method with a fmol DNA Cycle Sequencing System (Promega Corp.) using deoxyadenosine 5′-[α-[35S]thio]triphosphate (Amersham Corp.). Oligonucleotides were synthesized at the University of Wisconsin Biotechnology Center (Madison, WI). Plasmid transformation was carried out as described by Inoue et al. (24).
Construction of Cys Substitutions.
All plasmids carrying Cys mutations are derivatives of plasmid pDF163 (25), which carries wild-type uncBEFH genes (nucleotides 870-3216)‡. The single Cys mutations on uncB or uncE were initially generated by PCR oligonucleotide-directed mutagenesis (27) with oligonucleotides carrying the appropriate substitutions. The PCR-generated fragments with Cys substitutions in subunit a were cloned between the PstI/BsrGI (1561–1911) sites in pDF163 and fragments with Cys substitutions in subunit c were cloned between the BsrGI/HpaI (1911–2162) sites in pDF163. The a plus c double Cys-substituted plasmids were constructed by combinations of the PstI/BsrGI (1561–1911) or BsrGI/HpaI (1911–2162) fragments. The presence of the mutations was verified by sequencing the entire length of subcloned double-stranded DNA through the ligation junctions. Except for the desired base changes, the sequences were identical to that of the wild-type gene.
Construction of ΔuncBEFH deletion strain JWP109.
The ΔuncBEFH deletion plasmid, pJWP102, is a derivative of plasmid pAP55 (28), which carries the whole unc operon. The ΔuncBEFH deletion (nucleotides 1012–3202, from the stop codon of uncI to the stop codon of uncH) was generated by PCR mutagenesis (27) with an antisense primer (GTTGCATGCGCCAGTCCCCTTACCCTTTGTTGTTAA), where the underlined bases denote the stop codon. The PCR fragment was cut with MfeI and SphI and cloned into these sites in plasmid pAP55, at nucleotide 458 and 3216, respectively, to generate plasmid pJWP102. The chromosomally ΔuncBEFH deleted strain JWP109 was constructed by a cartridge eviction method (21). The strain, Sac-14 (21), which carries the sacRB–nptI cartridge between two BamHI sites in the uncB gene (nucleotides 1110–1727), is sensitive to sucrose and resistant to kanamycin. Plasmid pJWP102 was transformed into Sac-14 cells, where recombination of the chromosomal unc gene with pJWP102 (ΔuncBEFH)DNA resulted in loss of the sacRB–nptI cartridge and a sucrose-resistant and kanamycin-sensitive phenotype. The ΔuncBEFH operon was transduced with P1vir into strain MJM63(23) by cotransduction with Ilv+ to give strain JWP109. The chromosomal ΔuncBEFH deletion was confirmed by size analysis and DNA sequencing of a PCR-amplified product.
Comparative Growth Studies.
The transformant colonies were transferred to minimal medium plates containing 22 mM succinate, 2 mg/liter thiamine, 0.2 mM uracil, 0.2 mM l-arginine, 0.02 mM dihydroxybenzoic acid, and 0.1 mg/ml ampicillin and incubated at 37°C with scoring for growth after 1–5 days. Growth yields were measured as described using 0.04% glucose as carbon source (21).
Membrane Preparations and Assays.
Plasmid transformants of strain JWP109 were grown in M63 minimal medium containing 0.6% glucose, 2 mg/liter thiamine, 0.2 mM uracil, 0.2 mM l-arginine, 0.02 mM dihydroxybenzoic acid, and 0.1 mg/ml ampicillin, supplemented with 10% LB medium (20), and harvested in the late exponential phase of growth. Cells were suspended in TMG buffer (50 mM Tris⋅HCl, 5 mM MgCl2, 10% glycerol, pH 7.5) containing 1 mM phenylmethylsulfonyl fluoride and 0.1 mg/ml of DNase I and disrupted by passage through a French press at 124 MPa at 4°C and membranes prepared as described (22).
Cross-Linking Catalyzed by Cu(1,10-phenanthroline)2SO4 (CuP).
