Abstract
Cytochrome P450 3A4 is generally considered to be the most important human drug-metabolizing enzyme and is known to catalyze the oxidation of a number of substrates in a cooperative manner. An allosteric mechanism is usually invoked to explain the cooperativity. Based on a structure–activity study from another laboratory using various effector–substrate combinations and on our own studies using site-directed mutagenesis and computer modeling of P450 3A4, the most likely location of effector binding is in the active site along with the substrate. Our study was designed to test this hypothesis by replacing residues Leu-211 and Asp-214 with the larger Phe and Glu, respectively. These residues were predicted to constitute a portion of the effector binding site, and the substitutions were designed to mimic the action of the effector by reducing the size of the active site. The L211F/D214E double mutant displayed an increased rate of testosterone and progesterone 6β-hydroxylation at low substrate concentrations and a decreased level of heterotropic stimulation elicited by α-naphthoflavone. Kinetic analyses of the double mutant revealed the absence of homotropic cooperativity with either steroid substrate. At low substrate concentrations the steroid 6β-hydroxylase activity of the wild-type enzyme was stimulated by a second steroid, whereas L211F/D214E displayed simple substrate inhibition. To analyze L211F/D214E at a more mechanistic level, spectral binding studies were carried out. Testosterone binding by the wild-type enzyme displayed homotropic cooperativity, whereas substrate binding by L211F/D214E displayed hyperbolic behavior.
Human cytochrome P450 3A4 (P450 3A4) is one of the most abundant xenobiotic-metabolizing enzymes in the human liver (1) and intestine (2, 3) and is capable of oxidizing a wide range of structurally diverse drugs, including antineoplastic, antihistaminic, cardiac, psychotropic, analgesic, hormonal, and immunosuppresant agents (4). In addition to its broad substrate specificity and abundance, another important aspect of the metabolic activity of P450 3A4 is its apparent allosteric nature. P450 3A4 demonstrates homotropic cooperativity toward a number of substrates, including progesterone (5–7), testosterone (8), 17β-estradiol (8), aflatoxin B1 (8), and amitriptyline (8). In addition, the activity of the enzyme can be influenced heterotropically. For example, α-naphthoflavone (ANF) stimulates the oxidation of progesterone (5–7), testosterone (7), estradiol (9), aflatoxins (10, 11), polycyclic aromatic hydrocarbons (12, 13), carbamazepine (14), and acetaminophen (15). Similarly, progesterone can heterotropically stimulate carbamazepine metabolism by P450 3A4 (14), and both progesterone and testosterone stimulate estradiol metabolism (9).
The most extensively studied allosteric protein is probably hemoglobin. The molecular mechanisms of both the homotropic action of oxygen binding and the heterotropic control exerted by binding of CO2 were initially determined by a combination of x-ray crystallographic data and the availability of naturally occurring amino acid substitutions (16). The understanding of the cooperative nature of P450 3A4 to the same level of detail as hemoglobin has been hindered by the lack of both an x-ray crystal structure and amino acid substitution mutants that affect cooperativity. Three-dimensional (3D) computer models of P450 based on the premise of structural homology between the mammalian P450s and one or more of the four crystallized bacterial P450 enzymes have proven to be useful tools for understanding site-directed mutagenesis data. A homology model of P450 3A4 (17) suggests that the active site of the enzyme is very large and probably capable of accommodating more than one substrate at a time in the area of the active site. Studies using flavonoids as heterotropic stimulators of polycyclic aromatic hydrocarbon metabolism have suggested that the effector and substrate are probably both present in the active site at the same time (13). The presence of two different substrates simultaneously in the active site has also been used to explain the partial competitive inhibition of testosterone metabolism by erythromycin (18).
We had shown previously that alteration of P450 3A4 residues predicted to be in or very near the enzyme active site reduced heterotropic stimulation of steroid metabolism by ANF (7). One hypothesis for the mechanism of cooperativity that is consistent with the above data is that an additional effector binding site is located adjacent to the substrate binding site and that the effector exerts its influence on enzyme turnover by limiting the mobility of the substrate in the active site. To directly test this hypothesis we have modified P450 3A4 by substituting larger amino acid residues in a region that is suspected of comprising a portion of the effector binding site. In this report P450 3A4 residues Leu-211 and Asp-214 were changed to the larger Phe and Glu residues, respectively. These substitutions resulted in the elimination of both homotropic and heterotropic cooperativity in steroid hydroxylation reactions and in a reduced response to ANF. Spectral studies of testosterone binding revealed that the double mutant displayed a hyperbolic binding curve and behaved as if it possessed a single steroid binding site, whereas the wild-type P450 3A4 demonstrated a sigmoidal binding pattern indicative of more than one binding site.
