Abstract
The 1,3–1,4-β-glucanase from Bacillus macerans (wtGLU) and the 1,4-β-xylanase from Bacillus subtilis (wtXYN) are both single-domain jellyroll proteins catalyzing similar enzymatic reactions. In the fusion protein GluXyn-1, the two proteins are joined by insertion of the entire XYN domain into a surface loop of cpMAC-57, a circularly permuted variant of wtGLU. GluXyn-1 was generated by protein engineering methods, produced in Escherichia coli and shown to fold spontaneously and have both enzymatic activities at wild-type level. The crystal structure of GluXyn-1 was determined at 2.1 Å resolution and refined to R = 17.7% and R(free) = 22.4%. It shows nearly ideal, native-like folding of both protein domains and a small, but significant hinge bending between the domains. The active sites are independent and accessible explaining the observed enzymatic activity. Because in GluXyn-1 the complete XYN domain is inserted into the compact folding unit of GLU, the wild-type-like activity and tertiary structure of the latter proves that the folding process of GLU does not depend on intramolecular interactions that are short-ranged in the sequence. Insertion fusions of the GluXyn-1 type may prove to be an easy route toward more stable bifunctional proteins in which the two parts are more closely associated than in linear end-to-end protein fusions.
The jellyroll belongs to those domain folds (“superfolds”) that most frequently are found in globular proteins (1). It is characterized by two strictly antiparallel β-sheets forming a distorted β-barrel (2, 3). In a series of studies, we have demonstrated that the jellyroll fold as present in the 1,3–1,4-β-glucanases from Bacillus macerans and Bacillus licheniformis (4, 5) is insensitive to various changes of amino acid sequence. Proteins with conserved three-dimensional structure and the predicted enzymatic activity are obtained after site-directed mutagenesis of single residues (6), gene fusion leading to hybrid enzymes with parts derived from different wild-type proteins (7, 8), and circular permutation of the polypeptide chain (9, 10). The latter experiments suggested that the order of secondary structure elements and the identity of the N-terminal protein region are not crucial for in vivo foldability and the native state of the protein. They give rise to the hypothesis that the folding of the 1,3–1,4-β-glucanase jellyroll is dominated by intramolecular interactions that are long-ranged in the sequence and insensitive toward changes in sequential order. The present study aims at testing this hypothesis by fusing the B. macerans 1,3–1,4-β-glucanase (wtGLU) with the extracellular 1,4-β-xylanase of Bacillus subtilis (wtXYN).
Although it belongs to a different family of glycosyl hydrolases (11), wtXYN shares with the bacterial 1,3–1,4-β-glucanases the specificity toward β-1,4 pyranosyl linkages and the general folding topology. It differs in sequence only at one position from the Bacillus circulans 1,4-β-xylanase (12), the crystal structure of which is known (13). Genes encoding 1,3–1,4-β-glucanases and 1,4-β-xylanases sometimes are clustered in bacterial genomes (14), and a linear, bifunctional protein with both enzymatic activities has been found in Ruminococcus flavefaciens (15). Because glucans and xylans occur in large quantities in the biosphere as constituents of plant cell walls, enzymes capable of their degradation are of considerable interest.
CpMAC-57 is a circularly permuted variant of wtGLU with native-like properties (9). This protein has provided the basis for the molecular cloning of GluXyn-1, a bifunctional insertion fusion protein composed of two globular domains, GLU and XYN, described in this report. In GluXyn-1, the XYN domain is inserted into a surface loop of cpMAC-57 between the residues corresponding to the chain termini of wtGLU. GluXyn-1 displays both enzymatic activities at wild-type level. The three-dimensional structures of both domains are nearly indistinguishable from wtGLU and wtXYN. Thus, the GLU domain is able to fold to completion in vivo in spite of the insertion of the autonomously folding XYN domain. This result is taken as evidence that the jellyroll fold of the bacterial 1,3–1,4-β-glucanases is, indeed, dominated by long-range intramolecular contacts. In addition to its significance for protein folding, GluXyn-1 may serve as an example for the generation of stable bifunctional proteins by insertional fusion of domains.
MATERIALS AND METHODS
Plasmid Construction.
