Abstract
The phenylpropanoid pathway provides precursors for the biosynthesis of soluble secondary metabolites and lignin in plants. Ferulate-5-hydroxylase (F5H) catalyzes an irreversible hydroxylation step in this pathway that diverts ferulic acid away from guaiacyl lignin biosynthesis and toward sinapic acid and syringyl lignin. This fact led us to postulate that F5H was a potential regulatory step in the determination of lignin monomer composition. To test this hypothesis, we have used Arabidopsis to examine the impact of F5H overexpression. Arabidopsis is a useful model system in which to study lignification because in wild-type plants, guaiacyl and syringyl lignins are deposited in a tissue-specific fashion, while the F5H-deficient fah1 mutant accumulates only guaiacyl lignin. Here we show that ectopic overexpression of F5H in Arabidopsis abolishes tissue-specific lignin monomer accumulation. Surprisingly, overexpression of F5H under the control of the lignification-associated cinnamate-4-hydroxylase promoter, but not the commonly employed cauliflower mosaic virus 35S promoter, generates a lignin that is almost entirely comprised of syringylpropane units. These experiments demonstrate that modification of F5H expression may enable engineering of lignin monomer composition in agronomically important plant species.
Lignin is a phenolic polymer that imparts strength and decay resistance to the plant secondary cell wall and is thought to have been essential to the evolution of terrestrial plants (1, 2). Precursors for lignin biosynthesis are synthesized from l-phenylalanine via the phenylpropanoid pathway that provides ferulic acid (4-hydroxy-3-methoxycinnamic acid) and sinapic acid (3,5-dimethoxy-4-hydroxycinnamic acid) for the synthesis of guaiacyl- and syringyl-substituted lignin monomers, respectively. In angiosperms, lignin biosynthesis requires the activity of at least two cytochrome P450-dependent monooxygenases, cinnamate-4-hydroxylase (C4H) (3, 4), and ferulate-5-hydroxylase (F5H) (5), although the activity of F5H is required only for the synthesis of syringyl monomers (6).
The mechanism(s) by which plants control lignin monomer composition is of substantial interest (7). Lignin monomer composition has a significant effect on the ease with which lignin can be degraded during industrial pulping (8, 9), and also influences forage digestibility (10). If lignin monomer composition could be manipulated by biotechnological means, the cost of pulp production could be significantly decreased, and the value of animal feedstocks would be increased due to their improved nutritional value. These applications have inspired many research efforts directed toward lignin modification (11).
The balance between guaiacyl and syringyl units in lignin varies among plant species (12), within a given plant, and even within the wall of a single plant cell (13). Several mechanisms have been proposed for the control of lignin monomer composition, including enzyme substrate specificity and the transcriptional regulation of genes encoding enzymes of monomer-specific pathways (11); however, no conclusive evidence has been reported to support these hypotheses. Meyer et al. (14) recently cloned the gene encoding F5H, and we report here the results of a series of transgenic studies designed to evaluate whether the expression of the F5H gene regulates lignin monomer composition in Arabidopsis.
MATERIALS AND METHODS
Plant Material.
Arabidopsis thaliana was grown under a 16-hr light/8-hr dark photoperiod at 100 μE · m−2 · s−1 at 22°C, cultivated in ProMix potting mixture (Premier Horticulture, Red Hill, PA).
Analysis of Nucleic Acids.
RNA was extracted from plant tissues (15), electrophoretically separated, transferred to Hybond N+ membrane (Amersham), and hybridized with radiolabeled probes prepared from cDNA or genomic clones according to standard protocols. Sequence analysis was performed on plasmid DNA using the United States Biochemical Sequenase kit (version 2.0) using standard vector-based sequencing oligonucleotides or custom-synthesized oligonucleotides as appropriate.
Generation of Plant Transformation Constructs.