MATERIALS AND METHODS
Plasmids and Escherichia coli Strains Used.
The construct, pSE3A4, expressing human P450 3A4 was described previously (7). For enzyme purification, four histidine residues were added to the C terminus of the P450 3A4 coding sequence as described (19). Growth and induction of E. coli DH5α cells containing pSE3A4 or the indicated mutants was performed as previously reported (20). P450 expression levels ranged from 200–500 nmol/liter. Enzymes were purified from solubilized membrane preparations on Talon (CLONTECH) metal affinity columns by using conditions described previously (19). P450 was measured by reduced carbon monoxide difference spectra. The specific P450 content based on total protein determined by the method of Lowry (21) typically averaged ≈11 nmol/mg protein. P420 contamination was <5%.
Recombinant DNA Manipulations.
Site-directed mutagenesis of P450 3A4 was accomplished by PCR using primers synthesized by National Biosciences (Plymouth, MN). The primers incorporated a BamHI site used for cloning the amplified fragment directly into pSE3A4. The primer sequences, designed to hybridize in the reverse direction, were (L211F) 5′-CGGGATCCAAAAAATCAAATCTAAAAAGCTTCTTGG-3′ and (D214E) 5′-CGGGATCCAAAAATTCAAATCT-3′, with the substituted position underlined. PCR conditions were as described (7). The double mutant, L211F/D214E, was constructed by using the D214E primer and L211F plasmid as template.
Steroid Hydroxylase Assays.
Purified P450 3A4 proteins were reconstituted by preincubation of 5 pmol P450 with 20 pmol of rat NADPH–P450 reductase [expressed in and purified from E. coli as described previously (7)], 10 pmol of rat cytochrome b5, 0.4% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonic acid), and 100 μg/ml DOPC (dioleoyl phosphatidylcholine). Reactions proceeded for 5 min at 37°C in 100 μl of 15 mM MgCl2, 50 mM Hepes (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) buffer (pH 7.6), 100 μg/ml DOPC, and 0.04% CHAPS. Longer incubations revealed that reaction rates were linear with time for at least 15 min. 4-14C-labeled testosterone (Amersham) or progesterone (DuPont/NEN) stock solutions in methanol were used as substrate at the concentrations indicated. Reactions were started by the addition of 1 mM NADPH and stopped with 50 μl of tetrahydrofuran. Quantification of metabolites by TLC, autoradiography, and liquid scintillation counting was performed as described (20).
Spectral Binding Studies.
Binding spectra were recorded on a Beckman DU-7 spectrophotometer. P450 samples were diluted to 0.5 μM in 50 mM Hepes (pH 7.6), 0.1 mg/ml DOPC, and 0.05% CHAPS. The sample was divided into nine equal aliquots. A 1/100 vol of various dilutions of testosterone in methanol (or methanol alone) was added to each aliquot. The final methanol concentration in each sample was 1%. Difference spectra were recorded from 500 to 340 nm for each sample by using the protein sample containing methanol alone as the reference. The change in absorbance (ΔA) was determined by subtracting the absorbance at 419 nm (trough) from the absorbance at 388 nm (peak).
Data Analysis.
Nonlinear regression (Sigmaplot Windows, Jandel, San Rafael, CA) was used to determine Vmax and S50, by using the equation v = (VmaxSn)/(S50n + Sn) as described (6, 8). The Hill coefficient was determined from linear regression of the plot log[v/(Vmax − v)] vs. log S. In the case of spectral titrations, ΔAmax was determined by nonlinear regression of the plot of ΔA vs. S using the equation ΔA = ΔAmaxSn/Ksn + Sn or the equation ΔA = ΔAmaxS/Ks + S for wild-type and L211F/D214E, respectively.
RESULTS
Characterization of Testosterone Hydroxylase Activities.
