Abstract
G-protein-coupled receptors (GPCRs) are known to exist in dynamic equilibrium between inactive- and several active-state conformations, even in the absence of a ligand. Recent experimental studies on the β2 adrenergic receptor (β2AR) indicate that structurally different ligands with varying efficacies trigger distinct conformational changes and stabilize different receptor conformations. We have developed a computational method to study the ligand-induced rotational orientation changes in the transmembrane helices of GPCRs. This method involves a systematic spanning of the rotational orientation of the transmembrane helices (TMs) that are in the vicinity of the ligand for predicting the helical rotations that occur on ligand binding. The predicted ligand-stabilized receptor conformations are characterized by a simultaneous lowering of the ligand binding energy and a significant gain in interhelical and receptor-ligand hydrogen bonds. Using the β2AR as a model, we show that the receptor conformational state depends on the structure and efficacy of the ligand for a given signaling pathway. We have studied the ligand-stabilized receptor conformations of five different ligands, a full agonist, norepinephrine; a partial agonist, salbutamol; a weak partial agonist, dopamine; a very weak agonist, catechol; and an inverse agonist, ICI-115881. The predicted ligand-stabilized receptor models correlate well with the experimentally observed conformational switches in β2AR, namely, the breaking of the ionic lock between R1313.50 at the intracellular end of TM3 (part of the DRY motif) and E2686.30 on TM6, and the rotamer toggle switch on W2866.48 on TM6. In agreement with trp-bimane quenching experiments, we found that norepinephrine and dopamine break the ionic lock and engage the rotamer toggle switch, whereas salbutamol, a noncatechol partial agonist only breaks the ionic lock, and the weak agonist catechol only engages the rotamer toggle switch. Norepinephrine and dopamine occupy the same binding region, between TM3, TM5, and TM6, whereas the binding site of salbutamol is shifted toward TM4. Catechol binds deeper into the protein cavity compared to the other ligands, making contact with TM5 and TM6. A part of the catechol binding site overlaps with those of dopamine and norepinephrine but not with that of salbutamol. Virtual ligand screening on 10,060 ligands on the norepinephrine-stabilized receptor conformation shows an enrichment of 38% compared to ligand unbound receptor conformation. These results show that ligand-induced conformational changes are important for developing functionally specific drugs that will stabilize a particular receptor conformation. These studies represent the first step toward a more universally applicable computational method for studying ligand efficacy and GPCR activation.
INTRODUCTION
Ligands for G-protein-coupled receptors (GPCRs) vary in size from small molecules to proteins, and they elicit varied responses in GPCR-mediated cell signaling pathways. There is substantial evidence that GPCRs exist in multiple inactive and active conformational states that are in dynamic equilibrium (1–3). GPCR ligands exhibit different types and ranges of efficacies depending on the receptor and the signaling pathway it activates. Thus, we have
Full agonists that bind and activate a particular signaling pathway in the receptor.
Partial agonists that bind and activate the receptor to different degrees, typically less than that of the full agonist.
Neutral antagonists that bind but prevent activation of the receptor (these ligands, however, do not alter the constitutive activity of the receptor if any).
Inverse agonists that suppress the constitutive activity of the receptor and therefore do not activate the receptor.
The relative efficacies of the ligands should be compared for their response to the same signaling pathway. Adrenergic receptors (AR) constitute a very important subclass of GPCRs and have been studied well for the effect of ligand structure on receptor activation (4–17). Extensive site-directed mutational studies on β2AR for the purposes of drug design (18–21) point to the binding pocket of epinephrine, the endogenous agonist, being in the transmembrane (TM) domain defined by helices 3, 4, 5, and 6. However it is not clear which residues in the TM region lead to activation of the receptor. There are several types of ligands known for β2AR. Epinephrine and norepinephrine are full agonists for β2AR-mediated GDP/GTP exchange in the G-protein, whereas salbutamol and dopamine are partial agonists, and ICI-118551 is an inverse agonist that blunts the basal activity of β2AR. These molecules are highly related in structure and yet elicit very different responses for the same signaling pathway from the receptor. Norepinephrine and salbutamol have high structural similarity, but with slightly different functional groups and good binding affinities to β2AR, but norepinephrine is an agonist whereas salbutamol is a partial agonist to the receptor-mediated GDP/GTP exchange in the G-protein.
Using fluorescence spectroscopic techniques on purified β2AR protein, Kobilka and co-workers have shown that GPCR activation is a multistep process and that ligands with different chemical structures stabilize different receptor conformations (6,11). They have also studied the nature of conformational changes by probing into the interhelical contacts that are broken on ligand binding and activation. By attaching fluorescent probes at different locations within β2AR, Kobilka and co-workers showed that upon activation, norepinephrine and dopamine both break the ionic lock between R1313.50 at the intracellular end of TM3 (part of the DRY motif) and E2686.30 on TM6, and turn on the rotamer toggle switch on TM6, whereas salbutamol only engages the ionic lock, and catechol (which is a weak partial agonist) engages only the rotamer toggle switch (6,9–11). Here, we have used the Ballesteros residue numbering system for GPCRs (22). Swaminath et al. have shown that the aromatic ring of salbutamol occupies a different binding region than the aromatic rings of catecholamine agonists such as dopamine and epinephrine (11). Thus, there is a strong correlation between ligand efficacy and the stabilized conformational state. However, the detailed characteristic features of the different ligand-stabilized receptor conformations are still unknown.
Besides fluorescence measurements, conformational changes have been detected by measuring the solvent accessibilities of mutated cysteines at different points in the receptor. Javitch and co-workers (7) found a counterclockwise rotation of TM6, viewing from the extracellular side. Other studies have reported the importance of conserved aromatic residues on TM6, such as F2826.44 (16). It was found that mutating F2826.44 to a hydrophobic residue (Leu or Ala) increased the constitutive and norepinephrine-induced activities of β2AR.
A variety of techniques, such as fluorescence measurements (12,23), site-directed mutagenesis (16,17,24), cysteine accessibility (7,8,13), disulfide cross-linking (25,26), zinc cross-linking (23), EPR spectroscopy (26), spin labeling (27–29), circular dichroism (23), and x-ray crystallography (30), have been used to investigate the activation mechanism in GPCRs. Since bovine rhodopsin is the only GPCR whose crystal structure is available, the initial attempts in studying the activation process were mainly targeted toward rhodopsin (2,23,26–32). Using site-directed spin labeling (SDSL) and cysteine cross-linking measurements, Khorana and co-workers showed that the activation of rhodopsin involves a relative motion of TM6 with respect to TM3. The SDSL results indicated a counterclockwise rotation of TM6 when viewed from the extracellular side. Also, cross-linking the cysteine residues at the cytoplasmic ends of TM3 and TM6 prevented transducin activation, which showed that the intracellular ends of TM3 and TM6 move away from one another on activation (26). The role of the highly conserved DRY motif in rhodopsin activation was highlighted by Arnis et al. (33), who showed that charge-neutralizing mutations of the arginine D(E)RY increased the constitutive activity of rhodopsin. This result was later interpreted as the breaking of the ionic lock when the high-resolution crystal structure of inactive rhodopsin became available. Crystal structures of several photointermediates of rhodopsin have been reported recently (34,35), which allows us to observe their differences from the dark state of rhodopsin. In the crystal structure of metarhodopsin II, W2656.48 on TM6 was found to toggle its rotamer, showing evidence of a universal switching mechanism for GPCR activation (23,26). Recent modeling studies have used constrained molecular dynamics (MD) and conformational scanning techniques to develop structural models of active GPCRs such as rhodopsin and β2AR (36,37). Although an atomic-resolution structure of the active state of rhodopsin (or any other GPCR) is still not known, detailed experimental information is available for multiple ligands of varied efficacy, and their corresponding receptor conformations for β2AR thereby offer a good model system for studying GPCRs.
For better drug design, it is necessary to identify the different receptor conformations that are stabilized by various ligands (38,39). Computational methods in mapping the conformational changes in the receptor on ligand-binding are relatively few, mainly due to the long timescale of the conformational changes. MD simulation techniques have been used to observe short-timescale, initial events of activation in GPCRs (40–42). However, MD simulation timescales are still insufficient to simulate large-scale motions.