In survey experiments, membrane vesicles in TMG buffer were routinely treated with 1.5 mM CuP for 60 min at room temperature (22–24°C) to catalyze disulfide bond formation. In subsequent experiments, the temperature and time of incubation were varied. The cross-linking reaction was terminated by addition of Na2EDTA to a final concentration of 15 mM and N-ethylmaleimide to a final concentration of 20 mM. After 10 min at 22–24°C, treated membrane vesicles were mixed with 0.2 volumes of 6× SDS sample buffer (350 mM Tris⋅HCl, pH 6.8, 10% SDS, 30% glycerol, 0.12 mg/ml bromphenol) and incubated at 22–24°C for 1 h; 25 mM DTT or 2% (vol/vol) 2-mercaptoethanol was included for reduction of disulfide bonds. The solubilized membrane proteins (20 μg) were electrophoresed on a 15% polyacrylamide gel using the Tris-Tricine buffer described in the study of Schägger and von Jagow (29). After electrophoresis, proteins were electrophoretically transferred onto a polyvinylidene difluoride membrane for immunoblotting (30). Rabbit antisera to subunit a (31) and subunit c (32) were preabsorbed to ΔuncB-C membranes as described (31) and diluted 1:1000 into PBS (137 mM NaCl, 6.5 mM Na2HPO4, 1.5 mM NaH2PO4) containing 0.02% NaN3 and 2% BSA before use. Immunoblots were developed with multiple exposures, using the ECL System (Amersham Corp.), and scanned within a linear range of intensity using a flat bed scanner.
Effective termination of the cross-linking reaction, before solubilization of the sample in SDS, was critical in the screening for a–c cross-links formed within the membrane. Although N-ethylmaleimide is membrane permeable, and can react with most of the sulfydryl groups located in the membrane, it was ineffective in terminating the cross-linking reaction with aI221C/cG69C membranes. The aI221C/cG69C mutant pair was one of the most reactive of the 84 pairs generated. A 10-fold molar excess of EDTA relative to CuP proved to be effective in terminating the cross-linking reaction and was routinely used with all other mutants. The a–c products in all doubly substituted Cys pairs were reduced by 2% 2-mercaptoethanol or 25 mM DTT (1 h at 22–24°C in SDS sample buffer), except the aI221C/cG69C combination which required heating at 100°C for 10 min in SDS sample buffer containing 25 mM DTT.
RESULTS
ΔuncBEFH Deletion Strain JWP109 and Its Characteristics.
To conveniently express mutant subunits of the Fo complex, we constructed a chromosomal ΔuncBEFH deletion in strain JWP109 using the cartridge eviction method (19) and complemented it with the equivalent genes expressed from derivatives of plasmid pDF163.§ Strain JWP109 cannot grow on succinate since growth depends on a functional oxidative phosphorylation system. The pDF163 transformant of strain JWP109, strain JWP111, grows nearly as well on succinate minimal medium as the chromosomal wild-type strain (Fig. 1A). The relative growth yields of the strain JWP109 and strain JWP111 on glucose minimal medium were 62% and 87%, respectively, relative to the chromosomal wild-type strain MM180.
Figure 1.
Growth (function) of Cys-substituted mutants (A) and summary of a–c cross-linking results (B). (A) Cells were plated on succinate minimal medium and the colony size (mm) was measured after incubation at 37°C for 3 days as a measure of oxidative phosphorylation function. The chromosomal, wild-type control strain showed colonies of 2 mm after 3 days at 37°C. (B) Relative yield of a–c cross-linked dimer formed between Cys at the positions indicated. Membrane vesicles were treated with CuP at 10°C for 60 min. Relative yield of the a–c cross-link product: 0, none; −/+, ≤5%; +, 6–10%; ++, 11–20%; +++, 20–40%; ++++, >40%. 0* indicates Cys pair forming no cross-link at 10°C but significant cross-link (8%) at 20°C.
Effects of Cys Substitutions on Function.
Plasmids carrying Cys substitutions in subunit a, subunit c or combinations thereof were transformed into strain JWP109. The growth of transformant strains was tested on succinate minimal medium. The Cys substitutions in subunit a had little effect on growth, with the exceptions of aL207C and aG218C, which grew considerably slower than wild type (Fig. 1A). Many of the Cys substitutions in helix-2 of subunit c resulted in a modest slowing of growth on succinate minimal medium. The cG69C, cI55C, and cY73C mutants grew very poorly or not at all. Most of the a–c double Cys substitutions grew nearly as well as one of the respective single substitutions including the aN214C/cA62C, aN214C/cM65C, and aI223C/cL72C mutants, which proved to form high yield a–c cross-links. Approximately 30% of the double substitutions grew very poorly or not at all, including the aL207C/cI55C and aI221C/cG69C, aL224C/cY73C, and aI225V/cY73C mutants, which also proved to form high yield a–c cross-links. It is noteworthy that the aL207C/cI55C double mutant grew, albeit poorly, whereas the cI55C mutant did not grow at all.