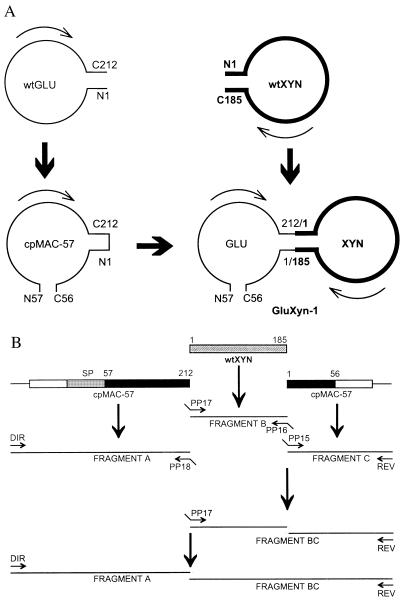
The cloned genes encoding the circularly permuted 1,3–1,4-β-glucanase cpMAC-57 (9) and wtXYN (12) were fused by splicing-by-overlap extension (16). The conditions for performing PCR were as described earlier (9). In separate primary amplifications (see Fig. 1), three overlapping DNA fragments, A, B, and C, were amplified by PCR.
Figure 1.
Design and construction of GluXyn-1. (A) Schematic structure of GluXyn-1 and the parental enzymes used for the construction of the bifunctional insertion fusion protein. The circularly permuted cpMAC-57 (9) starts with residue 57 of wtGLU whereas the original N and C termini are covalently linked leading to an enzyme whose C terminus is residue 56. cpMAC-57 and wtXYN (bold line) were linked by cutting the loop between residues 212 and 1 of cpMAC-57 (numbering according to GLU) and connecting those residues with the terminal residues of XYN. The resulting construct, GluXyn-1, starts with N57 and ends with C56 just as cpMAC-57, but contains in its central part the full-length sequence of mature XYN. For simplicity, only the sequences encoding the mature enzymes are shown. Amino acid residues in GluXyn-1 are numbered according to their sequence positions in the parent proteins, wtGLU and wtXYN, throughout this paper. (B) Construction of GluXyn-1 by PCR splicing of gene fragments. Fragment A covers the upstream regulatory sequence of the gene encoding the B. macerans 1,3–1,4-β-glucanase (open box) and sequences encoding the signal peptide (SP) and the mature 1,3–1,4-β-glucanase from residues 57 to 212 (solid box). Fragment B encodes the full-length mature XYN, and fragment C consists of the coding region for residues 1–56 of B. macerans β-glucanase (solid box) and a short segment of the 3′ noncoding region of cpMAC-57 (open box). Using sequence specific primers, the three fragments were amplified by splicing-by-overlap extension whereby terminal extensions complementary to the adjacent sequence in the resulting GluXyn-1-encoding construct (shaded box) were linked to the synthesized fragments. In the second (fragments B and C) and third step of amplification (fragments A and BC) the obtained fragments were used to amplify the full-length hybrid gene encoding GluXyn-1. Sequence specific primers PP15-PP18 are indicated by arrows. Direct (DIR) and reverse (REV) primers annealing with the flanking vector sequences also are shown.
Fragment A encoding the signal peptide and residues 57–212 of cpMAC-57 was amplified from plasmid pcpMAC-57 with primers DIR (5′-CGC CAG GGT TTT CCC AG) and PP18 (5′-CTG TGC TAG CAT TGC TCG TAT ATT TTA CCC), fragment B encoding the mature wtXYN from plasmid pMW7 with primers PP17 (5′-ATA CGA GCA ATG CTA GCA CAG ACT ACT GGC) and PP16 (5′-CAC ACT CCC CCA CAC CAC TGT TAC GTT AGA AC), and fragment C encoding residues 1–56 of cpMAC-57 from plasmid pcpMAC-57 with primer PP15 (5′-AAC AGT GTG GGG GAG TGT GTT CTG GGA ACC) and the reverse primer REV (5′-AGC GGA TAA CAA TTT CAC ACA GGA). The amplification products were separated on 1% (wt/vol) agarose and isolated with Prep-A-Gene (Bio-Rad). The fragments A, B, and C were then combined and amplified in two subsequent splicing-by-overlap extension reactions to give the entire hybrid gene GLUXYN. This fragment was used to replace the sequence between BstEII and HindIII in plasmid pTZ19R-MAC (7) to yield pFG1 harboring the 5′ noncoding (promoter) region of the B. macerans 1,3–1,4-β-glucanase gene (17) and the GluXyn-1-encoding region. Escherichia coli DH5α cells transformed with pFG1 were able to hydrolyze the mixed-linked β-glucan lichenan and xylan as demonstrated after plating on lichenan and xylan containing Luria–Bertani (LB) agar (10 g Bacto tryptone/5 g Bacto yeast extract/5 g NaCl/1 g substrate xylan or lichenan/15 g Difco agar per liter). Staining with 0.1% Congo Red revealed zones of hydrolysis surrounding recombinant E. coli colonies. Plasmid DNA isolated from such clones was used to verify the expected DNA sequence by the dideoxy chain termination reactions (18).
Gene Expression, Protein Purification, and Characterization.