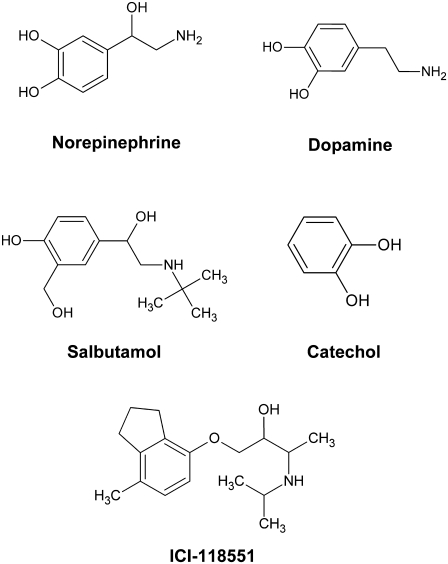
In this article, we have developed a computational method that involves systematic spanning of the conformational subspace of the TM helices in GPCRs, combined with energy minimization, to predict the conformational changes that occur on ligand binding. We call this method ligand-induced transmembrane rotational conformational changes (LITiCon), and it optimizes the ligand-stabilized receptor conformations for ligands with different efficacies. This method provides a model to predict the interhelical contacts made or broken in the vicinity of ligand binding, which is useful in designing residues to label for fluorescent experiments that delineate the conformational switches leading to the activation of GPCRs. We have validated LITiCon for β2AR by predicting the receptor conformations that are stabilized by norepinephrine (a full agonist), salbutamol (a noncatechol partial agonist), and dopamine (a partial agonist), and catechol (a very weak partial agonist). The structures of these compounds are shown in Fig. 1.
FIGURE 1.
Structures of β2AR ligands used in this study.
We have correlated the residue distances in the predicted ligand-stabilized receptor models with the available fluorescence or Trp-bimane fluorescence quenching experiments for β2AR (6,9–11). In the predicted structural models, we observe that norepinephrine and dopamine break the ionic lock and rotamer toggle switch, whereas salbutamol and catechol engage one of them but not both, as observed in experiments. Analysis of the binding sites reveals distinct similarities and differences in the residues present in the ligand-binding site that bring out the salient features of ligand-stabilized receptor conformations. For example, norepinephrine shows an equally strong interaction with both S2035.42 and S2045.43 on TM5, whereas salbutamol shows a preferentially stronger interaction with S2045.43. These salient differences among the binding modes of the various ligands will help us to distinguish strong agonists from weak and partial agonists. We also find that the interhelical contact between F2826.44 on TM6 and Y3267.53 on TM7 breaks on binding of norepinephrine, but not on that of salbutamol. On binding of norepinephrine, an interhelical hydrogen bond (HB) is formed between M2155.54 on TM5 and W2866.48 on TM6 which is absent for dopamine binding. This would allow us to distinguish norepinephrine-bound β2AR from the dopamine-bound conformation of β2AR. Finally, we discuss how the ligand-perturbed receptor model shows better discrimination for adrenergic ligands, and these models could be used in virtual ligand screening (VLS) for drug design in GPCRs.
COMPUTATIONAL METHODS
LITiCon
We have developed a computational procedure to map the perturbations in the helical rotational orientations induced by ligand binding in the TM region of GPCRs. This method consists of two steps.
We identify which of the TM helices get perturbed directly on ligand binding.
Once the helices have been identified, there is a systematic and simultaneous spanning of orientations on all the TMs involved in ligand binding. For each combination of the rotational orientations several properties are calculated as described below. This step generates the binding energy surface of the entire rotational space of the helices between the initial (ligand not bound) and final states. This energy surface will be used to identify the final ligand-stabilized conformational changes that the receptor undergoes and possibly the trajectory of the receptor conformational changes from the initial state to the final state.
The computational method described below is applicable to any starting structure or structural model of a GPCR. For example, one could start from the crystal structure, or the model of a GPCR generated by homology modeling techniques, or any other predicted model.
Step 1: ligand-bound rotational optimization
In this step, individual TM helices are rotated from −180° to 180° in increments of 5°. Details of the rotations and the rotational axes are provided in Supplementary Material. The following four steps are performed for each conformation generated:
Optimization of all side-chain conformations using SCWRL 3.0 (43).
Conjugate gradient minimization of the potential energy of the rotated TM region in the field of the rest of protein fixed until convergence of 0.1 kcal/mol-Å RMS deviation in force/atom is achieved.
Minimization of the entire protein for 1000 steps or until RMS in force/atom is 0.1 kcal/mol-Å.
- At each of these rotational steps we also calculate
- the ligand binding energy, defined as the difference of the potential energy of the ligand with protein fixed, and the potential energy of the free ligand with generalized Born solvation method (44).
- interhelical and ligand-receptor hydrogen bonds using HBPLUS 3.0 (45); and
- interhelical salt bridges.
This produces a map of the ligand-binding energies, along with other calculated properties for all the rotational conformations from 0° to 360° rotations for each TM helix. The change in binding energy and the interhelical hydrogen bonds plus the ligand-receptor hydrogen bonds are plotted as shown in Fig. 2. This would lead to identification of the TM helices that undergo conformational changes on ligand binding. For example, Fig. 2 shows the rotational angle optimization for TM3, TM5, and TM6, demonstrating the significant change in binding energies for norepinephrine bound to β2AR.
FIGURE 2.
(a) Plot of change in ligand-binding energy for various rotation angles of helices 3, 5, and 6 with norepinephrine bound. (b) Plot of interhelical hydrogen bonds. (c) Ligand receptor hydrogen bonds.
Step 2: calculation of the binding energy surface for systematic conformational subspace sampling of simultaneous rotational orientation of all the TM helices
The results of step 1 show which of the helices undergo helical rotations upon direct contact with the ligand. In this step, we perform simultaneous rotations of all the helices directly impacted by ligand binding, from −180° to 180° in steps of 5°. For example, in the case of norepinephrine-bound β2AR, simultaneous rotations of helices 3, 5, and 6 would be performed. This procedure ensures a systematic spanning of all combinations of rotational orientations of all the helices involved directly in ligand binding. Such an optimization procedure would allow us to go over barriers that MD simulations cannot overcome. Each rotational combinatorial conformation thus generated is subject to the four steps of calculation described above in step 1 of the LITiCon procedure. Since more than one TM helix is being rotated simultaneously, all the TM helices that get rotated are allowed to move during the conjugate gradient energy minimization in step 3. For each generated conformation, we calculate the binding energy of the ligand, the number of protein-ligand HBs, and the number of interhelical HBs (see Supplementary Material for a description of the methods used for identifying minima) and sort them by total number of hydrogen bonds and then by binding energy. The final ligand-stabilized receptor structure is selected based on low binding energy and high number of hydrogen bonds.
MD simulations of the ligand-stabilized receptor conformations
The two steps of the LITiCon method do not take into account the allosteric conformational changes in the TM regions that do not contact the ligand directly, or the conformational changes in the TM regions due to hinge bending and tilting of the helices and also the conformational changes in the loop regions that occur on ligand binding. To account for such conformational changes, MD simulations of the predicted ligand-stabilized receptor conformations are performed in an explicit lipid bilayer, water, and salt. We use the NAMD (46) MD simulation package for this purpose. The bilayer was constructed out of palmitoyloleoyl phosphatidylcholine lipid molecules packed around the TM barrel of the receptor. The total number of atoms in the system was ∼80,000. The CHARMM22 force field was used to model the interatomic interactions. A 15-ns NVT constant-temperature MD simulation was performed at 310 K (human body temperature) on the norepinephrine-bound β2AR and the apoprotein.
In this study, we applied the LITiCon method on β2AR bound to the agonists norepinephrine, salbutamol, dopamine, and catechol, as well as the inverse agonist ICI-118551. The helical rotational changes induced in the TM region on ligand binding are analyzed. The ligand-docked conformations of β2AR for each of the ligands was then used to pack a double layer of the lipid palmitoyloleoyl phosphatidylcholine around the ligand-docked protein structures using rigid-body molecular dynamics (47). Subsequently we performed steps 1–3 of the LITiCon procedure as described in detail previously in this section.
Calculation of the strength of the ionic lock between TM3 and TM6
We have calculated the strength of interaction between R1313.50 and E2686.30 using the following procedure. For each ligand-stabilized model of the receptor from LITiCon, we performed side-chain rotamer reassignment for residues R1313.50 and E2686.30 with and without the constraints of making this ionic lock using the side-chain reassignment program SCREAM (V. Kam and W. A. Goddard 3rd, unpublished). The best energy structure for the conformation with the ionic lock was compared to the best energy structure of the conformation without the ionic lock. The energy calculated is the potential energy computed with the CHARMM22 force field.