Disulfide Cross-Link Formation with CuP.
The initial screening experiments for a–c disulfide cross-link formation were done by combining substitutions of cM57C, cG58C, cL59C, cV60C, cA62C, and cA67C in the helix-2 of subunit c and aN214C, aA217C, aG218C, aL220C, aI221C, and aI223C in the helix-4 of subunit a. Among the 36 pairs, only the aN214C/cA62C double Cys mutant was observed to yield strong cross-links after CuP treatment for 1 h at 22–24°C. The mutant aN214C/cG58C formed a distinct a–c product, but in lower yield. To investigate the a–c interface in greater detail, 29 additional doubly substituted mutants (with Cys in helix-4 of subunit a and helix-2 of subunit c) were made and screened for cross-linking. Of the 65 total Cys–Cys substituted pairs, 23 formed detectable a–c cross-links in this initial screen with the intensity of a–c cross-linked product varying from ≤5–90% of the total immunopositive subunit a detected. A subsequent screening of 19 pairs with Cys substitutions in helix-1 yielded no positive cross-links. An immunoblot of various aI221C double Cys mutant membranes following CuP treatment at room temperature is shown in Fig. 2. The a–c cross-linked products were identified from immunoblots developed with antisera to subunits a and c, the subunit c immunoreactive product being more difficult to detect. The nonspecific immunoreactive bands (designated IA* in Fig. 2) were also present in membranes of the ΔuncBEFH deletion background and in wild-type membranes.
Figure 2.
Cross-linking of aI221C-substituted membranes in varying combinations with second Cys in subunit c. SDS-solubilized membranes were electrophoresed under nonreducing condition before immunoblotting. The blot was first probed with antiserum against subunit c. The blot was then stripped of bound antibodies by submerging it in a buffer containing 62.5 mM Tris⋅HCl (pH 6.8), 2% SDS, and 100 mM 2-mercaptoethanol for 30 min at 50°C and reprobed with antiserum against subunit a. The bands marked IA* are immunoblotting artifacts. Wild-type membranes are from pDF163- (uncBEFH+) transformed strain JWP109 (ΔuncB–H). Deletion membranes are from strain JWP109 (ΔuncB–H).
Additional experiments were carried out in an attempt to distinguish the more closely proximal residues from those that might be cross-linked due to thermal motion. Rates of cross-linking were measured over the interval of 5–180 min using a series of doubly Cys-substituted pairs that formed high and low yields of cross-linked product when incubated for 60 min at 22–24°C (i.e., aN214C/cG58C, aN214C/cA62C, aN214C/cM65C, aN214C/cI66C, aI221C/cG69C, and aL224C/cY73C). In the two pairs forming low yield cross-links (aN214C/cG58C and aN214C/cI66C), cross-links accumulated linearly at a low rate over the entire 3-h interval. The four pairs giving higher yield cross-links all showed biphasic kinetics, with cross-linked product accumulating most rapidly over the initial 10–30 min of the experiment and then in a slower phase continuing to 3 h. However, the initial rate data did not allow greater distinction between the mutant pairs than that given by differences in yield of cross-link from the 60-min incubation.