GluXyn-1 was synthesized in E. coli pFG1 cells growing for 15 hr in double-strength LB medium as described previously (19). Whole-cell extracts were prepared by ultrasonication in 1/10 of the culture volume in lysis buffer (10 mM Tris⋅HCl, pH 8/5 mM EDTA/1 mM phenylmethylsulfonyl fluoride). The protein samples were diaultrafiltrated against 50 mM NaOac, pH 8.5/5 mM CaCl2. The sample was applied to a column containing Q-Sepharose (Pharmacia) and eluted with a linear gradient of 0–300 mM NaCl. Fractions containing both 1,3–1,4-β-glucanase and 1,4-β-xylanase activities were further purified by gel filtration using Superdex G75 (Pharmacia). Protein concentrations were determined according to ref. 20. The proteins obtained were in the correct size and without visible impurities as checked by SDS/PAGE. Zymograms were prepared from polyacrylamide gels copolymerized with 0.25% of the appropriate substrates, lichenan or Remazole Brilliant Blue (RBB) xylan. After running, SDS was removed by repeated washing with ethanol/water (1:1, by vol) equilibrated in 40 mM NaOac, pH 6.0/CaCl2 and incubated at 37°C. Samples containing GluXyn-1 displayed the zone of substrate hydrolysis caused by the reactivated enzyme at 43 kDa (not shown). Enzymatic activities were determined in triplicate with either lichenan or oat spelt xylan (0.5%) as substrate (21) at 50°C in 50 mM NaOac buffer, pH 6.0/20 mM CaCl2.
Other biochemical analyses were performed as described (19). The half-life (t1/2) of enzymatic activity was determined at 60°C in 50 mM NaOac buffer, pH 5.0/10 mM CaCl2/50 μg ⋅ ml−1 BSA. Final products of enzymatic hydrolysis were determined after incubation of 1 unit of purified enzyme with 2.5 mg substrate (in 0.5 ml) at 50°C, pH 6, for 24 hr. Thin-layer chromatography was performed with silica plates (10 × 20 cm) in ethylacetate/acetic acid/distilled water (50:25:25, by vol). Spots were visualized with o-naphthol (0.8 g per liter of 17% sulfuric acid methanol).
Crystallization and Acquisition of X-Ray Diffraction Data.
Crystals of GluXyn-1 were obtained at room temperature by the standard vapor diffusion, hanging-drop method. A solution of 2.8 mg of GluXyn-1 per 1 ml of 20 mM Tris⋅HCl, pH 8.7/2 mM CaCl2 was mixed with an equal volume of 40 mM KP, pH 7.0/16% (by weight) PEG 8000. The final size of a crystal was 0.2 × 0.2 × 0.1 mm after 2 days.
The crystal was flash-frozen directly out of the hanging drop 1 hr after increasing the PEG concentration to 26%. Diffraction data were collected from a single flash-frozen crystal on beamline BM14 (λ = 1.008) at the European Synchrotron Radiation Facility, Grenoble, France. The complete data set was acquired on a 180-mm MAR-Research (Hamburg) imaging plate. The 119 images collected in 1.5° oscillations with a crystal-detector distance of 170 mm were processed with the program denzo (22) and then scaled with the program scala from the CCP4 program system (23). In space group P21 with a = 45.3 Å, b = 133.7 Å, c = 77.9 Å, and β = 99.8°, the presence of two molecules in the asymmetric unit is required to yield a packing density in the expected range (24), VM = 2.6 Å3 Da−1 corresponding to a solvent content of 46.4%. A total of 165,157 observations were reduced to 48,499 unique reflections. The resulting data set contained 91.3% of the reflections expected in the resolution range between 20 and 2.1 Å.
Crystal Structure Determination and Refinement.
The structure was solved by molecular replacement by using the 1.8 Å crystal structure of cpMAC-57 (Protein Data Bank entry 1CPN, ref. 9) and the 1.8 Å structure of the B. circulans 1,4-β-xylanase (entry 1BCX, ref. 13) as search models in amore (25). The sequence of the B. circulans 1,4-β-xylanase differs at position 147 (Thr) from wtXYN, where this residue is a Ser. Furthermore, the protein used in 1BCX was inactive because of a mutation of Glu-172 to Cys.