The C4H-F5H transcriptional fusion construct was generated by using a 2,897-bp fragment of the C4H promoter (16) and a 2,719-bp fragment of the F5H genomic sequence (14) fused 50-bp upstream of the inferred F5H ATG start codon. As a result, the C4H promoter drives the expression of the F5H gene using the C4H transcription start site and the termination signal present on the F5H genomic sequence. A small fragment of pGEM-7Zf(+) (Promega) polylinker sequence remains in this construct at the C4H:F5H fusion junction. This expression cassette was inserted into the T-DNA of the binary vector pGA482 (17) to give pGA482-C4H:F5H. The generation of the pGA482-35S:F5H construct has been described (14).
Plant Transformation.
Plant transformation constructs were introduced into Agrobacterium tumefaciens C58 pGV3850 (18) by electroporation (19). Stability of constructs was confirmed by restriction analysis of plasmid DNA isolated from A. tumefaciens. Cultures harboring the binary vectors were used to transform the fah1–2 mutant by vacuum infiltration (16). Kanamycin-resistant seedlings derived from independent infiltration experiments were grown in soil and permitted to set seed. Plants from seed stocks that segregated 3:1 for kanamycin-resistant progeny were again permitted to set seed, and homozygous transgenic lines were reselected in the next generation.
Histochemistry.
Staining of hand sections of Arabidopsis rachis internodes was conducted as described (6).
Nitrobenzene Oxidation.
For the determination of lignin monomer composition, stem tissue of mature, 5-week-old Arabidopsis plants was ground to a powder in liquid nitrogen and extracted with 20 ml of 0.1 M sodium phosphate buffer (pH 7.2) for 30 min at 37°C followed by three extractions with 80% ethanol at 80°C. The tissue was then extracted once with acetone and dried. Tissue was saponified by treatment with 1.0 M NaOH for 24 hr at 37°C, washed three times with water, once with 80% ethanol, once with acetone, and dried. Nitrobenzene oxidation of stem tissue samples was performed with a protocol modified from Iiyama and Lam (20). Samples of lignocellulosic material (5 mg each) were mixed with 500 μl of 2 M NaOH and 25 μl of nitrobenzene. This mixture was incubated in a sealed glass tube at 160°C for 3 hr. The reaction products were cooled to room temperature and 5 μl of a 20 mg ml−1 solution of 3-ethoxy-4-hydroxybenzaldehyde in pyridine was added as an internal standard before the mixture was extracted twice with 1 ml of dichloromethane. The aqueous phase was acidified with HCl (pH 2) and extracted twice with 900 μl of ether. The combined ether phases were dried with anhydrous sodium sulfate and the ether was evaporated in a stream of nitrogen. The dried residue was resuspended in 50 μl of pyridine, 10 μl of BSA [N,O-bis-(trimethylsilyl)-trifluoracetamide)] was added and 1 μl aliquots of the silylated products were analyzed using a Hewlett-Packard 5890 series II gas chromatograph equipped with a Supelco SPB I column (30 m × 0.75 mm). Lignin monomer composition was calculated from the integrated areas of the peaks representing the trimethylsilylated derivatives of vanillin, syringaldehyde, vanillic acid and syringic acid. Total nitrobenzene oxidation-susceptible guaiacyl units (vanillin and vanillic acid) and syringyl units (syringaldehyde and syringic acid) were determined after correction for recovery efficiencies for each of the products during the extraction procedure relative to the internal standard. The identity of each of the peaks used for quantitation of lignin monomer composition was confirmed using GC-electron impact MS by comparison to authentic compounds.
Analysis of Lignin Using the Derivatization Followed by Reductive Cleavage Method.
As an independent measure of lignin monomer composition, the buffer-extracted and saponified cell wall samples described above were also analyzed using the DFRC protocol (21).
RESULTS AND DISCUSSION
Characterization of Lignification in Wild-Type Arabidopsis.