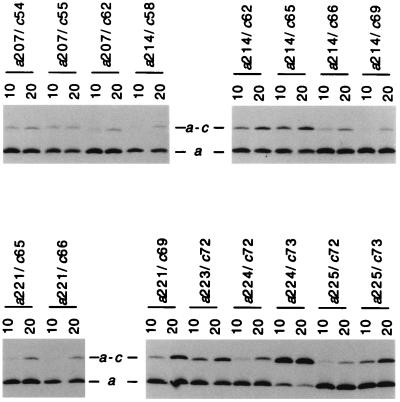
The doubly Cys-substituted mutant pairs did differ considerably in the temperature dependence of the cross-linking reaction. As shown in Fig. 3, a number of mutant pairs formed cross-links at 20°C, but not at 10°C. Examples showing these characteristics are aN214C/cG58C, aN214C/cG69C, aI221C/cM65C, and aI221C/cI66C. On the other hand, most mutant pairs forming high yield cross-links at 20°C also formed substantial cross-links at 10°C and also at 0°C (Table 1). Obvious examples include aN214C/cA62C, aN214C/cM65C, aI221C/cG69C, aI223C/cL72C, aL224C/cY73C, and aI225C/cY73C. The aL207C/cF54C and aL207C/cI55C mutants are notable in that nearly equal amounts of cross-links were observed at 10°C and 20°C. Both mutants also showed substantial cross-link formation at 0°C. The temperature dependence of cross-link formation for the 16 Cys–Cys pairs showing ≥10% cross-link formation at 20°C is summarized in Table 1. A number of pairs show substantial cross-link formation at 20°C and negligible cross-link formation at 10°C or 0°C, and cross-linking is likely to be at least partially dependent upon thermal movement of subunits a and c during cross-linking. On the other hand, the pairs showing substantial cross-link formation at 10°C or 0°C are more likely to be proximal in the complex. A survey of cross-link formation in mutant pairs at 10°C is shown in Fig. 4, and the results are summarized in tabular form in Fig. 1B.
Figure 3.
Comparison of a–c cross-link formation at 10 and 20°C with a series of doubly Cys-substituted membranes. A portion of an immunoblot of a SDS gel run under nonreducing conditions is shown following probing with antiserum to subunit a. The positions of the double Cys substitutions and temperature (10°C or 20°C) of the 1-h CuP cross-linking reaction are indicated.
Table 1.
Effect of temperature on the yield of a–c cross-links
| Position of Cys substituions |
a in cross-link, %
|
||
|---|---|---|---|
| 0°C | 10°C | 20°C | |
| a207/c54 | 7 ± 1 | 10 ± 3 | 19 ± 2 |
| a207/c55 | 11 ± 2 | 12 ± 3 | 16 ± 4 |
| a207/c62 | 3 ± 1 | 5 ± 2 | 15 ± 4 |
| a214/c58 | 0 | 3 ± 1 | 10 ± 3 |
| a214/c62 | 5 ± 0 | 21 ± 4 | 29 ± 4 |
| a214/c65 | 6 ± 0 | 25 ± 4 | 36 ± 2 |
| a214/c66 | 0 | 7 ± 3 | 16 ± 3 |
| a214/c69 | 0 | 3 ± 2 | 10 ± 1 |
| a221/c65 | 0 | 6 ± 1 | 26 ± 6 |
| a221/c66 | 0 | 1 ± 1 | 17 ± 4 |
| a221/c69 | 5 ± 1 | 18 ± 3 | 40 ± 4 |
| a223/c72 | 12 ± 0 | 32 ± 3 | 42 ± 3 |
| a224/c72 | 2 ± 1 | 9 ± 3 | 27 ± 7 |
| a224/c73 | 58 ± 0 | 65 ± 0 | 81 ± 8 |
| a225/c72 | 0 | 6 ± 2 | 11 ± 5 |
| a225/c73 | 3 ± 0 | 24 ± 2 | 35 ± 7 |
All mutant pairs forming ≥10% a–c cross-links at 20°C are shown. The percentage of immunopositive subunit a in the a–c cross-link is indicated (average ± SD of two to four experiments for each pair).
Figure 4.
Formation of a–c cross-links by CuP reaction at 10°C in a series of doubly Cys-substituted membranes. The conditions are as described in Fig. 3. The positions of the double Cys substitutions are indicated. Wild-type membranes are from a pDF163 transformant of strain JWP109 (ΔuncB–H). Deletion membranes are from strain JWP109 (ΔuncB–H).
Categories of a–c Cross-Linked Pairs.