Ligand docking methods
We have used the MembStruk4.0 method (49) to predict the structural model of human β2AR as the starting structure for LITiCon. We have predicted an ensemble of structural models for the apoprotein of human β2AR. The individual ligands shown in Fig. 1 were docked to the apoprotein model using the hierarchical docking procedure described in this section. We have docked ligands with various affinities and efficacies to the ensemble of receptor conformations of β2AR. We have used a DREIDING force field for the receptor and ligand and CHARMM22 charges for the protein. The ligands were built with the Maestro suite of software (Schrodinger, Portland, OR) and the quantum-mechanical ESP fitted charges were calculated in a solvent of low dielectric since this would mimic the interior of the GPCR TM barrel. The ScanBindSite algorithm (50,51) was performed individually for each ligand to scan the entire receptor surface and locate possible binding region(s).
We then performed ligand conformational sampling using the Monte Carlo sampling program in the Macromodel suite in Maestro. Each low-energy ligand conformation was docked into the putative binding site using Glide extraprecision docking (52). We kept 500 docked conformations for each ligand from Glide and sorted them by buried surface area of the ligand and interaction energy with the protein. The top 100 ligand docked conformations were minimized with protein fixed and the binding energies were calculated as the difference between the ligand energy in protein and the ligand energy in water. The ligand energy in water was calculated using the generalized Born module in Maestro. The final docked structures for all the ligands were also subjected to the induced-fit docking model from the Prime module in Maestro. In induced-fit docking, the side chains of all residues inside the binding pocket were made flexible for efficient sampling of the docked conformations. This ensured that the ligand-induced shape fluctuations of the binding pocket were taken into consideration during prediction of the docked structures.
Method used for virtual ligand screening of optimized receptor conformations
Virtual ligand screening is a stringent test of the usefulness of the ligand-stabilized receptor models for selecting specific drugs. The 10,000-ligand test set was picked randomly from 300,000 compounds in the National Cancer Institute database. Next, 60 known adrenergic ligands were added to the 10,000 compounds to create the final test set of 10,060 compounds. The set of adrenergic ligands is diverse enough to include antagonists, inverse agonists, full and partial agonists, and noncatechol compounds. These compounds were then docked to the receptor models using the HTVS mode of GLIDE. Both the receptor and the ligand vdW radii were scaled down by 50% to allow soft receptor conformations. The docked conformations were first sorted by total charge and then by protein-ligand interaction (sum of vdW and HB interaction). The percentage yield of adrenergic ligands for cutoff c is defined as
 |
Therefore, the yield represents the relative probability of selecting adrenergic ligands at a certain cutoff after filtering compared to randomly selecting ligands from the test set.
RESULTS AND DISCUSSION
Predicted apoprotein model
In the initial predicted structural model, the key conserved residues such as N511.50 on TM1, D792.50 on TM2, and N3227.49 on TM7 (part of the NPxxY motif) are all facing the protein core. These residues form a HB network in the predicted model, which is also observed in the crystal structure of rhodopsin. R1313.50 at the intracellular end of TM3 (part of the DRY motif) forms a salt bridge with E2686.30 on TM6 (the ionic lock). Using the ScanBindSite algorithm, we predicted the binding cavity for all the ligands studied here to be between TM3, TM5, and TM6, which is in agreement with published mutation studies (18,53,54). The key residues involved in ligand binding are all facing the binding cavity. These are D1133.32 on TM3, N2936.55 and the WxP motif on TM6. Among the serines on TM5 that are involved in binding, S2035.42 and S2075.46 are inside the binding cavity, and S2045.43 is outside, facing the membrane. Later in this work, we show that after optimizing the receptor conformations in the presence of the ligands, all three serines come inside the binding cavity, making contact with the ligands.
Perturbation of the ligand-binding site and the TM helices after ligand binding
The ligand-docked structural models of β2AR for each of the five ligands studied here were all subjected to the LITiCon procedure. The first step of the LITiCon procedure, to decipher which TM helices are affected by ligand binding, showed that helices 3, 5, and 6 undergo conformational changes (on ligand binding) for all the five ligands. This is demonstrated in Fig. 2 for norepinephrine. For example, it is seen from Fig. 2 a that TM3 shows preferences for specific receptor conformations around −45° to 45° rotations. The other four TM helices (TM1, TM2, TM4, and TM7) did not show any change in binding energy. In addition to TM3, TM5, and TM6, TM7 also shows a change in binding energy on rotation, for norepinephrine, salbutamol, and ICI-118551 (Supplementary Material, Fig S3). However, the preferred receptor conformation for TM7 is the starting conformation and therefore TM7 rotations were not considered for step 2 of the LITiCon procedure. The computations involved in simultaneous rotations of TM helices 3, 5, and 6 for all the five ligands were performed and the binding potential energy surfaces were calculated for various rotational orientations of TM3, TM5, and TM6. This is a four-dimensional surface, and the best ligand optimized receptor conformation was chosen after sorting the various minima in this binding energy surface by interhelical hydrogen bonds, ligand-receptor hydrogen bonds, and ligand binding energy.
Conformational switches triggered by ligand binding and correlation to experiments
The best ligand-stabilized receptor model for each ligand was validated by qualitative comparison (quantitative distances are not available from experiments) of the residue distances to those obtained from Trp-bimane quenching, FRET, and cysteine accessibility experiments in β2AR. Fig. 3 shows snapshots of the predicted conformations before and after the ligands norepinephrine and salbutamol bind to β2AR. The ionic lock between R1313.50 and E2686.30 is disrupted (distance over 10Å) in all norepinephrine-, dopamine-, and salbutamol-stabilized receptor models. For brevity, only the norepinephrine-bound conformation is shown in Fig. 3 b.
FIGURE 3.
Conformational switches in human β2AR. (a) Ionic lock in apoprotein. (b) Breaking of ionic lock by norepinephrine. (c) W2866.48 rotamer in the apoprotein conformation. The water mediated hydrogen bond between W2866.48 and D792.50 is highlighted. (d) Toggling of the W2866.48 rotamer by norepinephrine. (e) No change in the W2866.48 rotamer for the salbutamol-bound conformation (not toggled).
Fig. 3 also shows the change in the rotamer of W2866.48 on TM6 upon norepinephrine binding, which represents the rotamer toggle switch. In the apoprotein structure (Fig. 3 c), the nitrogen of W2866.48 is facing the protein core toward TM2, similar to the rhodopsin crystal conformation. In the crystal structure of inactive rhodopsin, the nitrogen of the conserved tryptophan on TM6 forms a water-mediated hydrogen bond with a conserved aspartate on TM2. During the MD simulations of the β2AR apoprotein structure, water moved into the binding cavity and formed a hydrogen-bond network connecting residues W2866.48 and D792.50. Fig. 3 c shows the apoprotein structure after 15 ns of MD simulation, along with the positions of the two water molecules that take part in the water-mediated hydrogen bond. In all the structures shown in Fig. 3, the atomic positions are averaged over a period of the last 1 ns. In the norepinephrine-bound structure shown in Fig. 3 d, W2866.48 switches to a different rotamer that places the nitrogen atom of W2866.48 away from the protein core and facing TM5. However, in the salbutamol and ICI-118551 bound structures, the rotamer conformations of W2866.48 are the same as in apoprotein. We have determined the preferred orientation of W2866.48 rotamer in each of the ligand-bound conformations (Supplementary Material, Fig. S5) and found that norepinephrine, dopamine, and catechol engage the rotamer toggle switch, whereas salbutamol and ICI-118551 do not. This is in agreement with a previously reported rotamer toggle switch for these ligands (6).