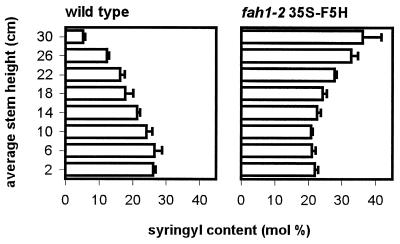
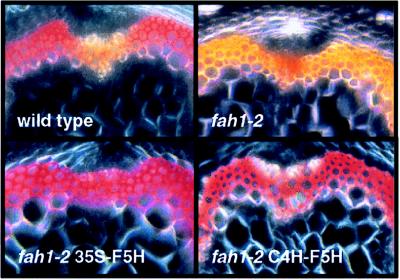
The lignin of the mature Arabidopsis rachis contains guaiacyl and syringyl residues in an overall ratio of ≈4:1; however, this ratio is not constant throughout plant development. The syringyl content of the rachis increases from <6 mol% within the apical 4 cm of the bolt to >26 mol% near the base of the inflorescence (Fig. 1). Histochemical staining of Arabidopsis rachis cross-sections indicates that syringyl lignin biosynthesis is also developmentally regulated in a tissue-specific manner (6, and Fig. 2). Syringyl lignin accumulation is restricted to the cells of the sclerified parenchyma that flank the vascular bundles while guaiacyl lignin is deposited only in the cells of the vascular bundle. The increase in syringyl lignin content during rachis development is a consequence of sclerified parenchyma maturation as these cells undergo secondary thickening after the vascular bundle has been formed from the cells of the procambium (22).
Figure 1.
Developmental changes in lignin monomer composition during development of the Arabidopsis rachis in wild type and 35S-F5H transgenics. Lignin monomer composition was determined by nitrobenzene oxidation of lignocellulosic material derived from 4-cm rachis segments from 5-week-old Arabidopsis plants.
Figure 2.
Histochemical staining for lignin monomer composition in Arabidopsis stem cross-sections. Lower rachis segments were hand sectioned, stained with the Mäule reagent and observed by light microscopy using cross-polarizing optics. Red staining indicates the presence of syringyl residues in the plant secondary cell wall.
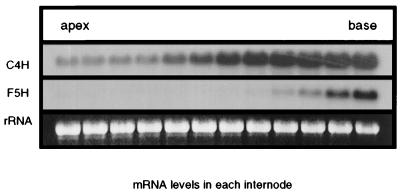
To evaluate whether transcriptional regulation of the F5H gene may play a role in the control of lignin monomer composition during rachis development, hybridization experiments were performed using RNA isolated from elongating rachis internodes. In these experiments, changes in the steady-state levels of the F5H transcript were compared with those of C4H, which was used as an internal measure of phenylpropanoid gene expression (Fig. 3). These studies revealed that the expression of F5H mRNA is significantly delayed relative to that of C4H. C4H mRNA is detected in even the uppermost internodes examined (≅2 mm long) whereas a significant level of F5H expression was not observed until the ninth internode.
Figure 3.
RNA blot analysis of F5H and C4H expression in Arabidopsis rachis internodes. RNA was isolated from the uppermost 12 internodes of 5-week-old Arabidopsis plants beginning near the top of the inflorescence with those internodes ≥2 mm in length. Blots were probed with cDNAs corresponding to the Arabidopsis C4H and F5H genes. Equal loading of lanes was verified by ethidium bromide staining of the 28S ribosomal RNA band.
Overexpression of F5H Using the Cauliflower Mosaic Virus (CaMV) 35S Promoter.