In summary (see Fig. 1B), 7 of 84 doubly Cys-substituted mutants formed significant (≥10%) a–c cross-linked product during CuP treatment for 1 h at 10°C, i.e., aS207C/cI55C, aN214C/cA62C, aN214C/cM65C, aI221C/cG69C, aI223C/cL72C, aL224C/cY73C, and aI225V/cY73C. Nine other combinations formed detectable products in lower yield at 10°C and significantly greater (≥10%) a–c cross-linked product at 20°C, i.e., aS206C/cI55C, aN214C/cG58C, aN214C/cI66C, aA217C/cM65C, aA217C/cI66C, aG218C/cM65C, aG218C/cI66C, aI221C/cM65C, aI221C/cI66C, and aL224C/cL72C. Mutants showing lower but detectable levels of cross-linked product (<10% of subunit a total) at 20°C, or in the initial room temperature survey, are also indicated in Fig. 1B. No cross-linking was observed between Cys–Cys pairs, where Cys was substituted in helix-1 of subunit c.
DISCUSSION
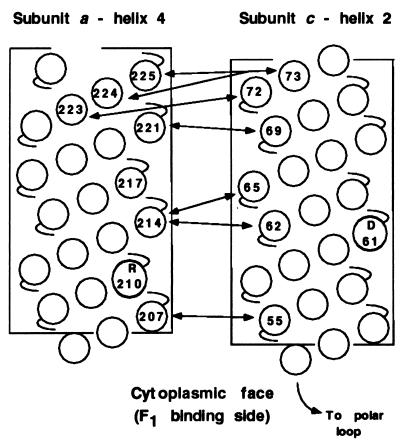
Neighboring residues in the second transmembrane helix of subunit c and the fourth transmembrane helix of subunit a were defined by sulfhydryl cross-linking after genetic introduction of Cys into both helices. A series of Cys–Cys cross-links, formed at 10° or 20°C, span a length of 19 residues in both helices and define the direction of packing of the helices. The periodicity of cross-link formation generally mirrors that expected for one face of an α-helix (Fig. 5). The initial set of Cys substitutions in subunit a were made based on a cluster of suppressor substitutions in residues 217, 221, and 224 of subunit a which optimized function of the cA24D/cD61G double mutant. A transmembrane α-helix with these three residues positioned on one face was postulated to interact with the transmembrane helix of subunit c anchoring the essential aspartyl residue (20). The potential of the genetic method in identifying transmembrane regions of protein–protein interaction is supported here by the finding that Cys substitution in two of the three positions yielded cross-links with appropriately Cys-substituted subunit c.
Figure 5.
Summary of major cross-links formed between helix-4 of subunit a and helix-2 of subunit c at 10°C. The circles numbered without specifying residue type indicate the position of the Cys substitutions in α-helical representations of the transmembrane segments. The positions of essential residues aR210 and cD61 are also indicated.
The pattern of high yield cross-links was used to orient the direction of packing of helices relative to each other. Lower yield cross-links generally support the suggested orientation but also indicate that there may be considerable mobility of subunits within Fo. Many of the lower yield cross-links only formed at temperatures ≥20°C, which suggests that thermal motion may be required for their formation. Clearly, some of the high- and low-yield cross-links could not form simultaneously due to spatial constraints. For example, the cM65C residue formed a high-yield cross-link with the aN214C residue but also cross-linked to a modest extent with the aI221C residue, the residues being separated by two potential turns of an α-helix. Conceivably, the vertical position of different subunits c relative to the transmembrane helix of subunit a could vary depending on their position in the c oligomer. As is discussed below, the formation of c–c dimers (see Fig. 2) is difficult to explain without a consideration of subunit mobility within Fo.
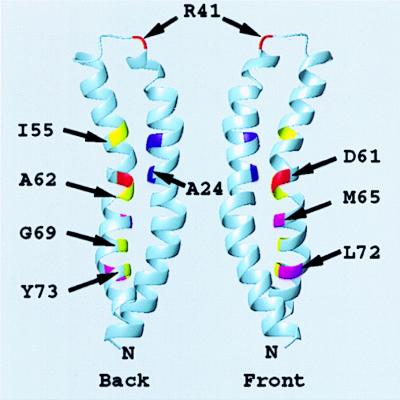
Girvin et al. (46) recently completed the NMR structure of monomeric subunit c. The side-to-side packing of the two helices of the subunit leads to formation of two flattened surfaces with Gly23 neighboring Asp61 in the middle of the front surface and the side chains of Ala24 and Ala62 neighboring each other in the middle of the rear surface (Fig. 6). The functional unit was suggested to be a dimer, wherein two subunits c are packed in a front-to-back arrangement. This would place the Asp61 side chain of one monomer between the side chains of Ala24 and Ala62 of a second monomer. The dimer would provide an explanation for the functional interchange of Asp between residues 61 and 24 (33), and the use of a Ser62 hydroxyl as a liganding group for Li+ in a mutant with altered cation specificity (34). Jones et al. (35), in an independent cross-linking study, have confirmed the front-to-back packing of subunit c and shown that it extends to generate an oligomeric ring. Importantly, Jones et al. (35) verify a number of the interactions predicted by the NMR model. Various ring-like arrangements have previously been suggested by others (7, 36–40).