A previous study of P450 3A4 indicated that alanine substitution mutations of amino acid residues 210 and 211 affected the modulation of steroid hydroxylase activity by ANF (7). In this study additional substitutions were engineered to assess further the contribution of these positions. The underlying strategy was to create mutations that increased the size of the residue, thereby potentially reducing the size of the effector binding site. When Leu at position 210 was changed to Phe, decreased ANF stimulation of the formation of minor metabolites of testosterone (2β-OH and 15β-OH) and progesterone (16α-OH) was noticed. However, enzyme expression levels in E. coli as compared with wild-type P450 3A4 were reduced (data not shown). When Leu-211 was changed to Phe, rates of production of each testosterone metabolite in the absence of ANF were higher than P450 WT (Table 1), and decreased stimulation by ANF was observed. From previous results (7) and from a model of the proposed structure of P450 3A4 (17) we predicted that increasing the size of residue 214 (Asp) might yield a similar phenotype to L211F. However, D214F had decreased expression levels in E. coli and lower enzymatic activity (data not shown). Because it had previously been demonstrated that conservation of a charged residue at position 290 was important to the structure and function of canine P450 2B11 (22, 23), the larger residue Glu was also engineered as a replacement for Asp-214 in P450 3A4. The resulting protein, D214E, was expressed at a level equivalent to the wild-type enzyme and exhibited higher enzymatic activity in the absence of effector and a decreased response to ANF (Table 1). Because each single mutant retained some residual stimulation by ANF, the double mutant, L211F/D214E, was constructed. The double mutant displayed higher basal activity in the absence of the effector and lower-fold stimulation by ANF than the wild-type enzyme or either single mutant, indicating that the effects were additive. The percentage of the total product constituted by the 6β-OH metabolite was very similar between WT P450 3A4 and L211F/D214E, suggesting that the active site was not drastically altered. However, as noted previously (7, 19), in reactions catalyzed by P450 3A4, ANF differentially stimulated steroid hydroxylation at the narrower ends (2β and 15β positions) over the middle (6β) position. The metabolite ratio, expressed as 6β-OH/2β-OH + 15β-OH, was 5.7 in the absence of ANF and 3.5 in the presence of ANF. Interestingly, in reactions catalyzed by L211F/D214E the metabolite ratio was 3.7, and was not altered by the presence of ANF.
Table 1.
Effect of ANF (25 μM) on testosterone hydroxylase activities of P450 3A4 wild-type and site-directed mutant enzymes using 25 μM testosterone as substrate
| 3A4 Sample | MeOH*
|
ANF*
|
Fold stimulation by ANF
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| 2β-OH | 6β-OH | 15β-OH | 2β-OH | 6β-OH | 15β-OH | 2β-OH | 6β-OH | 15β-OH | |
| WT | 0.4 (10) | 3.4 (85) | 0.2 (5) | 1.1 (14) | 6.0 (78) | 0.6 (8) | 2.8 | 1.8 | 3.0 |
| L211F | 0.8 (11) | 5.9 (83) | 0.4 (6) | 1.5 (15) | 7.2 (75) | 1.0 (10) | 1.9 | 1.2 | 2.5 |
| D214E | 1.1 (14) | 5.9 (76) | 0.8 (10) | 1.8 (16) | 8.4 (73) | 1.2 (11) | 1.6 | 1.4 | 1.5 |
| L211F/D214E | 1.1 (14) | 6.3 (79) | 0.6 (7) | 1.0 (12) | 6.7 (80) | 0.73 (8) | 0.9 | 1.1 | 1.2 |
Values are expressed as nanomoles of product formed per minute per nanomole of P450 and are the average of duplicate determinations. Numbers in parentheses represent the rate of metabolite formation as a percentage of the total (2β-OH + 6β-OH + 15β-OH testosterone).
Kinetics of Testosterone Metabolism.
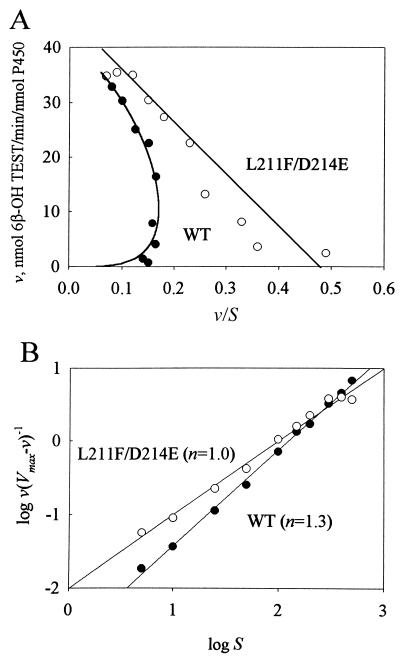
Previous work has illustrated the importance of using a wide range of substrate concentrations to understand P450 3A4 activity (5, 8, 18). Kinetic analysis of testosterone metabolism was performed on WT P450 3A4 and L211F/D214E. In the absence of heterotropic effector (ANF) the kinetics of WT P450 3A4 testosterone 6β-hydroxylation were sigmoidal, as demonstrated previously (8). For wild-type P450 3A4, the Vmax was 40 ± 1 nmol 6β-OH testosterone per min/nmol P450 and the S50 value was 132 ± 7 μM. For L211F/D214E the Vmax was 44 ± 2 nmol 6β-OH testosterone per min/nmol P450 and the S50 value was 100 ± 12 μM. Eadie–Hofstee plots (Fig. 1A) indicated nonlinear kinetics for wild-type P450 3A4 and linear kinetics for L211F/D214E. Linear regression without weighting of the plot of log[v/(Vmax − v)] vs. log S (Fig. 1B) was used to determine the Hill coefficient (n), an indicator of the degree of cooperativity. For the wild-type enzyme, n = 1.3, showing positive cooperativity with increased substrate concentration. For P450 L211F/D214E the Hill coefficient was 1.0, which indicates no cooperativity.