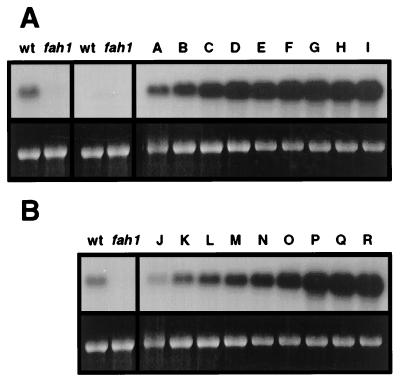
The above findings are consistent with the hypothesis that syringyl lignin accumulation is regulated at the level of F5H transcription. To test this hypothesis in transgenic plants, we generated a chimaeric F5H construct (35S-F5H) in which regulatory sequences of the F5H gene were replaced with the strong constitutive CaMV 35S promoter (14). This construct was transformed into fah1–2 mutant plants in which F5H transcript levels are below detectable limits (14). This strategy was employed to maximize levels of F5H mRNA expression while avoiding epigenetic gene silencing effects that were observed in a wild-type genetic background (not shown). CaMV 35S promoter-driven F5H gene expression resulted in steady-state levels of F5H mRNA that were 10–60 times higher than those found in wild-type plants (Fig. 4). The syringyl content of rachis tissue in these plants was found to be as high as 29 mol% and was positively correlated with the abundance of the F5H transcript in each line (Table 1). In comparison, the lignin of wild-type rachis tissue was found to contain 18 mol% of syringyl residues. In addition, histochemical staining of rachis cross sections indicated that the tissue specificity of syringyl lignin deposition was abolished in transgenic lines ectopically expressing F5H mRNA (Fig. 2). Syringyl unit deposition was no longer restricted to the cells of the sclerified parenchyma but was also found in the lignin deposited by the cells of the vascular bundle. This indicates that cells of the vascular bundle are competent to synthesize, secrete, and polymerize monolignols derived from sinapic acid if they are made competent to express an active F5H gene. Taken together, these data demonstrate that F5H gene expression not only determines lignin monomer composition in a quantitative fashion but also the tissue specificity of syringyl lignin deposition.
Figure 4.
Impact of promoter choice on F5H overexpression in transgenic Arabidopsis. Stem tissue from 5-week-old plants of the wild type, the fah1–2 mutant, and nine independent fah1–2 lines homozygous for the 35S-F5H transgene (A) was harvested and used for RNA isolation. Blots were probed with the F5H cDNA and were exposed to film for 24 hr to visualize the level of F5H expression in the wild type and the fah1–2 mutant (Left), and for 2 hr to evaluate F5H expression in the 35S-F5H transgenics (Right). Identical analyses were carried out on 5-week-old stem tissue from the wild type, the fah1–2 mutant, and nine fah1–2 lines homozygous for the C4H-F5H transgene (B). Blots were probed with the F5H cDNA and were exposed to film for 12 hr to visualize the level of F5H expression.
Table 1.
Impact of 35S promoter-driven F5H overexpression on lignin monomer composition in transgenic Arabidopsis
| Line | Total G units* (μmol g−1 d.w.) | Total S units† (μmol g−1 d.w.) | Total G+S units (μmol g−1 d.w.) | Mol% S |
|---|---|---|---|---|
| Wild type | 168 ± 16 | 37.5 ± 4.5 | 205 ± 21 | 18.4 ± 0.91 |
| fahl-2 | 272 ± 23 | ND | 272 ± 23 | – |
| A | 332 ± 38 | 17.5 ± 2.0 | 350 ± 40 | 5.06 ± 0.17 |
| B | 211 ± 18 | 33.5 ± 3.5 | 244 ± 21 | 13.7 ± 0.55 |
| C | 204 ± 17 | 48.5 ± 3.0 | 253 ± 19 | 19.2 ± 0.56 |
| D | 187 ± 10 | 46.5 ± 2.5 | 233 ± 11 | 19.9 ± 0.86 |
| E | 270 ± 24 | 79.5 ± 9.0 | 349 ± 33 | 22.7 ± 0.82 |
| F | 287 ± 30 | 98.0 ± 16 | 385 ± 45 | 25.3 ± 1.23 |
| G | 193 ± 16 | 67.0 ± 5.5 | 260 ± 20 | 25.8 ± 0.78 |
| H | 161 ± 15 | 59.0 ± 6.5 | 220 ± 22 | 28.8 ± 0.92 |
| I | 173 ± 11 | 69.5 ± 8.5 | 253 ± 19 | 27.5 ± 1.80 |
Stem tissue from the plants described in Fig. 4 was used for lignin monomer composition by nitrobenzene oxidation. Average values of 10 replicates and their SDs are shown.
*Sum of vanillin + vanillic acid.
Sum of syringaldehyde + syringic acid.
ND, not detectable.