Figure 6.
Ribbon depiction of the folding of subunit c derived from the NMR structure. The “front” and “back” face of two subunits c are indicated. A functional c2 dimer is proposed to be formed by packing the front face of one monomer against the back face of a second monomer (46), i.e., with D61 of one monomer packed between A24 and A62 of a second monomer. The positions of the backbone atoms of key residues discussed in the text are indicated. The illustration was done in the program molmol (45).
Subunits a and b are now thought to lie outside the subunit c oligomer, rather than in the center (36), on the basis of evidence from electron microspectroscopic imaging of negatively stained Fo (41) and atomic force microscopy of native Fo (42, 43). The cross-linking results reported here indicate that the first and second helices of subunit c must lie, respectively, on the inside and outside of the ring. Helix-4 of subunit a was easily cross-linked with the C-terminal helix of subunit c, whereas no cross-linking was observed with the N-terminal helix (Fig. 1B).
In the model described above, the Asp61 carboxyl lies between packed subunits rather than at the periphery as suggested by others (37, 39, 40). If subunit a participates in binding or release of protons from Asp61 in wild-type subunit c, it is likely that transmembrane helix-4 packs on the outer surface of the c-oligomeric ring at the interface between two subunits c, with side chains of aL207C, aN214C, aI221C, and aL224C lying on the packing face of a continuous α-helix. Of the Cys-substituted positions in subunit c forming high-yield cross-links, the cM65C and cL72C side chains lie toward the middle of the front flattened face, whereas the cI55C and cA62C side chains lie toward the middle of the back flattened face (Fig. 6). The α-carbons of cG69C and cY73C lie toward the interface of the two interacting helices. The aN214C side chain must therefore be positioned such that it can interact with either half of the subunit c dimer to form the two high-yield cross-links, i.e., aN214C-cM65C or aN214C-cA62C. The interacting ridge of helix-4 of subunit a may have to insert itself between the two c subunits to do this.
The formation of c–c dimers from Cys at identical positions in helix-2 of two subunits c (see Fig. 2) is not easily reconciled by an oligomeric ring model with front-to-back type packing. These homodimers also form in the singly substituted subunit c mutants. In the cases of cA62C or cM65C, the subunits would have to reorient and come face-to-face (i.e., back-to-back or front-to-front) to form such homodimers. Conceivably, a transient reorientation of a fraction of the subunit c in the oligomer could occur during the movements related to rotation. A transient reorientation of the packing of helix-2 relative to helix-1 within a subunit c monomer also needs to be considered. A rotation of up to 180° of helix-2 in one subunit c of the oligomer would place identical residues in the neighboring subunit c close to each other and permit dimer formation. Such a rotation would also expose Asp61 to the periphery of the ring, as is hypothesized by others (37, 39, 40), and cause residues forming high-yield a–c cross-links to relocate more toward the periphery where they would be more obviously accessible to the cross-linkable residues in subunit a. No cross-links were observed between Cys in helix-4 of subunit a and residues predicted to be at the periphery of the oligomeric ring, i.e., aN214C/cV60C and aI221C/cA67C (Fig. 1B). It is possible that the NMR model depicts c subunits in the oligomer that are not interacting with helix-4 of subunit a.