Figure 1.
Kinetic analysis of testosterone hydroxylation by P450 3A4 (•) and L211F/D214E (○). Assays were performed as described by using nine different substrate concentrations ranging from 5 to 500 μM. (A) Eadie–Hofstee analysis of testosterone hydroxylase activities. Wild-type data points were fit to a plot of the Hill equation. Linear regression was used to fit the L211F/D214E data. (B) Linear regression of the plots of log[v/(Vmax − v)] vs. log S.
Kinetics of Progesterone Metabolism.
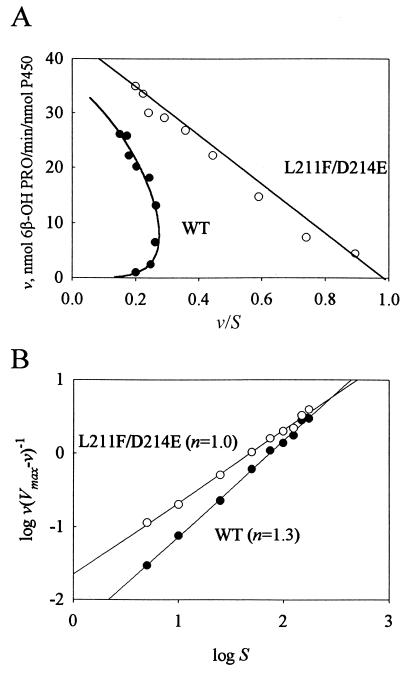
Kinetic analysis of progesterone hydroxylation also revealed a nonlinear Eadie–Hofstee plot for wild-type P450 3A4 and a linear plot for L211F/D214E (Fig. 2A). Hill coefficients were n = 1.3 and n = 1.0 (Fig. 2B) for WT P450 3A4 and L211F/D214E, respectively, indicating a lack of homotropic cooperativity by the L211F/D214E mutant. For wild-type P450 3A4 and L211F/D214E the Vmax values were 35 ± 3 and 44 ± 1 nmol 6β-OH progesterone per min/nmol P450, and the S50 values were 75 ± 13 and 49 ± 1 μM, respectively.
Figure 2.
Kinetic analysis of progesterone hydroxylation by P450 3A4 (•) and L211F/D214E (○). Assays were performed as described by using nine different substrate concentrations ranging from 5 to 175 μM. (A) Eadie–Hofstee analysis of progesterone hydroxylase assays. Wild-type data points were fit to a plot of the Hill equation. Linear regression was used to fit the L211F/D214E data. (B) Linear regression of the plots of log[v/(Vmax − v)] vs. log S.
Spectral Binding Analyses.
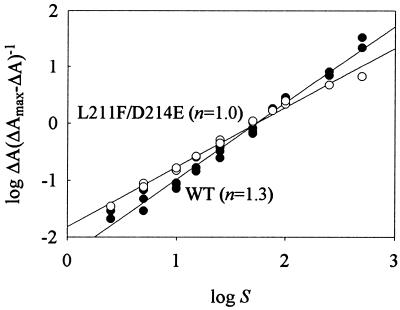
To assess whether the kinetic differences between wild-type P450 3A4 and L211F/D214E were a result of differences in substrate binding, spectral binding studies were performed. The interaction of testosterone with both wild-type P450 3A4 and L211F/D214E resulted in a type I binding spectrum with a peak at 388 nm and a trough at 419 nm (data not shown). A plot of ΔA vs. S demonstrated sigmoidal binding by P450 3A4 and hyperbolic binding by L211F/D214E (data not shown). Nonlinear regression of the testosterone binding data by using the equation ΔA = ΔAmaxSn/Ksn + Sn or the equation ΔA = ΔAmaxS/Ks + S for wild-type and L211F/D214E, respectively, was used to derive ΔAmax and Ks values. (The Hill equation can give a value for the substrate concentration that produces half-maximal binding and not a Ks value per se, but the term is used here for the sake of simplicity). For wild-type P450 3A4 ΔAmax was 0.080 ± 0.001 μM−1 and the Ks value was 56 ± 3 μM. For L211F/D214E the ΔAmax was 0.112 ± 0.003 μM−1 and the Ks value was 51 ± 3 μM. By using a difference extinction coefficient of 126 mM−1⋅cm−1 (24), the data illustrate that >60% of the P450 could be converted from the low to high spin state for both wild-type P450 3A4 and L211F/D214E. Plots of log ΔA(ΔAmax − ΔA)−1 vs. log S (Fig. 3) gave values for the Hill coefficient of 1.3 and 1.0 for testosterone binding by wild-type P450 3A4 and L211F/D214E, respectively.