Using cpMAC-57 as a search model in the cross-rotation function, the best two solutions showed up at 7.9 σ and 7.8 σ above the mean. They were clearly discriminated from the next, incorrect solution at 4.3 σ. The XYN model yielded maxima of the rotation function at 7.0 σ and 5.6 σ above the mean. Starting with cpMAC-57, the search models subsequently could be positioned inside the unit cell in a stepwise procedure by calculating the translation function for one model at a time and fixing correct solutions found in previous steps. In the resolution shell from 8.0 to 3.5 Å, the four search fragments yielded an R value of 35.3% and a correlation coefficient of 0.654. Inspection of the packing model revealed complete absence of stereochemical clashes and immediate juxtaposition of N and C termini of the GLU and XYN parts as required in GluXyn-1. Appropriate manual model changes guided by clear Fo − Fc difference electron density yielded the starting model for refinement.
At the beginning of the refinement, 5% of all measured reflections were set apart for subsequent monitoring of the free R value (26). Restraints for noncrystallographic symmetry had to be set separately for the GLU and XYN domains. Several rounds of slow-cooling, simulated-annealing refinement (27) against diffraction data of gradually increasing resolution resulted in R and R(free) values of 31.8 and 35.0%, respectively, for all data in the resolution range of 20.0 to 2.1 Å. Structure analysis was completed with maximum-likelihood refinement as implemented in refmac (28) by using a bulk solvent correction and an overall anisotropic temperature factor for the model. As indicated by R(free), noncrystallographic symmetry restraints were gradually loosened and completely removed toward the end of refinement. Solvent molecules were incorporated into the model after R dropped to 26.4% [R(free) 29.0%], filling peaks higher than 3.0 σ in the sigmaa-weighted difference density map (29) that had reasonable hydrogen-bonding geometry. They were removed from the model if their temperature factors exceeded 50 Å2. The refinement was considered complete when parameter shifts had converged and the difference density map was without interpretable features.
RESULTS
Design, Construction, Stability, and Enzymatic Activity of GluXyn-1.
The design and the strategy to construct the bifunctional hybrid enzyme GluXyn-1 are outlined in Fig. 1. GluXyn-1 results from the end-to-end ligation of wtGLU and wtXYN and the creation of novel chain termini in a surface loop of GLU, rendering Phe-57 of GLU the N terminus of GluXyn-1 and Ser-56 the C terminus. Alternatively, GluXyn-1 may be regarded as a fusion product of the circularly permuted 1,3–1,4-β-glucanase cpMAC-57 (9) and wtXYN through insertion of XYN into a surface loop of cpMAC-57. GluXyn-1 is based on the simplest possible design by using only cut-and-paste manipulations without deletions and insertions.
The construction of GluXyn-1 started from the genes encoding cpMAC-57 (9) and wtXYN (12). On the gene level, the N terminus of XYN was linked with the sequence consisting of the upstream regulatory region, the signal peptide, and residues 57–212 of cpMAC-57 (see Fig. 1B). The C terminus was fused with the remaining part of cpMAC-57 spanning residues 1 to 56. As a result, the mature bifunctional enzyme starts with Phe-57 of GLU (like cpMAC-57) after gene expression, export of the preprotein into the E. coli periplasm, and in situ proteolytic removal of the signal peptide. The XYN sequence forms the central part connecting the N-terminally located GLU domain 57–212 with the C-terminal part comprising residues 1–56 of GLU.
The properties of the purified bifunctional protein obtained from the periplasm of recombinant E. coli cells were compared with those of both parental enzymes. The data compiled in Table 1 demonstrate that substrate specificities (Km), pH for optimal activity, and, most important, enzymatic activities (kcat) of GluXyn-1 are essentially the same as in the two parental enzymes, wtGLU and wtXYN. The decrease in Vmax is explained easily by the higher molecular weight of the hybrid. A slight decrease in the temperature for optimal activity was observed, which is paralleled by a reduction of thermal stability at 60°C. The main products of enzymatic hydrolysis of lichenan (cellobiosylglucose) and oat spelt xylan (xylobiose, xylotriose, and xylotetraose) were identical in parental enzymes and the hybrid enzyme (results not shown).
Table 1.
Enzymatic activity and stability of the bifunctional insertion fusion protein GluXyn-1 compared with 1,3-1,4-β-glucanase of B. macerans and 1,4-β-xylanase of B. subtilis
| GluXyn-1 (1,3-1,4-β-glucanase activity) | wtGLU | GluXyn-1 (1,4-β-xylanase activity) | wtXYN | |
|---|---|---|---|---|
| Km, mg/ml | 0.87 ± 0.01 | 0.94 ± 0.07 | 3.36 ± 0.25 | 3.17 ± 0.25 |
| kcat, s−1 | 1,012 ± 1 | 1,040 ± 26 | 582 ± 20 | 705 ± 25 |
| Vmax, units/mg | 1,380 ± 1 | 2,399 ± 60 | 794 ± 28 | 2,015 ± 72 |
| pH for optimal activity | 6–7.5 | 6–7.5 | 6 | 6 |
| Temperature for optimal activity, °C | 55–60 | 65–70 | 50 | 55 |
| Thermostability t1/2 (min) at 60°C, pH 5 | 135 | 100% residual activity after 240 min | 118 | 90% residual activity after 240 min |
Crystal Structure Determination and Quality of the Model.