It should be clear that cross-link formation brought about by the packing of aL207C close to cI55C, and of aN214C close to cA62C or cM65C, would also place the aArg210 residue close to cGly58 and to cAsp61. Proton release from Asp61 could be promoted by the positively charged guanidino group and transient salt bridge formation between aArg210 and cAsp61 carboxylate. If the interaction involved an insertion of helix-4 of subunit a between front-to-back packed subunit c monomers or a rotation of helix-2 relative to that seen in the NMR structure, then the interaction could easily disrupt proton or cation binding between monomers. In the case of the Na+-translocating enzyme from Propiogenium modestum (44), similar interactions could disrupt the liganding of Na+ among the side chains of Gln, Glu, and Ser, at positions equivalent to residues 28, 61, and 62 in E. coli, to promote Na+ release.
Acknowledgments
This paper is dedicated to the memory of Professor Yu Wang (1910–1997). We thank Francis Valiyaveetil and Phil Jones of this laboratory for kindly donating some of the subunit c mutants used in this study. This work was supported by U.S. Public Health Service Grant GM23105 from the National Institutes of Health.
ABBREVIATION
- CuP
Cu(1,10-phenanthroline)2SO4
Footnotes
The uncBEFH genes, respectively, code for subunits a, c, b, and δ.
The unc DNA numbering system corresponds to that used by Walker et al. (26).
References
- 1.Boyer P D. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 2.Deckers-Hebestreit G, Altendorf K. Annu Rev Microbiol. 1996;50:791–824. doi: 10.1146/annurev.micro.50.1.791. [DOI] [PubMed] [Google Scholar]
- 3.Fillingame R H. J Exp Biol. 1997;200:217–224. doi: 10.1242/jeb.200.2.217. [DOI] [PubMed] [Google Scholar]
- 4.Fillingame R H. The Bacteria. Vol. 12. New York: Academic; 1990. pp. 345–391. [Google Scholar]
- 5.Abrahams J P, Leslie A G, Lutter R, Walker J E. Nature (London) 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 6.Weber J, Senior A E. Biochim Biophys Acta. 1997;1319:19–58. doi: 10.1016/s0005-2728(96)00121-1. [DOI] [PubMed] [Google Scholar]
- 7.Duncan T M, Bulygin V V, Zhou Y, Hutcheon M L, Cross R L. Proc Natl Acad Sci USA. 1995;92:10964–10968. doi: 10.1073/pnas.92.24.10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabert D, Engelbrecht S, Junge W. Nature. 1996;381:623–625. doi: 10.1038/381623a0. [DOI] [PubMed] [Google Scholar]
- 9.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Nature. 1997;386:249–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 10.Aggeler R, Haughton M A, Capaldi R A. J Biol Chem. 1996;270:9185–9191. doi: 10.1074/jbc.270.16.9185. [DOI] [PubMed] [Google Scholar]
- 11.Aggeler R, Capaldi R A. J Biol Chem. 1996;271:13888–13891. doi: 10.1074/jbc.271.23.13888. [DOI] [PubMed] [Google Scholar]
- 12.Watts S D, Tang C, Capaldi R A. J Biol Chem. 1996;271:28341–28347. doi: 10.1074/jbc.271.45.28341. [DOI] [PubMed] [Google Scholar]
- 13.Ogilvie I, Aggeler R, Capaldi R A. J Biol Chem. 1997;272:16652–16656. doi: 10.1074/jbc.272.26.16652. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Fillingame R H. J Biol Chem. 1995;270:24609–24614. [PubMed] [Google Scholar]
- 15.Watts S D, Zhang Y, Fillingame R H, Capaldi R A. FEBS Lett. 1995;368:235–238. doi: 10.1016/0014-5793(95)00658-v. [DOI] [PubMed] [Google Scholar]
- 16.Schneider E, Altendorf K. EMBO J. 1985;4:515–518. doi: 10.1002/j.1460-2075.1985.tb03658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn S D. J Biol Chem. 1992;267:7630–7636. [PubMed] [Google Scholar]