Figure 3.
Analysis of testosterone binding by spectral titration. P450 (0.5 μM) 3A4 wild-type (•) or L211F/D214E (○) was titrated with concentrations of testosterone ranging from 2.5 to 500 μM as described. ΔA was determined by difference spectroscopy by using a base of 419 nm and a peak of 388 nm (type I difference spectrum). ΔAmax was determined by nonlinear regression of ΔA vs. S using the Hill (wild-type) or Michaelis–Menten (L211F/D214E) equations. Hill coefficients (n) were determined from the slopes of the linear regression lines. Data represent three independent titrations of wild-type P450 3A4 and two titrations of L211F/D214E.
Heterotropic Cooperativity.
Previous studies have presented evidence for a single common binding site for the effectors ANF and progesterone (5, 19). The lack of positive homotropic cooperativity displayed by L211F/D214E suggested that it might also display a lack of positive heterotropic cooperativity when hydroxylation of one steroid was measured in the presence of another. The production of 6β-OH progesterone by P450 3A4 wild-type and L211F/D214E was measured by using 14C-labeled progesterone as substrate in the presence of 0, 25, and 100 μM nonradiolabeled testosterone (Table 2). At the progesterone concentrations tested testosterone acted as a positive heterotropic effector of the P450 3A4 wild-type 6β-hydroxylase activity. In contrast, L211F/D214E displayed inhibition of progesterone hydroxylase activity by testosterone.
Table 2.
Effect of testosterone on the progesterone 6β-hydroxylase activities of P450 3A4 wild-type and the site-directed mutant L211F/D214E
| Substrate [[14C]Pro], μM | P450 3A4 [Test], μM
|
L211F/D214E [Test], μM
|
||||
|---|---|---|---|---|---|---|
| 0 | 25 | 100 | 0 | 25 | 100 | |
| 5 | 0.9* | 1.4 (149) | 1.7 (189) | 2.2 | 2.0 (91) | 1.6 (73) |
| 10 | 2.1 | 2.7 (129) | 3.1 (148) | 4.3 | 4.0 (93) | 2.9 (67) |
| 25 | 5.7 | 6.4 (112) | 6.9 (121) | 8.9 | 8.3 (93) | 5.9 (66) |
| 50 | 10.9 | 12.4 (114) | 11.5 (106) | 13.4 | 13.6 (101) | 10.7 (80) |
Values are expressed as nanomoles of 6β-OH progesterone formed per minute per nanomole of P450 and are the average of duplicate determinations. Numbers in parentheses represent the rate of 6β-OH progesterone formation as a percentage of the rate of product formation in the absence of added testosterone.
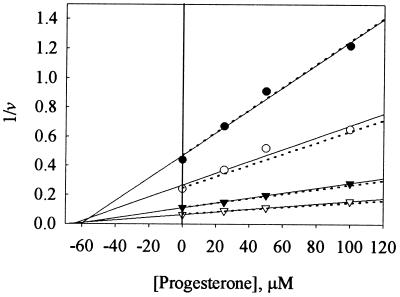
To characterize further the inhibition of steroid hydroxylase activity of L211F/D214E by a second steroid, the combination of steroid and effector was reversed, using testosterone as a substrate and progesterone as an effector. A Dixon analysis (25) of the results (Fig. 4) showed that L211F/D214E behaved in a manner consistent with competitive inhibition. Linear regression lines drawn through the experimental data points defined a Ki = 58 μM; the intersection of the lines above the abscissa signify competitive inhibition (25). The dotted lines in Fig. 4 were created from the equation v = VmaxS/Km(1 + I/Ki) + S (using the testosterone kinetic values: Vmax = 44 nmol/min per nmol, Km = 100 μM) and Ki = 58 μM. The calculated Ki for progesterone as an inhibitor (Ki = 58 μM) was approximately equivalent to the Km determined for progesterone as a substrate (Km = 44 μM).