TM helical rotations in β2AR with norepinephrine bound
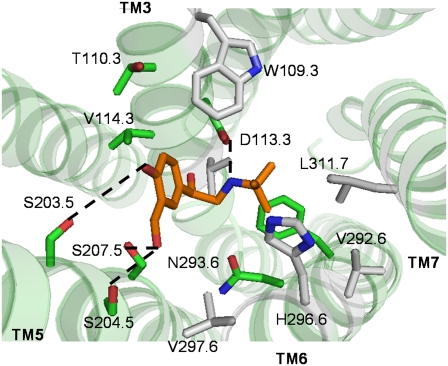
Fig. 4 shows the binding site of norepinephrine in the ligand optimized conformation. In the apoprotein model, the protonated amine nitrogen of norepinephrine forms a salt bridge with D1133.32 on TM3 (a distance of 2.87 Å) and the β-OH forms a HB with N2936.55 on TM6 (a distance of 2.88 Å). These contacts are preserved in the ligand-stabilized conformation as well. From site-directed mutagenesis studies, mutations of D1133.32 to Asn (18) and N2936.55 to Leu (55) reduce the binding affinity of full agonists by 8000- and 36-fold, respectively. Also, experimentally, mutations of all the three serines, Ser-2035.42 (24), Ser-2045.43, and Ser-2075.46 (54) on TM5 have been shown to reduce the binding affinity of norepinephrine by 25, 33, and 39 times, respectively. In the ligand-stabilized model, the two serines, S2045.43 and S2075.46, move into the binding pocket. Both S2035.42 and S2045.43 form HBs with the catechol para OH (4.5 Å and 4 Å), whereas S2075.46 forms a HB with the meta OH (2.9 Å). Besides the experimentally verified residues, we observe that F2085.47 on TM5 and W1093.28 on TM3 move closer to norepinephrine (<7 Å) in the ligand-stabilized receptor conformation. The ligand optimized model is also characterized by the disruption of several interhelical HBs, as well as the formation of new ones. The HB between N3227.49 on TM7 and D792.50 on TM2 is preserved in both the apoprotein and the ligand optimized models. However, the HB between N3187.45 on TM7 and C2856.47 on TM6 is broken in the ligand optimized model. A new HB is formed between W2866.48 on TM6 and M2155.54 on TM5. The toggled rotamer of W2866.48 results in the formation of a HB with M2155.54, thereby stabilizing this rotamer in the norepinephrine optimized model.
FIGURE 4.
Predicted binding site in the norepinephrine optimized β2AR conformation. The residues within 5 Å of the ligand are shown in gray. The residues having strong interaction with the ligand are in green.
TM helical rotations in β2AR with dopamine bound
Fig. 5 shows the binding site in the ligand optimized receptor model for dopamine. TM3 shows a helical rotation similar to that of norepinephrine, whereas TM5 and TM6 show slightly different rotational orientations. Dopamine-bound β2AR shows a TM5 rotation that is 10° higher than for norepinephrine. D1133.32 on TM3 makes a salt-bridge contact (distance of 2.9 Å) with the protonated primary amine group of dopamine. Unlike the norepinephrine-stabilized state of β2AR, the dopamine-stabilized model of β2AR shows a HB between S2045.43 and S2075.46 and catechol hydroxyl groups (distances between heavy atoms of 5.2 Å and 2.9 Å, respectively). S2035.42 has no HB with dopamine (distance 6.4 Å). This is in agreement with site-directed mutagenesis results for dopamine that show that the S204C mutation has a stronger effect (a sevenfold reduction in binding) compared to S203C (a 2.5-fold reduction in binding) (54). TM6 shows a preference for a smaller rotation angle in the dopamine-bound conformation compared to the norepinephrine-bound conformation, and this helical rotation still turns on the rotamer toggle switch. The interhelical HB between W2866.48 and M2155.54 is not formed in the dopamine-bound conformation, whereas this HB is formed in the norepinephrine-bound conformation. Due to the smaller rotation of TM6, the side chain of W2866.48 in the dopamine-bound structure is not sufficiently close to M2155.54, as it is in the norepinephrine-bound structure. Besides this, the HB between N3187.45 and C2856.47 is preserved in the dopamine-bound state, whereas this HB is broken in the norepinephrine-bound state. These two interhelical HBs can be used to differentiate the norepinephrine-bound state from the dopamine-bound state. They can be tested experimentally to delineate the role of the β-OH group in norepinephrine compared to that in dopamine. Dopamine shows a smaller change in binding energy after conformational changes to its optimized receptor conformation compared to norepinephrine.
FIGURE 5.
Predicted binding site in the dopamine optimized β2AR conformation. The residues within 5 Å of the ligand are shown in gray. The residues having strong interaction with the ligand are in green.
TM helical rotations in β2AR with catechol bound
Due to its smaller size, catechol buries deeper into the binding pocket than norepinephrine, salbutamol, or dopamine. Catechol induces almost the same helical rotation in TM5 as the other three agonists. In the predicted model of the catechol-stabilized receptor conformation (Fig. 6), all the TM5 serines are inside the binding pocket. One of the -OH groups of catechol forms a HB with S2075.46 on TM5 (HB distance, 4.1 Å). The other -OH group forms a HB with N2936.55 on TM6 (HB distance, 3.9 Å). Catechol induces a much smaller rotation in TM3 compared to norepinephrine, dopamine, or salbutamol. Unlike the other ligands, catechol does not have a protonated amine group and hence does not form any salt bridge with D1133.32 on TM3. Catechol shows favorable interactions with several hydrophobic residues on TM3, such as V1143.33 and V1173.36. The rotation of TM3 results in optimizing the interactions of these residues with catechol. Catechol interacts strongly with the aromatic residues on TM6, such as F2906.52 and W2866.48, and also with N2936.55. Although TM6 shows no major backbone change in the presence of catechol, the rotamer of W2866.48 is toggled by catechol.
FIGURE 6.
Predicted binding site in the catechol optimized β2AR conformation. The residues within 5 Å of the ligand are shown in gray. The residues having strong interaction with the ligand are in green.
TM helical rotations in β2AR with salbutamol bound
Salbutamol is a noncatechol strong partial agonist that is structurally different compared to both norepinephrine and dopamine. Fig. 7 shows the predicted binding site in the salbutamol-stabilized β2AR receptor conformation. Salbutamol has a protonated secondary amine nitrogen with a tertiary butyl group attached to it, and a β-OH group similar to norepinephrine, and instead of two OH groups on the aromatic ring, it has one OH group and another -CH2OH group. From our calculations, all three TMs—TM3, TM5, and TM6—in the salbutamol-bound structure show smaller rotations compared to norepinephrine. TM3 rotates by a smaller angle, so that W1093.28 makes a weaker contact with the ligand compared to the norepinephrine-bound structure. D1133.32 makes a strong salt-bridge contact with the protonated amine (distance, 2.9 Å). Due to the long -CH2OH group of salbutamol, a smaller rotation of TM5 places the serines in close proximity to the hydroxyl groups of salbutamol compared to norepinephrine. Unlike norepinephrine, N2936.55 does not form a HB with the β-OH group of salbutamol. Instead the β-OH group makes a HB with D1133.32. Mutation studies have shown that unlike norepinephrine, the β-OH group in noncatecholamine agonists does not interact strongly with N2936.55 (55). Mutating N2936.55 to Leu reduced the binding of the − isomer of norepinephrine 11-fold, whereas the reduction in binding was only fourfold for the + isomer. Therefore, the characteristics of the predicted binding site agree well with these mutation studies that bring out the differences in the recognition of the stereoisomers. Due to the presence of a bulky alkyl -(CH3)3 group at one end, salbutamol also interacts with TM7, although there is no significant change in the rotational orientation observed in the calculations presented here. L3117.38 shows favorable van der Waals interaction with the alkyl group of salbutamol.
FIGURE 7.
Predicted binding site of salbutamol in the optimized β2AR conformation. The residues within 5 Å of the ligand are shown in gray. The residues having strong interaction with the ligand are in green.
TM helical rotations in β2AR with ICI-118551 bound
ICI-118551 is an inverse agonist to β2AR, which has been shown to reduce the basal activity of wild-type β2AR. Fig. 8 shows the predicted binding site for ICI-118551 in the ligand-stabilized receptor conformation. According to our calculations, the ICI-118551-bound structure shows rotations of all three TMs (3, 5, and 6) and these rotations are smaller in magnitude compared to the agonist-stabilized states, thereby preserving the ionic lock between TM3 and TM6. The ligand-stabilized structural model for ICI-118551-bound β2AR is therefore closer to the starting β2AR conformation compared to the agonist-stabilized state. TM3 and TM6 rotate in the same directions as the agonist-stabilized conformations. However, TM5 rotates in the opposite direction, thereby placing the three serines away from the binding pocket and more toward the lipid bilayer. Similar to the agonists, ICI-118551 has a protonated secondary amine group, which makes a strong salt bridge (distance, 2.8 Å) with D1133.32 on TM3. ICI-118551 also has a hydroxyl group attached to the main carbon chain. However unlike norepinephrine, the hydroxyl group is not located at the β-position and placed further away from the aromatic group. In our predicted structure, the hydroxyl group of ICI-118551 forms a hydrogen-bond network with two residues on TM6, N2936.55 (distance, 2.9 Å) and H2966.58 (distance, 3.1 Å). We propose that these two hydrogen bonds with the ligand stabilize the receptor in an inactive state. The bulky aromatic group of ICI-118551 strongly interacts with the aromatic residues on TM6, F2896.51 and F2906.52. Unlike the agonists, the aromatic group of ICI-118551 does not have any hydroxyl group and forms no polar contact with the serine residues on TM5. The two methyl groups present near the tertiary amine of ICI-118551 show favorable hydrophobic contacts with Y1995.38 on TM5. Y1995.38 has been shown to interact with the antagonist carazolol using fluorescence quenching studies (56). The methyl groups on ICI-118551 also interact favorably with T1103.29 and V1143.33 on TM3.