The crystal structure of GluXyn-1 was solved by molecular replacement by using the known structures of the parent proteins, wtGLU and wtXYN (4, 13), as search models in the rotation and translation functions. Because there are two molecules of GluXyn-1 per asymmetric unit of the crystal and the search models are of approximately equal size, both GLU and XYN represent only about one-fourth each of the atoms in the asymmetric unit. The refined structural model of GluXyn-1 contains 787 residues in two chains, 2 calcium ions, and 312 water molecules per asymmetric unit of the crystal. Structure refinement converged at an R value of 17.7% at 2.1 Å resolution (Table 2). The protein model yielded a reasonable R(free) of 22.4% and had proper stereochemistry as indicated by procheck (30). Most (88.1%) of the residues were in the most favorable regions of the Ramachandran diagram (not shown). The rms deviations from ideal geometry were 0.012 Å for bond lengths, 0.031 Å for the bond angle distances, and 0.031 Å for planar 1–4 distances.
Table 2.
X-ray diffraction data and refinement statistics for GluXyn-1
| Crystal data | |
| Space group | P21 |
| Cell axes, Å | 45.27 133.70 77.95 |
| Cell angles, ° | 90 99.76 90 |
| No. of molecules per asymmetric unit | 2 |
| Data statistics* | |
| Resolution limits, Å | 20.0–2.1 |
| Resolution limits, outer shell, Å | 2.17–2.1 |
| No. of observations | 165,157 |
| No. of unique reflections | 48,499 |
| Average multiplicity | 3.4 (3.2) |
| Completeness, % | 91.3 (80.0) |
| R(merge)†, % | 5.8 (23.0) |
| 〈I/σ(I)〉 | 11.6 (3.3) |
| Reflections with I ≥ 3 σ(I), % | 81.6 (62.3) |
| Refinement and model statistics | |
| No. of protein nonhydrogen atoms | 6,219 |
| No. of solvent molecules | 312 |
| No. of reflections | 48,305 |
| No. of reflections, test set | 2,440 |
| Degrees of freedom | 26,132 |
| R(cryst), % | 17.7 |
| R(free), % | 22.4 |
| Mean B, all protein atoms, Å2 | 21.2 |
| Mean B, solvent, Å2 | 25.6 |
| Ramachandran residues in most favored regions, % | 88.1 |
| Ramachandran outliers | None |
| rms deviation from target values‡ | |
| Bond lengths, Å | 0.012 |
| Bond angle distances, Å | 0.031 |
| Planar 1–4 distances, Å | 0.031 |
Numbers in parentheses refer to data in outer resolution shell.
R(merge) = 100 × (Σh,i|Ih,i − Ih |/Σh,i Ih,i), where the summation is over all observations Ih,i contributing to the reflection intensities Ih.
With respect to the parameters of Engh and Huber (41).
The crystal structure is well defined throughout the polypeptide chain with the exception of residues 112–124 (XYN) of molecule A of the asymmetric unit, which show high B-factors and also low electron density. In the other molecule this area has normal B-factors <50 Å2, and the electron density is defined clearly. In addition, the C-terminal amino acid residues, four from molecule A and three from molecule B of the asymmetric unit, are not seen in the electron density because of disorder. The mean B-factor is 21.2 Å2 for all protein atoms after refinement and 25.6 Å2 for the 312 water molecules of the asymmetric unit (maximal B = 50.7 Å2). An upper limit for the mean coordinate error may be obtained from a Luzzati (31) plot based on R(free). This procedure yields mean coordinate errors of 0.2–0.3 Å. In molecule A, difference electron density suggests two alternative orientations for the side chain of Trp-185 (XYN), which are modeled with equal occupancy. The cation-binding site in GLU is occupied by a Ca2+ ion in octahedral coordination essentially as in wtGLU (4).
Tertiary Structure.
The three-dimensional structure of GluXyn-1 is shown schematically in Fig. 2 in an orientation that permits a front view into the active-site channel of the GLU domain (green and yellow) and a side view through the active-site channel of the XYN domain (blue). Both domains of GluXyn-1 adopt the same fold as the proteins from which they are derived, wtGLU and wtXYN. The active-site channels of the GLU and XYN domains are oriented at approximately right angles against each other. The catalytic glutamic acid residues of both face the solvent, explaining the observed enzymatic activity toward both substrates, lichenan and xylan.