Figure 4.
Dixon plots of testosterone hydroxylase activities of L211F/D214E in the presence of progesterone. Four concentrations of 14C-labeled testosterone (•, 5 μM; ○, 10 μM; ▴, 25 μM; and ▵, 50 μM) as substrate were incubated in the presence of four concentrations of progesterone as effector. Solid lines represent linear regression of the data; dotted lines represent plots of the reciprocal of v = VmaxS/Km(1 + [Pro]/Ki) + S vs. [Pro].
DISCUSSION
Despite the fact that cooperativity in P450 3A4-catalyzed reactions has been known for years, limited progress has been made in understanding this important phenomenon. The large active site of P450 3A4, which is capable of recognizing a wide range of substrates, stands in contrast to other cooperative enzymes. These tend to have very well-defined substrates and very tight binding affinities. Such enzymes catalyze a limited number of reactions and are theorized to have evolved cooperative regulation as a way of affording additional control over enzymatic activity. P450s on the other hand have probably evolved to recognize a large and diverse array of varied compounds.
Our approach to understanding P450 3A4 cooperativity has been to use a combination of 3D molecular modeling, mutagenesis, and biochemical characterization as a means of elucidating structure-activity relationships. A key component to the study of the interaction between substrate, effector, and protein is the availability of mutated enzymes that display altered responsiveness to effector action. Based on previous studies of alanine-substitution mutants at positions 210–216 (7) and on a 3D homology model of P450 3A4 (17), Leu-211 and Asp-214 were changed to larger residues to mimic effector binding. We believe that additional amino acid substitutions will be required to completely abolish the cooperativity elicited by ANF. Molecular modeling studies suggest that two ANF molecules can simultaneously occupy the active site when progesterone is docked in a 6β-binding orientation (6). Our initial observations with a single concentration of ANF as effector demonstrated that cooperativity was reduced, but not completely eliminated. Additional studies using a range of concentrations of ANF (0–150 μM) demonstrated mild stimulation of progesterone hydroxylation at ANF concentrations up to 25 μM and slight inhibition at the highest ANF concentrations (data not shown). For these reasons, this study has focused mainly on the use of steroids as substrates or effectors, where homotropic and heterotropic cooperativity was abolished in the double mutant L211F/D214E.
From this and previous studies, a clearer picture of P450 3A4 cooperativity is starting to emerge. The preponderance of evidence suggests that there are at least two distinct sites in P450 3A4 for substrate and effector binding. There are two major questions to be answered. The first relates to the location of the effector binding site—i.e., is it in the active site or at a separate allosteric site (8)? The second question, the mechanism underlying effector and substrate interaction, can only be addressed after the first question has been answered. Our model for cooperativity (Fig. 5A) agrees with the Shou model (13), which theorizes that substrate and effector bind in adjacent sites that are both part of a large binding cavity. To test this model our approach has been to mutagenize residues that we believe define part of the active site. In L211F/D214E, this cavity would be artificially narrowed (Fig. 5B), reducing the kinetics of steroid binding or hydroxylation to that of a single-site enzyme. Residues 211 and 214 are proposed (26) to be in a region of the enzyme that is equivalent to substrate recognition site 2 of P450 family 2 enzymes (27). The P450 2B1 residue 206 from this region has been shown to play an important role in determining substrate specificity (28). Based on these results, it seems unlikely that the effector binding site is at a separate allosteric site and that our bulky substitutions are merely mimicking a proposed conformational change normally induced by effector binding. Our results strongly support the suggestion (13) that the effector site is part of the active site and that ANF, testosterone, and progesterone bind at this site.
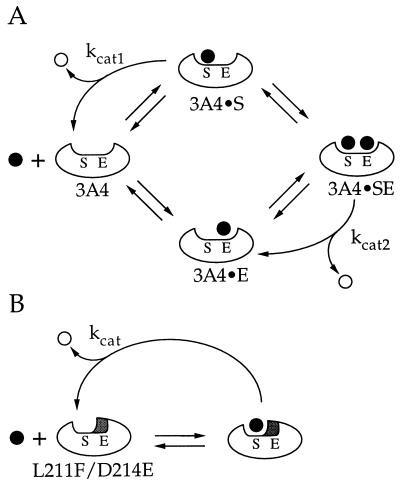
Figure 5.