Although overexpression of the F5H gene led to an increase in syringyl lignin content in our transgenic plants, we were surprised to find that a significant proportion of their lignin was still derived from ferulic acid despite the high levels of F5H gene expression that were obtained (Fig. 4, Table 1). The nine transgenic lines described in detail here were identified among a total of 100 independent transgenic lines all of which were screened with respect to the syringyl content of their lignin. None of the lines examined accumulated a lignin with a syringyl content that exceeded 35 mol%. Similar experiments were conducted with constructs in which an enhanced version of the CaMV 35S promoter (23) was employed or in which the F5H cDNA was ectopically expressed using the CaMV 35S promoter and the nopaline synthase termination signal. These experiments provided similar results excluding the possibility that the 3′ or intronic sequences of the genomic F5H sequence were responsible for the limited efficacy of the CaMV 35S-F5H construct. These data indicated that some other factor limits syringyl monomer deposition in the 35S-F5H transgenic plants.
To explore what might cause this limitation and to further characterize the impact of CaMV 35S-driven F5H expression, developmental changes in lignin monomer composition were determined in the transgenic line H, which was homozygous for the 35S-F5H transgene (Fig. 1). Ectopic expression of the F5H gene led to a high syringyl content in apical rachis segments that gradually decreased in more mature internodes. This contrasts sharply with the pattern seen in the wild type where syringyl content was low near the apex and increased basipetally.
Overexpression of F5H Using the C4H Promoter.
The above observations led us to generate several hypotheses regarding what factor(s) might limit syringyl content in F5H-overexpressing transgenic lines. First, another enzyme required for syringyl lignin biosynthesis may become rate-limiting in the context of F5H overexpression. Second, the genes encoding hydroxycinnamoyl CoA ligase or enzymes of the so-called “alternative pathway” (24–25) may be more highly expressed later in rachis development and may permit phenylpropanoid precursors to escape hydroxylation by F5H. Similarly, the intracellular ferulate pools may have greater accessibility to the soluble hydroxycinnamoyl CoA ligase than to the membrane-localized F5H, thereby limiting the impact of F5H overexpression. And third, although the CaMV 35S promoter is often used as a strong, non-tissue-specific promoter, it may fail to promote high levels of F5H gene expression in the specialized cells generating the precursors for lignin biosynthesis.
To test the third hypothesis, we reasoned that a promoter whose role is to target gene expression to lignifying cells might be a better choice for manipulation of F5H expression than the CaMV 35S promoter. The C4H promoter was chosen for these experiments because C4H is expressed at high levels early in rachis development (Fig. 3), and because we had previously shown that a 2.9-kb fragment of the C4H promoter is capable of directing reporter gene expression to lignifying cells of the Arabidopsis rachis (16). We thus generated a new construct in which F5H transcription was driven by regulatory sequences of the C4H gene and this chimaeric F5H gene was again transformed into fah1–2 mutant plants. Lignin analysis of transgenic stem tissue revealed that expression of the F5H gene under the control of the C4H promoter resulted in the production of a lignin with a syringyl content that greatly exceeded that observed in the 35S-F5H transgenics (Table 2), despite the fact that the levels of F5H mRNA in these transgenic lines were substantially lower than those in the 35S-F5H transgenics (Fig. 4). In several of the transgenic lines (Fig. 4 and Table 2, lines N, Q, R), the lignin was almost solely comprised of syringyl residues. As in the 35S-F5H transgenics, the tissue-specificity of syringyl lignin deposition was abolished in plants carrying the C4H-F5H transgene (Fig. 2). Interestingly, many of the C4H-F5H transgenics contained some tracheary elements that failed to stain with the Mäule reagent (Fig. 2). When grown under the same controlled conditions, the C4H-F5H transgenic plants showed no obvious phenotypic differences that distinguish them from wild-type plants.
Table 2.