Figure 2.
Schematic drawing of the crystal structure of GluXyn-1. Both polypeptide chain termini are in the GLU domain (Upper) of the insertion fusion protein. In the molscript (40) diagram, the N-terminal part of the GLU domain is colored in green, the XYN domain (Lower) is in blue, and the C-terminal part of the GLU domain, after XYN in the sequence, is in yellow. β-Sheets are shown as curved arrows, α-helices are shown as red, wound ribbons, the calcium ion bound to the GLU domain is shown as a purple sphere half concealed by a β-strand, and the disulfide bridge of the GLU domain is shown in ball-and-stick representation (gold).
The GLU domain of GluXyn-1 has a conformation closely similar to wtGLU (4) and its circularly permuted variant cpMAC-57 (9). It is characterized by two curved β-sheets forming a sandwich-like jellyroll fold with a deep active-site channel known to bind the polysaccharide substrate and to harbor the catalytic residues (8). The disulfide bond between Cys-30 and Cys-59 is closed, and a calcium ion is bound to a site that is remote from the active site of the GLU domain by coordination to the same protein residues as in wtGLU and in cpMAC-57. Least-squares superpositions between the GLU domains and wtGLU or cpMAC-57 yield rms displacements between equivalent α-carbon atoms comparable to those obtained for the fit of the two independent GLU domains in the crystal structure of GluXyn-1 (0.3–0.4 Å), confirming that this domain has adopted a native-like fold in spite of the insertion of the complete XYN domain.
The XYN domain of GluXyn-1 has a conformation closely similar to the B. circulans 1,4-β-xylanase (13). It consists of two antiparallel β-sheets whose curvature gives rise to the substrate-binding channel. In contrast to GLU, which contains only single-turn fragments of α-helix, the XYN domain, as the protein from which it is derived, contains one α-helix of three complete turns. The least-squares superposition of the two XYN domains of the asymmetric unit with wtXYN (13) yields rms displacements between 0.3 and 0.4 Å. As before, these values agree with the superposition of the two XYN domains of the GluXyn-1 crystal (0.3 Å) and confirm that the protein domains are identical within the limits of accuracy of the structure determination.
Interdomain Orientation.
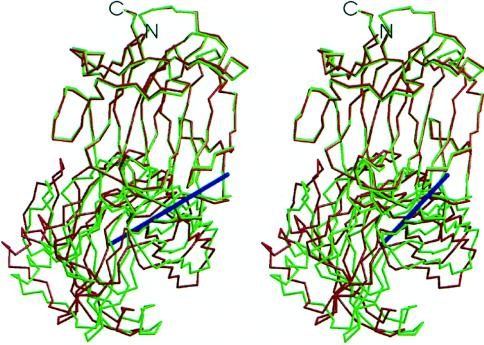
Least-squares superposition of both GLU domains in the asymmetric unit of the crystal yields a displacement of the XYN domains corresponding to a rotation of 14.6° (Fig. 3). The variability in interdomain orientation is limited by the design of GluXyn-1 where the polypeptide chain crosses between the domains twice. Both connections are well ordered and unambiguously defined in electron density (not shown). Depending on the conformation of Trp-185 of the XYN domain, the solvent-accessible surface buried in the domain interface of molecule A is 513 Å2 or 610 Å2 (467 Å2 in molecule B). In both independent molecules, this Trp-185 contributes the largest contact surface to the domain interface. There is a small number of specific interdomain contacts stabilizing the orientation of the two domains relative to each other. The most prominent contact is a hydrogen bond from Lys-208 (GLU domain) to Asn-141 (XYN domain). Another contact stabilizing the domain interface is the double hydrogen bond between the side chains of Gln-150 and the carboxyl-terminal Asn-212 of the GLU domain. The relatively small solvent-accessible surface buried between the domains and the predominantly hydrophilic nature of the side chains involved are more typical for crystal lattice contacts than for the packing of subunits in oligomeric proteins (32, 33).
Figure 3.
Interdomain orientation in the insertion fusion protein GluXyn-1. Superposition of the GLU domains (top) of the two independent molecules of the asymmetric unit of the crystal (red and green) are shown as a stereographic α-carbon trace. The axis of rotation relating the two XYN domains (bottom) is shown in blue. The orientation is similar to that in Fig. 2.
The domain motion within GluXyn-1 can be attributed almost complete to a change in peptide orientation by 80° of the bond linking Gly-1 and Ser-2 of the GLU domain. No other main-chain torsion angle in the interface region changes by more than 20°. Thus, a very sharply localized conformational transition leads to a major domain rearrangement in GluXyn-1.