FIGURE 8.
Predicted binding site of ICI-118551 in the optimized β2AR conformation. The residues within 5 Å of the ligand are shown in gray. The residues having strong interaction with the ligand are in green.
Helical rotation similarities and differences among agonists and inverse agonist
The predicted ligand-stabilized receptor models exhibit similarities with and differences from one another, as evidenced by the fluorescence experiments. Thus, for the agonist and the three partial agonists studied here, our predictions show a clockwise rotation of TM3 and counterclockwise rotations of TM5 and TM6, when viewed from the extracellular side. For the inverse agonist, TM5 prefers a clockwise rotation. It should be noted that the directions of the rotations are relative to the starting structure, which is the apoprotein structure in this case. In both bovine rhodopsin and β2AR, strong experimental evidence (7,26) points toward a counterclockwise rotation of TM6, which is in agreement with our observations. TM3, TM5, and TM6 show rotational changes for norepinephrine, dopamine, and salbutamol, whereas only TM3 and TM5 show rotational changes for catechol-bound β2AR. The above observations indicate that the driving force for TM5 rotation is to bring all the three serines inside the ligand-binding cavity to form HBs with the ligand -OH groups. Thus, the TM5 movement is common to all the four agonists, although to different extents. The inverse agonist ICI-118551 does not form any HB with the TM5 serines and hence does not cause any significant rotation of TM5. Fig. 9 shows the different steps in norepinephrine-induced helical rotations. The shape of the binding pocket is represented by the blue surface. The movement of TM5 and subsequent reshaping of the binding cavity as shown in Fig. 9 b leads to pulling of the ligand closer to TM5. Therefore, TM3 and TM6 undergo conformational changes to move closer to the ligand to optimize the distances between the protonated amine and β-OH groups. This leads to breaking of the ionic lock between TM3 and TM6. For the weak partial agonist catechol, the only major movement observed is in TM5, which improves the hydrogen-bond interactions between the serines and catechol OH groups; there is very little rotational movement of TM6.
FIGURE 9.
Different steps in the receptor conformational change induced by norepinephrine. The shape of the binding cavity is represented by the blue surface. Note that in b, the shape of the binding cavity is shifted toward TM5, which makes TM3 and TM6 rotate.
Interhelical hydrogen bond differences among catechol agonists and noncatechol agonists
The structural differences among the various adrenergic agonists are reflected in the receptor conformations stabilized by these ligands. The receptor conformations differ in 1), the position of the ligand inside the binding cavity; and 2), the interhelical HB contacts. Norepinephrine and dopamine, the catecholamine agonists, occupy almost the same position inside the binding cavity, whereas salbutamol (a noncatechol partial agonist) is shifted more toward TM4. In addition, norepinephrine and dopamine reflect a similar pattern of interhelical HB contacts, which is different from that of salbutamol. The HB between T1183.37 and T1644.56 is broken by salbutamol, but is preserved in the norepinephrine- and dopamine-bound structures. In the apoprotein state, the side-chain sulfur of C1253.44 on TM3 hydrogen bonds with the backbone oxygen of V2185.57 at the intracellular end of TM5. This HB is broken by both norepinephrine and dopamine, but preserved by salbutamol. These two examples indicate that the catecholamine agonists follow a common trend in breaking or preserving the same interhelical HBs. Moreover, all of the interhelical HBs that distinguish the salbutamol-bound conformation from the norepinephrine- or dopamine-bound conformations involve TM3. We find that the rotation of TM3 in the case of salbutamol is 50% less compared to those in norepinephrine and dopamine, which accounts for the differences among the interhelical HBs involving TM3. This is because TM5 does not have to rotate much to make HBs with the longer hydroxyl groups of salbutamol. On the other hand, the HB contacts that distinguish the full agonist norepinephrine from the other agonists all involve TM6. One of them is the HB between W2866.48 and M2155.54, which is only present in the norepinephrine-bound state. In the apoprotein state, N3187.45 hydrogen-bonds with the backbone sulfur of C2856.47. This HB is broken by norepinephrine and salbutamol, and not by dopamine. In the case of norepinephrine, the rotation of TM6 is 15° more than the rotation for dopamine and salbutamol, which explains the differences among the TM6 contacts.
Conformational switches
It has been proposed in the literature that the active states of GPCRs are characterized by breaking and making conformational switches (1,6). These conformational switches are turned on by ligand binding and allow the receptor to change conformation toward activation. Two such switches identified in rhodopsin and β2AR are the ionic lock between TM3 and TM6 and the rotamer toggle switch on TM6 (discussed in the Introduction) (6,13,33). All the ligands engage different switches or combinations of switches, and this has been experimentally demonstrated for ligands of different efficacy for β2AR. The strong agonists, such as norepinephrine, turn on both ionic lock and rotamer toggle switches, partial agonists turn on one or the other switch, whereas inverse agonists and antagonists do not engage any of these switches (6). We have investigated the status of these two conformational switches in our ligand-stabilized receptor conformations.
Ionic lock between TM3 and TM6
For the ionic lock, we compared the energy of the conformations with and without the ionic lock. For the apoprotein state, the structure with the ionic lock is 5 kcal/mol more stable than the conformation without the ionic lock. This shows that the ionic lock is maintained in the apoprotein state. In the predicted models, norepinephrine, dopamine, and salbutamol stabilize helical rotations where the ionic lock is broken, whereas catechol does not break the ionic lock. Although norepinephrine, dopamine, and salbutamol induce conformational changes in both TM3 and TM6, catechol leads to a conformational change only in TM3. It follows that simultaneous movements of both TM3 and TM6 are required for breaking the ionic lock. This is in agreement with the experiments reported in Yao et al. (6).
Rotamer toggle switch on TM6
We also investigated the status of the rotamer toggle switch in our predicted structural models. In the various GPCRs studied so far, the tryptophan in the WxP motif on TM6 is found to toggle its rotamer in response to conformational changes in its environment. The rotamer of W2866.48 is determined by two factors, 1), movement of TM6, and 2), position of the ligand aromatic moiety inside the binding cavity. The interaction between the ligand and W2866.48 is mediated by F2906.52, which is located between the ligand and the W2866.48 rotamer (Supplementary Material, Fig. S5). Both norepinephrine and dopamine engage the rotamer toggle switch because the aromatic ring is close to the WxP motif on TM6. On ligand binding and subsequent TM6 movement, F2906.52 alters its rotamer conformation in order to maximize the pi-pi interaction with the ligand. Conformational change in F2906.52 rotamer leads to the toggling of the W2866.48 rotamer, which is located right below F2906.52 and interacts with the F2906.52 rotamer. In case of salbutamol, the aromatic ring of the ligand is located further away from TM6. As a result, the interaction between salbutamol and the TM6 residues is too weak to induce a rotamer toggling. The very weak agonist catechol binds very close to the WxP motif of TM6 and thereby engages the rotamer toggle switch without causing any conformational change to TM6.