- 18.Hatch L P, Cox G B, Howitt S M. J Biol Chem. 1995;270:29407–29412. doi: 10.1074/jbc.270.49.29407. [DOI] [PubMed] [Google Scholar]
- 19.Valiyaveetil, F. I. & Fillingame, R. H. (1998) J. Biol. Chem., in press. [DOI] [PubMed]
- 20.Fraga D, Hermolin J, Fillingame R H. J Biol Chem. 1994;269:2562–2567. [PubMed] [Google Scholar]
- 21.Valiyaveetil F I, Fillingame R H. J Biol Chem. 1997;272:32635–32641. doi: 10.1074/jbc.272.51.32635. [DOI] [PubMed] [Google Scholar]
- 22.Mosher M E, White L K, Hermolin J, Fillingame R H. J Biol Chem. 1985;260:4807–4814. [PubMed] [Google Scholar]
- 23.Miller M J, Fraga D, Paule C R, Fillingame R H. J Biol Chem. 1989;264:305–311. [PubMed] [Google Scholar]
- 24.Inoue H, Nojima H, Okayama H. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 25.Fraga D, Fillingame R H. J Biol Chem. 1989;264:6797–6803. [PubMed] [Google Scholar]
- 26.Walker J E, Saraste M, Gay N J. Biochim Biophys Acta. 1984;768:164–200. doi: 10.1016/0304-4173(84)90003-x. [DOI] [PubMed] [Google Scholar]
- 27.Barik S. In: Methods in Molecular Biology. Trower M K, editor. Vol. 57. Totawa, NJ: Humana; 1996. pp. 203–215. [DOI] [PubMed] [Google Scholar]
- 28.Brusilow W S A, Proter A C G, Simoni R D. J Bacteriol. 1983;263:1265–1270. doi: 10.1128/jb.155.3.1265-1270.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schägger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 30.Towbin H, Staehelin T, Gordon J. Proc Natl Acad Sci USA. 1979;81:7279–7283. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hermolin J, Fillingame R H. J Biol Chem. 1995;270:2815–2817. doi: 10.1074/jbc.270.6.2815. [DOI] [PubMed] [Google Scholar]
- 32.Girvin M E, Hermolin J, Pottorf R, Fillingame R H. Biochemistry. 1989;28:4340–4343. doi: 10.1021/bi00436a032. [DOI] [PubMed] [Google Scholar]
- 33.Miller M J, Oldenburg M, Fillingame R H. Proc Natl Acad Sci USA. 1990;87:4900–4904. doi: 10.1073/pnas.87.13.4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Fillingame R H. J Biol Chem. 1995;270:87–93. doi: 10.1074/jbc.270.1.87. [DOI] [PubMed] [Google Scholar]
- 35.Jones, P. C., Jiang, W. & Fillingame, R. H. (1998) J. Biol. Chem., in press. [DOI] [PubMed]
- 36.Howitt S M, Rodgers A J W, Hatch L P, Gibson F, Cox G B. J Bioenerget Biomemb. 1996;28:415–420. doi: 10.1007/BF02113983. [DOI] [PubMed] [Google Scholar]
- 37.Vik S B, Antonio B J. J Biol Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- 38.Groth G, Walker J E. FEBS Lett. 1997;410:117–123. doi: 10.1016/s0014-5793(97)00529-2. [DOI] [PubMed] [Google Scholar]
- 39.Engelbrecht S, Junge W. FEBS Lett. 1997;414:485–491. doi: 10.1016/s0014-5793(97)00997-6. [DOI] [PubMed] [Google Scholar]
- 40.Elston T, Wang H, Oster G. Nature. 1998;391:510–513. doi: 10.1038/35185. [DOI] [PubMed] [Google Scholar]
- 41.Birkenhäger R, Hoppert M, Deckers-Hebestreit G, Mayer F, Altendorf K. Eur J Biochem. 1995;230:58–67. [PubMed] [Google Scholar]
- 42.Takeyasu K, Omote H, Nettikadan S, Tokumasu F, Iwamoto-Kihara A, Futai M. FEBS Lett. 1996;392:110–113. doi: 10.1016/0014-5793(96)00796-x. [DOI] [PubMed] [Google Scholar]
- 43.Singh S, Turina P, Bustamante C J, Keller D J, Capaldi R A. FEBS Lett. 1996;397:30–34. doi: 10.1016/s0014-5793(96)01127-1. [DOI] [PubMed] [Google Scholar]
- 44.Kaim G, Wehrle F, Gerike U, Dimroth P. Biochemistry. 1997;36:9185–9194. doi: 10.1021/bi970831q. [DOI] [PubMed] [Google Scholar]
- 45.Koradi R, Billeter M, Würthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 46.Girvin, M. E., Rastogi, V. K., Abildgaard, F., Markley, J. L. & Fillingame, R. H. (1998) Biochemistry, in press. [DOI] [PubMed]