DISCUSSION
The bifunctional fusion protein GluXyn-1 was produced to test whether the B. macerans 1,3–1,4-β-glucanase jellyroll can adopt a native-like structure in vivo in spite of the insertion of an autonomous folding unit, here derived from the B. subtilis 1,4-β-xylanase. Earlier circular permutation experiments (9, 10) had shown that the sequential order of secondary structure elements is not crucial for in vivo folding. By proving native-like folding of both domains of GluXyn-1 and demonstrating unchanged enzymatic activities we can show here that, in addition, the spacing of structural elements does not determine the folded state of the jellyroll domain as long as crucial intramolecular contacts are stereochemically allowed to form. Although some intradomain interactions that were short ranged in the sequence of wtGLU, or its circularly permuted variant cpMAC-57, are now separated by the 185 additional amino acid residues of the XYN domain, the tertiary structure and function of the GLU domain are completely preserved.
Circular permutations of polypeptide chains recently have been identified in several natural protein sequences (34, 35) and are accepted as common protein engineering tools (36). However, engineered permutants mainly have been generated with small, single-domain proteins. In contrast, the random circular permutation of genes encoding the catalytic chains of aspartate transcarbamoylase (37) has led to functional protein variants in which the chain termini were found in either of the two globular domains. These proteins are equivalent to GluXyn-1 in the sense that, in each variant, the folding of the domain harboring the termini is interrupted by the other globular domain. This protein, however, has evolved as a two-domain polypeptide with a domain interface that probably is not changed dramatically by the sequence permutations.
In spite of the slightly different interdomain orientations described above, GluXyn-1 is a very stable fusion protein as proven by its efficient in vivo folding, full enzymatic activity at high temperature and easy crystallizability. In addition, GluXyn-1 is inaccessible to proteolytic attack in vivo, and aggregation of misfolded protein is not observed. This is noteworthy in view of often-limited yield in E. coli of correctly folded multidomain proteins, which has been attributed to posttranslational folding (38). The observed stability of GluXyn-1 may in part be because of tight constraints on the relative orientation of the two domains imposed by the backbone crossing twice between them. This stabilization will be absent in linear, end-to-end fusions and may be exploited in certain biotechnological applications. Fusions of glycosyl hydrolases combining different substrate specificities offer the possibility to generate single, tailor-made enzymes highly efficient in hydrolyzing the complex components of plant cell walls, for example. By necessity, insertional fusions are restricted to polypeptides with chain termini located close enough to each other on the molecular surface to be inserted into a surface loop of a recipient protein. This may not be a very serious limitation, however, because it can be shown (39) that the ends can be brought to close proximity by motions of short, solvent-accessible peptide fragments in the large majority of globular proteins.
Acknowledgments
We are grateful to the staff of the European Synchrotron Radiation Facility in Grenoble, France, for help with diffraction data collection and to Y. A. Muller and H. Welfle (Max-Delbrück-Centrum, Berlin) for helpful comments on the manuscript. This study was made possible by grants from the Deutsche Forschungsgemeinschaft to U.H. (He 1318/9-3) and to R.B. (Bo 1113/1-3) and with support from the Fonds der Chemischen Industrie.
ABBREVIATIONS
- wtGLU
1,3–1,4-β-glucanase from B. macerans
- wtXYN
1,4-β-xylanase from B. subtilis
- GluXyn-1
bifunctional insertion fusion protein composed of domains GLU and XYN, which are derived from wtGLU and wtXYN
- cpMAC-57
circularly permuted form of wtGLU resulting from a genetic rearrangement that links N and C termini of wtGLU by a peptide bond and creates new termini at sequence positions 57 and 56
Footnotes
Data deposition: Atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11973 (entry code 1AXK).