Comparison of the binding sites of norepinephrine, dopamine, salbutamol, and catechol
Fig. 10 a shows the binding sites for all four ligands superimposed on one another. Although the aromatic rings of norepinephrine and dopamine occupy similar locations, the aromatic ring of salbutamol is shifted more toward TM4. Catechol binds deeper into the binding pocket compared to the other three ligands. Fluorescence lifetime measurements (11) indicate that catechol competes with both norepinephrine and dopamine for binding to β2AR, and does not compete with salbutamol. A part of the catechol binding site overlaps with both norepinephrine and dopamine, but not with salbutamol (Fig. 10 b). As discussed earlier, the binding site of salbutamol is slightly different compared to norepinephrine and dopamine. The different orientation of the aromatic ring of salbutamol is due to the presence of the -CH2OH group on the aromatic ring, which pushes the aromatic part of salbutamol more toward TM4 to avoid steric clashes between the CH2OH group and the TM5 residues. Therefore, in our models, catechol shares a common binding site with both norepinephrine and dopamine, but not with salbutamol. This explains why catechol competes with norepinephrine for binding, and not with salbutamol (11).
FIGURE 10.
Comparison among the binding sites of norepinephrine, dopamine, salbutamol, and catechol. (a) Superposition of all four binding sites. (b) Orientations of norepinephrine, salbutamol, and catechol.
Analysis of the residues in the binding cavities indicates that there are several common residues that interact with the ligands norepinephrine, dopamine, and salbutamol. These include D1133.32, W1093.28, and V1173.36 on TM3; S2035.42, S2045.43, and S2075.46 on TM5; N2936.55 and H2966.58 on TM6; and L3117.38 on TM7. D1133.32 (conserved among all biogenic amine receptors) forms a salt bridge with the protonated amine nitrogen of the ligands (18). N2936.55 (present only in β2AR) on TM6 forms a hydrogen bond with the β-OH group of norepinephrine. The hydrogen bond interaction of N2936.55 with norepinephrine depends on the position of the β-OH group at the stereocenter. Among the two isomers of norepinephrine, the (−) isomer shows a better binding affinity compared to the (+) isomer, because only the (−) isomer is capable of forming a hydrogen bond with N2936.55. This accounts for the stereospecificity of the (−) norepinephrine in binding to β2AR (55). Among the three serines on TM5 that interact with the ligands, the interaction of S2075.46 is the strongest. Norepinephrine shows equal interaction with the other two serines, S2035.42 and S2045.43, whereas for dopamine the interaction with S2035.42 is weaker than that with S2045.43. Mutation results show that S207A has a larger effect on norepinephrine binding compared to S204A (54), whereas for dopamine, the effect of mutation on S2045.43 is higher than that on S2035.42 (54). This is in agreement with our predicted models. In a different study, it was shown that the S204A mutation has a larger effect on salbutamol binding compared to S207A (57). In our model, the distance between the S2045.43 side chain and the salbutamol meta-OH group is shorter (3.4 Å) compared to the distance between S2045.43 and the norepinephrine para-OH group (4 Å). The longer -CH2OH group of salbutamol places the oxygen atom of the OH closer to the S2045.43 side chain compared to the oxygen atom of norepinephrine. This may explain why the S204A mutation has a larger effect on salbutamol than does S207A, whereas the opposite is true with norepinephrine.
Norepinephrine, dopamine, and salbutamol all interact with W1093.28 on TM3. For dopamine, this interaction is 2.5 kcal/mol stronger compared to the other two ligands. Interestingly, W1093.28 moves into the binding pocket as a result of rotation of TM3. Besides D1133.32, the ligands also interact with several hydrophobic residues on TM3, which include V1143.33 and V1173.36. These residues form the base of the binding cavity in β2AR. Among the three ligands, only the strong agonist norepinephrine shows a favorable interaction with F2085.47 on TM5. Dopamine also interacts with F2085.47, but the strength of this interaction is weaker compared to norepinephrine, whereas salbutamol does not interact with F2085.47. This residue is conserved among all biogenic amine receptors and may help to distinguish the binding mode of full agonists from that of the partial agonists. Due to its small size, catechol binds deeper into the protein cavity compared to the other three ligands. Unlike the other three ligands, catechol does not show any interaction with D1133.32 on TM3. One of the OH groups on catechol forms a hydrogen bond with N2936.55 on TM6, and the other OH group forms a hydrogen bond with S2075.46 on TM5. The aromatic ring of catechol strongly interacts with the aromatic cluster on TM6 comprising F2906.52 and W2866.48 and a few hydrophobic residues such as I2946.56 and V2976.59.
Potential conformational switches distinguishing full from partial agonists: role of F2826.44 on TM6
F2826.44 is a highly conserved residue (conserved among the biogenic amine receptors) in TM6 one turn below the tryptophan of the WxP motif. Mutating F2826.44 to hydrophobic residues such as alanine and leucine has been shown to increase the constitutive activity of the β2AR (16). In our apoprotein model, F2826.44 interacts with another aromatic residue Y3267.53 on TM7, which is part of the GPCR conserved NPxxY motif. This π-π aromatic interaction contributes to the stability of the apoprotein conformation of the receptor. Mutating F2826.44 to hydrophobic residues will eliminate this interaction with Y3267.53 and result in constitutive activity. Comparing the conformations of F2826.44 in the different ligand optimized states, as shown in Fig. 11, we find that norepinephrine causes F2826.44 to move away from Y3267.53 (distance increases from 6 Å to 7.6 Å), whereas for salbutamol, the distance between F2826.44 and Y3267.53 decreases to 4.3 Å. In the case of dopamine, the distance between these two residues is 6.9 Å, and for catechol, the distance does not change. Therefore, according to our predictions, the full agonist norepinephrine completely breaks the contact between F2826.44 and Y3267.53, and the partial agonist dopamine weakens this interaction. Salbutamol (the noncatecholamine partial agonist) stabilizes the interaction between these two residues. This could be tested out experimentally to distinguish full agonists from partial agonists.
FIGURE 11.
Relative orientations of F282 and Y326 in the apoprotein and ligand-bound structures. (a) Apoprotein conformation. (b) Norepinephrine-stabilized state. (c) Salbutamol-stabilized state.
Role of F2085.47 on TM5 in mediating the rotamer toggle switch on TM6
In our predicted ligand-bound conformations, F2085.47 on TM5 shows a strong interaction with the bound agonists. F2085.47 is conserved among the biogenic amine receptors. In the apoprotein state, F2085.47 is initially outside the binding cavity, but it moves in once the ligand binds and causes a conformational change to the receptor. The interaction between the agonists and F2085.47 is stronger by 1.4 kcal/mol compared to the interaction with F2906.52. According to the rotamer toggle switch model proposed in the literature, F2906.52 directly interacts with the ligand and transmits a conformational signal to the W2866.48 rotamer. In our ligand-bound conformations, the F2085.47 rotamer is positioned between the catechol ring and F2906.52 (Fig. 12) and shows a strong interaction with both the ligand and F2906.52. Therefore, F2085.47 is likely to play a role in the rotamer toggle switch, possibly by mediating the conformational signal from the agonist to F2906.52. This suggests a potential role of TM5 in rotamer toggling, in addition to the role of TM6.
FIGURE 12.
Location of F208 on TM5 in the binding pocket of norepinephrine, shown along with the WxP motif on TM6. Norepinephrine is shown in pink.
A potential molecular switch for distinguishing the role of the β-hydroxy group in norepinephrine from that in dopamine
In the norepinephrine-bound conformation, toggling of the W2866.48 rotamer places the imidazole nitrogen facing TM5. This facilitates the formation of a hydrogen bond with M2155.54 on TM5 (Fig. 12), which is partially conserved among the adrenergic receptors. This hydrogen bond is not present in any other agonist-stabilized structure. For the partial agonists salbutamol and dopamine, the rotation of TM6 is not sufficient to bring the W2866.48 side chain close to M2155.54 for the formation of the hydrogen bond. Norepinephrine and dopamine turn on both the ionic lock and the rotamer toggle switches, but they stabilize distinctly different receptor conformations, as observed in the fluorescence experiments. Therefore the question arises whether there is a third molecular switch that can distinguish the norepinephrine-bound structure from the dopamine-bound structure. The possibility that the HB between W2866.48 and M2155.54 could potentially serve as this new molecular switch can be verified experimentally.