Impact of C4H promoter-driven F5H overexpression on lignin monomer composition in transgenic Arabidopsis
| Line | Total G units* (μmol g−1 d.w.) | Total S units† (μmol g−1 d.w.) | Total G+S units (μmol g−1 d.w.) | Mol% S |
|---|---|---|---|---|
| Wild type | 241 ± 31 | 59.0 ± 14 | 300 ± 43 | 19.6 ± 2.3 |
| fahl-2 | 314 ± 64 | ND | 314 ± 63 | – |
| J | 62.0 ± 12 | 296 ± 72 | 358 ± 84 | 82.5 ± 0.97 |
| K | 199 ± 36 | 180 ± 30 | 378 ± 66 | 47.5 ± 0.96 |
| L | 213 ± 33 | 173 ± 24 | 385 ± 55 | 44.8 ± 1.7 |
| M | 116 ± 17 | 277 ± 23 | 392 ± 36 | 70.6 ± 1.9 |
| N | 43.0 ± 8.0 | 384 ± 64 | 426 ± 70 | 90.1 ± 0.26 |
| O | 73.0 ± 9.0 | 253 ± 17 | 326 ± 22 | 77.6 ± 2.0 |
| P | 65.0 ± 5.0 | 375 ± 34 | 440 ± 38 | 85.2 ± 0.76 |
| Q | 27.0 ± 3.5 | 308 ± 47 | 334 ± 50 | 92.1 ± 0.42 |
| R | 41.0 ± 6.5 | 369 ± 80 | 410 ± 86 | 90.0 ± 0.50 |
Stem tissue from the plants described in Fig. 4 was used for lignin monomer composition by nitrobenzene oxidation. Average values of five replicates and their SDs are shown.
*Sum of vanillin + vanillic acid.
Sum of syringaldehyde + syringic acid.
ND, not detectable.
Lignin Analysis Using the DFRC Method.
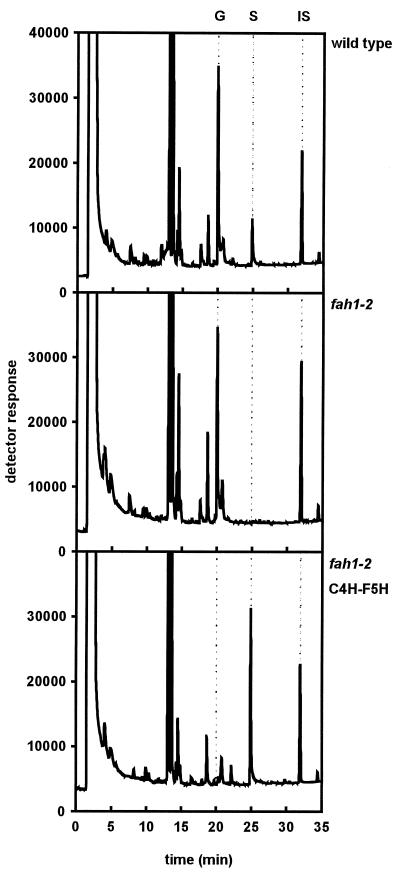
Because lignin is insoluble and resistant to chemical degradation, its accurate characterization in plant samples is technically challenging (2). For this reason, it is often argued that the analysis of lignin is best approached by using several complementary methods. To determine whether the impact of F5H overexpression could be verified by an independent method of lignin analysis, we used the recently developed DFRC protocol (21) to analyze the lignin monomer composition in cell walls of wild type, fah1, and a C4H-F5H transgenic (line Q) (Fig. 5). This method efficiently cleaves α- and β-aryl ethers to provide high yields of hydroxycinnamyl alcohol acetates that can be quantified by gas chromatography. In addition, these products are only generated from β-etherified precursors, thus this method is specific for lignin. DFRC analysis completely supported our nitrobenzene oxidation results. This independent lignin analysis method showed that wild-type Arabidopsis accumulates a lignin that is dominated by guaiacyl units, verified the absence of syringyl units in the fah1 mutant lignin, and again showed that F5H overexpression can lead to the accumulation of a lignin that appears to be derived almost solely from syringyl units (Fig. 5).
Figure 5.