References
- 1.Orengo C A, Jones D T, Thornton J M. Nature (London) 1994;372:631–634. doi: 10.1038/372631a0. [DOI] [PubMed] [Google Scholar]
- 2.Stirk H J, Woolfson D N, Hutchinson E G, Thornton J M. FEBS Lett. 1992;308:1–3. doi: 10.1016/0014-5793(92)81036-l. [DOI] [PubMed] [Google Scholar]
- 3.Chelvanayagam G, Heringa J, Argos P. J Mol Biol. 1992;228:220–242. doi: 10.1016/0022-2836(92)90502-b. [DOI] [PubMed] [Google Scholar]
- 4.Hahn M, Olsen O, Politz O, Borriss R, Heinemann U. J Biol Chem. 1995;270:3081–3088. doi: 10.1074/jbc.270.7.3081. [DOI] [PubMed] [Google Scholar]
- 5.Hahn M, Pons J, Planas A, Querol E, Heinemann U. FEBS Lett. 1995;374:221–224. doi: 10.1016/0014-5793(95)01111-q. [DOI] [PubMed] [Google Scholar]
- 6.Heinemann U, A, Gaiser O, Müller J J, Ponnuswamy M N. Biol Chem. 1996;377:447–454. [PubMed] [Google Scholar]
- 7.Olsen O, Borriss R, Simon O, Thomsen K K. Mol Gen Genet. 1991;225:177–185. doi: 10.1007/BF00269845. [DOI] [PubMed] [Google Scholar]
- 8.Keitel T, Simon O, Borriss R, Heinemann U. Proc Natl Acad Sci USA. 1993;90:5287–5291. doi: 10.1073/pnas.90.11.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn M, Piotukh K, Borriss R, Heinemann U. Proc Natl Acad Sci USA. 1994;91:10417–10421. doi: 10.1073/pnas.91.22.10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aÿ J, Hahn M, Decanniere K, Piotukh K, Borriss R, Heinemann U. Proteins Struct Funct Genet. 1998;30:155–167. doi: 10.1002/(sici)1097-0134(19980201)30:2<155::aid-prot5>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 11.Henrissat B, Bairoch A. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf M, Geczi A, Simon O, Borriss R. Microbiology. 1995;141:281–290. doi: 10.1099/13500872-141-2-281. [DOI] [PubMed] [Google Scholar]
- 13.Wakarchuk W W, Campbell R L, Sung W L, Davoodi J, Yaguchi M. Protein Sci. 1994;3:467–475. doi: 10.1002/pro.5560030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gosalbes M J, Perez-Gonzalez A, Gonzalez R, Navarro A. J Bacteriol. 1991;173:7705–7710. doi: 10.1128/jb.173.23.7705-7710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flint H J, Martin J, McPherson J, Daniel S, Zhang J-X. J Bacteriol. 1993;175:2943–2951. doi: 10.1128/jb.175.10.2943-2951.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton R M, Huni H D, Ho S N, Pullen J K, Pease L R. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 17.Borriss R, Büttner K, Mäntsälä P. Mol Gen Genet. 1990;222:278–283. doi: 10.1007/BF00633829. [DOI] [PubMed] [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Politz O, Simon O, Olsen O, Borriss R. Eur J Biochem. 1993;216:829–834. doi: 10.1111/j.1432-1033.1993.tb18204.x. [DOI] [PubMed] [Google Scholar]
- 20.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 21.Borriss R, Olsen O, Thomsen K K, von Wettstein D. Carlsberg Res Commun. 1989;54:41–54. doi: 10.1007/BF02907584. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z. DENZO: An Oscillation Data Processing Program for Macromolecular Crystallography. New Haven, CT: Yale Univ. Press; 1993. [Google Scholar]
- 23.Collaborative Computational Project, Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 24.Matthews B W. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 25.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 26.Brünger A T. Nature (London) 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 27.Brünger A T, Krukowski A, Erickson J W. Acta Crystallogr A. 1990;46:585–593. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov G N, Vagin A A, Dodson E J. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Read R J. Acta Crystallogr A. 1986;42:140–149. [Google Scholar]
- 30.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 31.Luzzati V. Acta Crystallogr. 1952;5:802–810. [Google Scholar]
- 32.Jones S, Thornton J M. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dasgupta S, Iyer G H, Bryant S H, Lawrence C E, Bell J A. Proteins Struct Funct Genet. 1997;28:494–514. doi: 10.1002/(sici)1097-0134(199708)28:4<494::aid-prot4>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 34.Heinemann U, Hahn M. Trends Biochem Sci. 1995;20:349–350. doi: 10.1016/s0968-0004(00)89073-8. [DOI] [PubMed] [Google Scholar]
- 35.Lindqvist Y, Schneider G. Curr Opin Struct Biol. 1997;7:422–427. doi: 10.1016/s0959-440x(97)80061-9. [DOI] [PubMed] [Google Scholar]
- 36.Heinemann U, Hahn M. Prog Biophys Mol Biol. 1996;64:121–143. doi: 10.1016/0079-6107(95)00013-5. [DOI] [PubMed] [Google Scholar]
- 37.Graf R, Schachman H K. Proc Natl Acad Sci USA. 1996;93:11591–11596. doi: 10.1073/pnas.93.21.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Netzer W J, Hartl F U. Nature (London) 1997;388:343–349. doi: 10.1038/41024. [DOI] [PubMed] [Google Scholar]
- 39.Christopher J A, Baldwin T O. J Mol Biol. 1996;257:175–187. doi: 10.1006/jmbi.1996.0154. [DOI] [PubMed] [Google Scholar]
- 40.Kraulis J P. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 41.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]