Insight into GPCR activation
In the predicted receptor models, the conformational states stabilized by each ligand for the same receptor are distinct from one another. The five ligands studied here differ in the rotation of TM5 and TM6. Fig. 13, a and b, shows the section of the ligand-binding energy landscape for the inverse agonist ICI-118551 and the full agonist norepinephrine, respectively, for rotations of TM5 and TM6 keeping TM3 at a fixed angle. The location of the starting structural model and the final ligand-stabilized model for each ligand are shown in Fig. 13. The ICI-118551-stabilized state is located close to the initial state on the binding energy landscape. The agonist-stabilized states are in the blue region in Fig. 13 a, showing that the inverse agonist has favorable binding energies for these receptor conformations as well. However, there is a significant energy barrier between the inverse-agonist- and agonist-stabilized states, as shown in Fig. 13 a. This makes the agonist-bound conformations inaccessible to the inverse agonist. Examining the receptor structural models, we find that the energy barrier could arise due to a steric clash between F2085.47 on TM5 and the aromatic part of ICI-118551, which restricts TM5 rotations. We propose that mutating F2085.47 to alanine will reduce the inverse agonistic effect of ICI-118551, thereby serving as a possible validation for this structural model. As seen in Fig. 13 b, positive rotation angles for TM6 are associated with unfavorable binding energies for both the agonist and the inverse agonist. The rest of the binding energy landscape for norepinephrine (Fig. 13 b) shows smaller energy barriers compared to the inverse agonist landscape. The receptor states stabilized by all the ligands are accessible from an agonist-bound state. The highest energy barrier in the norepinephrine landscape is ∼5 times lower compared to the highest barrier in the ICI-118551 landscape. The flat energy landscape of norepinephrine (and also the other agonists not shown) reflects the increased conformational flexibility of an agonist-bound receptor as opposed to an inverse-agonist-bound receptor. These calculations provide a quantitative model for the intuitive protein energy landscapes discussed by Kobilka and co-workers (58).
FIGURE 13.
Binding energy landscapes (as a function of TM5 versus TM6 rotation) for two adrenergic ligands. (a) Inverse agonist ICI-118551. (b) Full agonist norepinephrine. The red regions represent unfavorable binding energies and the blue regions represent favorable binding energies. The various ligand-stabilized states are marked by dark circles. The white circles represent the possible inverse agonist stabilized states.
Virtual ligand screening of ligand optimized receptor conformations
To demonstrate the use of the ligand optimized receptor conformation compared to the apoprotein conformation for drug design, we carried out a VLS of 10,060 ligands on the apoprotein and the norepinephrine-bound conformations. For both receptor conformations, all the adrenergic ligands scored among the top 40% after sorting by protein-ligand interaction, as described in the Methods section. Fig. 12 shows the distribution of the adrenergic ligands in the sorted ligand list for the apoprotein and the norepinephrine-bound conformations. In Fig. 14, each datapoint represents the probability of finding adrenergic ligands within a certain percent-cutoff range. For example, the point corresponding to the 15% cutoff represents the probability of finding adrenergic ligands between 12.5% and 17.5% cutoffs. For both conformations, the distributions show a high concentration of adrenergic ligands around a 10% cutoff. The norepinephrine-bound conformation shows a sharper, narrower peak compared to the apoprotein conformation, which indicates a higher localization of adrenergic ligands toward lower percent cutoffs for the norepinephrine-bound conformation. Next, we compare the yield of adrenergic ligands in the two conformations (Table 1). At low percent cutoffs, the norepinephrine-bound conformation shows consistently higher yields compared to the apoprotein conformation. For a filter cutoff of 10%, which is a common percent cutoff in commercial drug screening, the apoprotein conformation shows a 26% yield of adrenergic ligands, whereas for the norepinephrine-bound conformation, the yield is 36%. Therefore, using the norepinephrine-bound conformation improves the overall yield by 38% compared to apoprotein. Above 25%, the yields for both the conformations become comparable to one another.
FIGURE 14.
Distribution of adrenergic ligands in the sorted ligand list obtained from VLS. The distribution curves are shown after Gaussian smoothing and noise removal. The total number of ligands in the test database was 10,060. The optimum enrichment obtained corresponded to a 10% cutoff.
TABLE 1.
Overall yields for VLS of adrenergic ligands at different percent cutoffs
| % Cutoff | % Yield (apoprotein) | % Yield (norepinephrine-bound conformation) |
|---|---|---|
| 0 | 0 | 0 |
| 3.5 | 6.7 | 15.5 |
| 5.5 | 11.7 | 20.7 |
| 10 | 26.7 | 36.2 |
| 13 | 46.7 | 60.3 |
| 15 | 53.3 | 67.2 |
| 20 | 70 | 75.9 |
| 21 | 71.7 | 77.6 |
| 25 | 78.3 | 79.3 |
CONCLUSION
We applied the LITiCon procedure for examining the perturbations caused by ligand binding to GPCRs, by a systematic spanning of helical rotational orientations. The method was tested on human β2AR, for which experimental information on the activation kinetics and conformational changes is available. Starting from a model of the β2AR receptor, predicted using MembStruk4.0, and ligand docking, using Glide, we predicted the binding sites for five adrenergic ligands: norepinephrine (full agonist), salbutamol (partial agonist), dopamine (weak partial agonist), catechol (very weak partial agonist), and ICI-118551 (inverse agonist). We calculated the changes in rotational orientation induced by ligand binding for each helical domain for the five ligands. The binding energy surfaces for various helical rotational orientations were calculated. Experimentally, the different ligand-stabilized conformations are characterized by breaking of the ionic lock between R1313.50 on TM3 and E2686.30 on TM6 and the rotamer toggle switch of W2866.48 on TM6. Norepinephrine and dopamine engage both of these switches, but salbutamol only breaks the ionic lock, and catechol only activates the rotamer toggle switch. These results are in agreement with the predictions made using our method. We also investigated the binding cavity positions of the five ligands and described the similarities and differences among them. Norepinephrine and dopamine occupy the same position inside the binding cavity, and the binding site of salbutamol is shifted toward TM4 and away from TM6. The catechol binding site overlaps with those of norepinephrine and dopamine, but not with salbutamol. This is in agreement with the competition studies involving catechol, norepinephrine, and salbutamol. The inverse agonist ICI-118551 binds deep into the binding pocket and stabilizes the receptor in a conformation close to the starting structural model. The inverse agonist does not form any HBs with the serines on TM5, but forms a HB with H2966.58 and N2936.55 on TM6. The predictions made for the agonists bring out the similarities and differences between the binding sites of the ligands, in full agreement with the mutation experiments.
One of the important predictions of this method is a potential conformational switch that distinguishes catechol agonists from a noncatechol agonist like salbutamol. We find that the full agonist norepinephrine breaks the contact between F2826.44 on TM6 and Y3267.53 on TM7, whereas the partial agonist salbutamol stabilizes this interaction. This can be tested experimentally and could potentially serve as a distinguishing feature among full and partial agonists. Another prediction is that a conformational switch defines the role of the β-hydroxyl group in norepinephrine that makes it a stronger agonist than dopamine. The interhelical HB between W2866.48 and M2155.54 is formed upon activation by norepinephrine, whereas this is not present for the dopamine-stabilized state. This could potentially serve as a new molecular switch, which can be verified experimentally.
The binding energy surface for the inverse-agonist-bound β2AR shows that the agonist-stabilized conformations are inaccessible to the inverse agonist, because it is separated from them by a large energy barrier. However, the binding energy surface of the agonist-bound β2AR shows that other conformations are accessible for the agonist-bound state. The highest energy barrier in the norepinephrine landscape is about five times lower than the highest barrier in the ICI-118551 landscape, thus showing that the agonist-bound conformation is flexible. These calculations provide a quantitative basis for the existing intuitive models for GPCR activation (58).
Since each ligand stabilizes a slightly different receptor conformation, this computational method is very useful for drug design. We have shown that VLS of large ligand libraries leads to a substantial enrichment in the hit rate at 10% cutoff for the ligand-stabilized conformations. LITiCon can be applied to any starting structure, be it a crystal structure, a homology model of the receptor with the ligand docked, or any other model. Moreover, the LITiCon method can be used to derive models that can guide experiments in the study of activation processes in class A GPCRs using biophysical techniques. The method does not take into account the allosteric conformational changes that occur in the ligand-bound receptor. However, using a ligand-stabilized receptor conformation from this study, one could perform MD simulations for mapping the initial events leading to allosteric conformational changes.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
The authors thank the Beckman Research Institute of the City of Hope for funding this research. We also thank Dr. Brian Kobilka for insightful and helpful discussions.
Supriyo Bhattacharya and Spencer E. Hall contributed equally to this work.
Editor: Jennifer Linderman.