(A) Model showing cooperativity of P450 3A4-catalyzed steroid hydroxylation. The enzyme active site is proposed to contain a substrate (S) and effector (E) binding site. Product (○) formation can result from substrate (•) bound in the S-site (3A4•S or 3A4•SE), but not when bound in the E-site (3A4•E). (B) Model showing reactions catalyzed by L211F/D214E. Substitution of Phe and Glu for residues Leu-211 and Asp-214, respectively, occupies a portion of the active site that normally binds effector. This renders the mutant (L211F/D214E) functionally equivalent to the 3A4•E complex in A.
The more difficult question is the mechanism of cooperativity. For simplicity, our model proposes that substrate oxidation occurs only when the substrate is in the S site and does not result from binding in the E site. This is in contrast to the Shou model (13), which proposes that there are two catalytic sites, each site with access to the reactive oxygen. To distinguish between these two models experiments are currently being conducted to measure the oxidation of effector molecules by L211F/D214E and other mutants. According to our model, occupation of the effector site could produce positive cooperativity by increasing Vmax, decreasing Km, or both. In the case of wild-type enzyme, binding of the effector could limit the size of the active site, which would increase the frequency with which the substrate is placed into an orientation that results in product formation. Positive cooperativity would be evidenced if rate constant kcat2 is greater than kcat1 (Fig. 5A), and would manifest itself as a Vmax effect. This type of interpretation is consistent with the observations by Shou et al. (13) that ANF dramatically increased the Vmax for phenanthrene metabolism with little or no change in the Km.
Alternatively, binding of effector could increase the affinity of the substrate for the active site. Positive cooperativity would be manifested if the Km for product formation from the 3A4•SE complex (Fig. 5A) is lower than the Km for 3A4•S. The effector could influence substrate binding by providing direct substrate–effector contacts, or by changing the shape of the substrate binding site. The latter interpretation is supported by the observation that metabolite profiles are altered by the presence of effector (13, 19) or with increased substrate concentration (19). The spectral titrations presented here (Fig. 3), which do not involve product formation, demonstrated cooperativity of testosterone binding by wild-type P450 3A4. The data suggest that cooperativity of steroid hydroxylation may be more a result of an altered Km, consistent with the finding that progesterone decreases the Ks for the binding of a type II ligand to rabbit P450 3A6 (29). However, it is conceivable that the sigmoidal type I binding of testosterone to P450 3A4 reflects the efficiency of displacement of the sixth axial ligand rather than binding per se.
If both substrate and effector are in the active site and can interact with each other it may be difficult to distinguish residues that directly affect effector binding from residues that indirectly affect effector action. For example, alteration of a residue in the substrate binding site can change the orientation of binding of substrate. In turn, this can cause an alteration of the interaction with effector and consequently an apparent altered response to effector. Recent results from our laboratory indicate that residues that are believed to directly contact the substrate can also alter the apparent action of the effector (6, 19). We believe that residues 211 and 214 help define a second site for binding of steroid as an effector rather than substrate based on the following two criteria: (i) in the absence of effector, L211F/D214E had an activity and metabolite profile equivalent to that of wild-type P450 3A4 in the presence of effector and (ii) whereas wild-type P450 3A4 kinetics are sigmoidal in the absence of effector and become linear in the presence of effector (5–8, 19), kinetics of L211F/D214E are linear in the absence of effector.
Other residues suggested by 3D modeling to be in the vicinity of positions 211 and 214 are currently being targeted for mutagenesis. Further analysis of L211F/D214E and other mutants using additional substrates and/or effectors will be required to fully map the effector site. Detailed knowledge of the location and structural requirements of the substrate binding and effector sites of P450 3A4 should prove invaluable in rationalizing and predicting interactions among the multitude of compounds that bind to P450 3A4.
Acknowledgments
We thank Dr. Grazyna D. Szklarz for expert assistance in interpreting the P450 3A4 molecular model. This work was supported by Grant GM 54995 and Center Grant ES 06694 from the National Institutes of Health.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: P450, cytochrome P450; ANF, α-naphthoflavone; 3D, three-dimensional.