GC analysis of DFRC lignin degradation products from representative lines described in Fig. 4 and Table 2. Retention times of trans-coniferyl alcohol diacetate (G), trans-sinapyl alcohol diacetate (S), and the tetracosane internal standard (IS) are indicated.
Conclusion.
Our results demonstrate that the composition of the lignin polymer is dictated by the temporal and tissue-specific expression pattern of the F5H gene in Arabidopsis. It is important to note that in none of our plants have we been able to directly measure the enzymatic activity of F5H; however, based on the phenotype of our transgenic plants it seems clear that this strategy must lead to a substantial increase in enzymatic activity. The evaluation of this hypothesis must await the development of appropriate enzymatic assays or antibody probes.
We have also shown that the CaMV 35S promoter, which frequently has been used in transgenic studies aimed at the modification of lignin biosynthesis (26–30), fails to promote high levels of F5H gene expression in cells undergoing or providing precursors for lignification. The promoter of the C4H gene used in this study is far more efficient in this regard and will be a very valuable tool in transgenic studies addressing plant lignification in the future. These data also indicate that the use of other endogenous promoters in biotechnological applications may enhance not only tissue-specificity but also tissue-efficacy of transgene expression when compared with non-specific ectopic promoters such as the CaMV 35S promoter.
Finally, we have shown that it is possible to genetically engineer plants to accumulate lignin that is highly enriched in syringyl residues, a type of lignin that is otherwise rare in nature (31). These lignins are thought to be more readily degraded during the pulping process and are less inhibitory to ruminant digestion of lignocellulosic feedstocks (8–10). The availability of Arabidopsis lines with pure guaiacyl (fah1), mixed guaiacyl/syringyl (wild type and 35S-F5H), and highly syringyl-enriched (C4H-F5H) lignins permits the investigation of the importance of lignin monomer composition on tracheary element function and lignin degradability. The apparently unaltered morphology of tracheary elements and sclerified parenchyma in transgenic plants depositing lignin highly enriched in syringyl units suggests that this lignin still provides lignified cells with sufficient rigidity to function normally in water conduction and mechanical support. Thus, it seems possible to increase the syringyl content of crop species and trees, thereby generating lignins that are easier to digest or extract without detrimental consequences on agricultural performance.
Acknowledgments
We thank Drs. Jo Ann Banks and Malcolm Campbell for careful review of this manuscript, and Dr. John Ralph for supplying authentic standards for DFRC analysis. This is journal paper no. 15700 of the Purdue University Agricultural Experiment Station. This work was supported by a grant from the Division of Energy Biosciences, United States Department of Energy to C.C., and Postdoctoral Fellowships from the Swiss National Science Foundation and the Alexander von Humboldt Foundation (Feodor Lynen Fellowship) to K.M.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: C4H, cinnamate-4-hydroxylase; F5H, ferulate-5-hydroxylase; CaMV, cauliflower mosaic virus; DFRC, derivatization followed by reductive cleavage.
References
- 1.Higuchi T. In: Plant Carbohydrates II. Tanner W, Loewus F A, editors. New York: Springer; 1981. pp. 194–224. [Google Scholar]
- 2.Lewis N G, Yamamoto E. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:455–496. doi: 10.1146/annurev.pp.41.060190.002323. [DOI] [PubMed] [Google Scholar]
- 3.Teutsch H-G, Hasenfratz M P, Lesot A, Stoltz C, Garnier J-M, Jeltsch J-M, Durst F, Werck-Reichhardt D. Proc Natl Acad Sci USA. 1993;90:4102–4106. doi: 10.1073/pnas.90.9.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizutani M, Ward E, Ohta D, Ryals J, Sato R. Biochem Biophys Res Commun. 1993;190:875–880. doi: 10.1006/bbrc.1993.1130. [DOI] [PubMed] [Google Scholar]
- 5.Grand C. FEBS Lett. 1984;169:7–11. [Google Scholar]
- 6.Chapple C C S, Vogt T, Ellis B E, Somerville C R. Plant Cell. 1992;4:1413–1424. doi: 10.1105/tpc.4.11.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whetten R, Sederoff R. Forest Ecol Management. 1991;43:301–316. [Google Scholar]
- 8.Chiang V L, Puumala R J, Takeuchi H, Eckert R E. Tappi J. 1988;71:173–176. [Google Scholar]
- 9.Chiang V L, Funaoka M. Holzforschung. 1990;44:309–313. [Google Scholar]
- 10.Jung H G, Deetz D A. In: Forage Cell Wall Structure and Digestibility. Jung H G, Buxton D R, Hatfield R D, Ralph J, editors. Madison, WI: Am. Soc. Agron./Crop Sci. Soc. Am./Soil Sci. Soc. Am. Press; 1993. pp. 315–346. [Google Scholar]
- 11.Campbell M M, Sederoff R R. Plant Physiol. 1996;110:3–13. doi: 10.1104/pp.110.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarkanen K V, Chang H-M, Allan G G. Tappi J. 1967;50:587–590. [Google Scholar]
- 13.Monties B. In: Annual Proceedings of the Phytochemical Society of Europe. van Sumere C F, Lea PE, editors. Vol. 25. Oxford, U.K.: Clarendon; 1985. pp. 161–181. [Google Scholar]
- 14.Meyer K, Cusumano J C, Somerville C, Chapple C C S. Proc Natl Acad Sci USA. 1996;93:6869–6874. doi: 10.1073/pnas.93.14.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldsbrough P B, Cullis C A. Nucleic Acids Res. 1981;9:1301–1309. doi: 10.1093/nar/9.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell-Lelong D A, Cusumano J C, Meyer K, Chapple C. Plant Physiol. 1997;113:729–738. doi: 10.1104/pp.113.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An G. Methods Enzymol. 1987;153:292–305. [Google Scholar]
- 18.Zambrisky P, Joos H, Genetello C, Leemans J, van Montagu M, Schell J. EMBO J. 1983;2:2143–2150. doi: 10.1002/j.1460-2075.1983.tb01715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer K, Leube M, Grill E. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- 20.Iiyama K, Lam T B T. J Sci Food Agric. 1990;51:481–491. [Google Scholar]
- 21.Lu F, Ralph J. J Agric Food Chem. 1997;45:2590–2592. [Google Scholar]
- 22.Turner S R, Somerville C R. Plant Cell. 1997;9:689–701. doi: 10.1105/tpc.9.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kay R, Chan A, Daly M, McPherson J. Science. 1987;236:1299–1302. doi: 10.1126/science.236.4806.1299. [DOI] [PubMed] [Google Scholar]
- 24.Ye Z-H, Kneusel R E, Matern U, Varner J E. Plant Cell. 1994;6:1427–1439. doi: 10.1105/tpc.6.10.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye Z-H, Varner J E. Plant Physiol. 1995;108:459–467. doi: 10.1104/pp.108.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atanassova R, Favet N, Martz F, Chabbert B, Tollier M-T, Monties B, Fritig B, Legrand M. Plant J. 1995;8:465–477. [Google Scholar]
- 27.Van Doorsselaere J, Baucher M, Chognot E, Chabbert B, Tollier M-T, Petit-Conil M, Leplé J-C, Pilate G, Cornu D, Monties B, et al. Plant J. 1995;8:855–864. [Google Scholar]
- 28.Dwivedi U N, Campbell W H, Yu J, Datla R S S, Bugos R C, Chiang V L, Podila G K. Plant Mol Biol. 1994;26:61–71. doi: 10.1007/BF00039520. [DOI] [PubMed] [Google Scholar]
- 29.Halpin C, Knight M E, Foxon G A, Campbell M M, Boudet A M, Boon J J, Chabbert B, Tollier M-T, Schuch W. Plant J. 1994;6:339–350. [Google Scholar]
- 30.Ni W, Paiva N L, Dixon R A. Transgenic Res. 1994;3:120–126. [Google Scholar]
- 31.Ralph J. J Natural Products. 1996;59:341–342. [Google Scholar]