References
- 1.Guengerich F P. Chem Res Toxicol. 1990;3:363–371. doi: 10.1021/tx00016a015. [DOI] [PubMed] [Google Scholar]
- 2.McKinnon R A, Burgess W M, Hall P M, Roberts-Thomson S J, Gonzalez F J, McManus M E. Gut. 1995;36:259–267. doi: 10.1136/gut.36.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolars J C, Lown K S, Schmiedlin-Ren P, Ghosh M, Fang C, Wrighton S A, Merion R M, Watkins P B. Pharmacogenetics. 1994;4:247–259. doi: 10.1097/00008571-199410000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Guengerich F P. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. Ortiz de Montellano P R, editor. New York: Plenum; 1995. [Google Scholar]
- 5.Schwab G E, Raucy J L, Johnson E F. Mol Pharmacol. 1988;33:493–499. [PubMed] [Google Scholar]
- 6.He Y A, He Y Q, Szklarz G D, Halpert J R. Biochemistry. 1997;36:8831–8839. doi: 10.1021/bi970182i. [DOI] [PubMed] [Google Scholar]
- 7.Harlow G R, Halpert J R. J Biol Chem. 1997;272:5396–5402. doi: 10.1074/jbc.272.9.5396. [DOI] [PubMed] [Google Scholar]
- 8.Ueng Y-F, Kuwabara T, Chun Y-J, Guengerich F P. Biochemistry. 1997;36:370–381. doi: 10.1021/bi962359z. [DOI] [PubMed] [Google Scholar]
- 9.Kerlan V, Dreano Y, Bercovici J P, Beaune P H, Floch H H, Berthou F. Biochem Pharmacol. 1992;44:1745–1756. doi: 10.1016/0006-2952(92)90068-t. [DOI] [PubMed] [Google Scholar]
- 10.Ueng Y, Shimada T, Yamazaki H, Guengerich F P. Chem Res Toxicol. 1995;8:218–225. doi: 10.1021/tx00044a006. [DOI] [PubMed] [Google Scholar]
- 11.Shimada T, Guengerich F P. Proc Natl Acad Sci USA. 1989;86:462–465. doi: 10.1073/pnas.86.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koley A P, Buters J T M, Robinson R C, Markowitz A, Friedman F K. J Biol Chem. 1997;272:3149–3152. doi: 10.1074/jbc.272.6.3149. [DOI] [PubMed] [Google Scholar]
- 13.Shou M, Grogan J, Mancewicz J A, Krausz K W, Gonzalez F J, Gelboin H V, Korzekwa K R. Biochemistry. 1994;33:6450–6455. doi: 10.1021/bi00187a009. [DOI] [PubMed] [Google Scholar]
- 14.Kerr B M, Thummel K E, Wurden C J, Klein S M, Kroetz D L, Gonzalez F J, Levy R H. Biochem Pharmacol. 1994;47:1969–1979. doi: 10.1016/0006-2952(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Wang E, Patten C J, Chen L, Yang C. Drug Metab Dispos. 1994;22:566–571. [PubMed] [Google Scholar]
- 16.Perutz M. Nature (London) 1970;228:726–739. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- 17.Szklarz G D, Halpert J R. J Comput Aided Mol Des. 1997;11:265–272. doi: 10.1023/a:1007956612081. [DOI] [PubMed] [Google Scholar]
- 18.Wang R W, Newton D J, Scheri T D, Lu A Y H. Drug Metab Dispos. 1997;25:502–507. [PubMed] [Google Scholar]
- 19.Domanski T L, Liu J, Harlow G R, Halpert J R. Arch Biochem Biophys. 1998;350:223–232. doi: 10.1006/abbi.1997.0525. [DOI] [PubMed] [Google Scholar]
- 20.John G H, Hasler J A, He Y, Halpert J R. Arch Biochem Biophys. 1994;314:367–375. doi: 10.1006/abbi.1994.1455. [DOI] [PubMed] [Google Scholar]
- 21.Lowry O H, Rosebrough N J, Farr A L, Randall R J. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 22.Harlow G R, He Y A, Halpert J R. Biochim Biophys Acta. 1997;1338:258–266. doi: 10.1016/s0167-4838(96)00209-9. [DOI] [PubMed] [Google Scholar]
- 23.Harlow G R, Halpert J R. Arch Biochem Biophys. 1996;326:85–92. doi: 10.1006/abbi.1996.0050. [DOI] [PubMed] [Google Scholar]
- 24.Gibson G, Cinti D, Sligar S, Schenkman J. J Biol Chem. 1980;255:1867–1873. [PubMed] [Google Scholar]
- 25.Dixon M. Biochem J. 1953;55:170–171. doi: 10.1042/bj0550170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasemann C A, Kurumbail R G, Boddupalli S S, Peterson J A, Deisenhofer J. Curr Biol. 1995;2:41–62. doi: 10.1016/s0969-2126(01)00134-4. [DOI] [PubMed] [Google Scholar]
- 27.Gotoh O. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 28.Luo Z, He Y, Halpert J R. Arch Biochem Biophys. 1994;309:52–57. doi: 10.1006/abbi.1994.1083. [DOI] [PubMed] [Google Scholar]
- 29.Johnson E F, Schwab G E, Vickery L E. J Biol Chem. 1988;263:17672–17677. [PubMed] [Google Scholar]