References
- 1.Kobilka, B. K. 2007. G protein coupled receptor structure and activation. Biochim. Biophys. Acta. 1768:794–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hubbell, W. L., C. Altenbach, C. M. Hubbell, and H. G. Khorana. 2003. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv. Protein Chem. 63:243–290. [DOI] [PubMed] [Google Scholar]
- 3.Bartfai, T., J. Benovic, J. Bockaert, R. A. Bond, M. Bouvier, A. Christopoulos, O. Civelli, L. A. Devi, S. R. George, A. Inui, B. K. Kobilka, R. Leurs, R. Neubig, J.-P. Pin, R. Quirion, B. P. Roques, T. P. Sakmar, R. Seifert, R. E. Stenkamp, and P. G. Strange. 2004. The state of GPCR research in 2004. Nat. Rev. Drug Discov. 3:577–626. [DOI] [PubMed] [Google Scholar]
- 4.Gether, U., S. Lin, P. Ghanouni, J. A. Ballesteros, H. Weinstein, and B. K. Kobilka. 1997. Agonists induce conformational changes in transmembrane domains III and VI of the β2 adrenoceptor. EMBO J. 16:6737–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghanouni, P., J. J. Steenhuis, D. L. Farrens, and B. K. Kobilka. 2001. Agonist-induced conformational changes in the G-protein-coupling domain of the β2 adrenergic receptor. Proc. Natl. Acad. Sci. USA. 98:5997–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao, X., C. Parnot, X. Deupi, V. R. P. Ratnala, G. Swaminath, D. L. Farrens, and B. K. Kobilka. 2006. Coupling ligand structure to specific conformational changes in the β2-adrenoceptor. Nat. Chem. Biol. 2:417–422. [DOI] [PubMed] [Google Scholar]
- 7.Javitch, J. A., D. Fu, G. Liapakis, and J. Chen. 1997. Constitutive Activation of the β2 Adrenergic Receptor Alters the Orientation of Its Sixth Membrane-spanning Segment. J. Biol. Chem. 272:18546–18549. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen, S. G. F., A. D. Jensen, G. Liapakis, P. Ghanouni, J. A. Javitch, and U. Gether. 1999. Mutation of a highly conserved aspartic acid in the β2 adrenergic receptor: constitutive activation, structural instability, and conformational rearrangement of transmembrane segment 6. Mol. Pharmacol. 56:175–184. [DOI] [PubMed] [Google Scholar]
- 9.Ghanouni, P., Z. Gryczynski, J. J. Steenhuis, T. W. Lee, D. L. Farrens, J. R. Lakowicz, and B. K. Kobilka. 2001. Functionally different agonists induce distinct conformations in the G protein coupling domain of the β2 adrenergic receptor. J. Biol. Chem. 276:24433–24436. [DOI] [PubMed] [Google Scholar]
- 10.Swaminath, G., Y. Xiang, T. W. Lee, J. Steenhuis, C. Parnot, and B. K. Kobilka. 2004. Sequential binding of agonists to the β2 adrenoceptor: kinetic evidence for intermediate conformational states. J. Biol. Chem. 279:686–691. [DOI] [PubMed] [Google Scholar]
- 11.Swaminath, G., X. Deupi, T. W. Lee, W. Zhu, F. S. Thian, T. S. Kobilka, and B. K. Kobilka. 2005. Probing the β2 adrenoceptor binding site with catechol reveals differences in binding and activation by agonists and partial agonists. J. Biol. Chem. 280:22165–22171. [DOI] [PubMed] [Google Scholar]
- 12.Vilardaga, J.-P., R. Steinmeyer, G. S. Harms, and M. J. Lohse. 2005. Molecular basis of inverse agonism in a G protein-coupled receptor. Nat. Chem. Biol. 1:25–28. [DOI] [PubMed] [Google Scholar]
- 13.Shi, L., G. Liapakis, R. Xu, F. Guarnieri, J. A. Ballesteros, and J. A. Javitch. 2002. β2 Adrenergic receptor activation. Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. J. Biol. Chem. 277:40989–40996. [DOI] [PubMed] [Google Scholar]
- 14.Granier, S., S. Kim, A. M. Shafer, V. R. P. Ratnala, J. J. Fung, R. N. Zare, and B. K. Kobilka. 2007. Structure and conformational changes in the C-terminal domain of the β2-adrenoceptor: insights from fluorescence resonance energy transfer studies. J. Biol. Chem. 282:13895–13905. [DOI] [PubMed] [Google Scholar]
- 15.Gether, U., F. Asmar, A. K. Meinild, and S. G. F. Rasmussen. 2002. Structural basis for activation of G-protein-coupled receptors. Pharmacol. Toxicol. 91:304–312. [DOI] [PubMed] [Google Scholar]
- 16.Chen, S., F. Lin, M. Xu, R. P. Riek, J. Novotny, and R. M. Graham. 2002. Mutation of a single TMVI residue, Phe282, in the β2 adrenergic receptor results in a structurally distinct activated receptor conformation. Biochemistry. 41:6045–6053. [DOI] [PubMed] [Google Scholar]
- 17.Gabilondo, A. M., K. Cornelius, and M. J. Lohse. 1996. Mutations of Tyr326 in the β2-adrenoceptor disrupt multiple receptor functions. Eur. J. Pharmacol. 307:243–250. [DOI] [PubMed] [Google Scholar]
- 18.Strader, C. D., I. S. Sigal, M. R. Candelore, E. Rands, W. S. Hill, and R. A. F. Dixon. 1988. Conserved aspartic acid residues 79 and 113 of the β-adrenergic receptor have different roles in receptor function. J. Biol. Chem. 263:10267–10271. [PubMed] [Google Scholar]
- 19.Strader, C. D., T. M. Fong, M. R. Tota, D. Underwood, and R. A. Dixon. 1994. Structure and function of G protein-coupled receptors. Annu. Rev. Biochem. 63:101–132. [DOI] [PubMed] [Google Scholar]
- 20.Dixon, R. A. F., I. S. Sigal, E. Randa, R. B. Register, M. R. Candelore, A. D. Blake, and C. D. Strader. 1987. Ligand binding to the β-adrenergic-receptor involves its rhodopsin-like core. Nature. 326:73–77. [DOI] [PubMed] [Google Scholar]
- 21.Dixon, R. A. F., I. S. Sigal, M. R. Cadelore, R. B. Register, W. Scattergood, E. Rands, and C. D. Strader. 1987. Structural features required for ligand binding to the β adrenergic receptor. EMBO J. 6:3269–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ballesteros, J. A., A. D. Jensen, G. Liapakis, S. G. F. Rasmussen, L. Shi, U. Gether, and J. A. Javitch. 2001. Activation of the β2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. J. Biol. Chem. 276:29171–29177. [DOI] [PubMed] [Google Scholar]
- 23.Dunham, T. D., and D. L. Farrens. 1999. Conformational changes in rhodopsin. Movement of helix F detected by site-specific chemical labeling and fluorescence spectroscopy. J. Biol. Chem. 274:1683–1690. [DOI] [PubMed] [Google Scholar]
- 24.Liapakis, G., J. A. Ballesteros, S. Papachristou, W. C. Chan, X. Chen, and J. A. Javitch. 2000. The forgotten serine. A critical role for Ser-2035.42 in ligand binding to and activation of the β2-adrenergic receptor. J. Biol. Chem. 275:37779–37788. [DOI] [PubMed] [Google Scholar]
- 25.Ward, S. D. C., F. F. Hamdan, L. M. Bloodworth, and J. Wess. 2002. Conformational changes that occur during M3 muscarinic acetylcholine receptor activation probed by the use of an in situ disulfide cross-linking strategy. J. Biol. Chem. 277:2247–2257. [DOI] [PubMed] [Google Scholar]
- 26.Farrens, D., C. Altenbach, K. Yang, W. Hubbell, and H. G. Khorana. 1996. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 274:768–770. [DOI] [PubMed] [Google Scholar]
- 27.Altenbach, C., J. Klein-Seetharaman, K. Cai, H. G. Khorana, and W. L. Hubbell. 2001. Structure and function in rhodopsin: mapping light-dependent changes in distance between residue 316 in helix 8 and residues in the sequence 60–75, covering the cytoplasmic end of helices TM1 and TM2 and their connection loop CL1. Biochemistry. 40:15493–15500. [DOI] [PubMed] [Google Scholar]
- 28.Altenbach, C., J. Klein-Seetharaman, J. Hwa, H. G. Khorana, and W. L. Hubbell. 1999. Structural features and light-dependent changes in the sequence 59–75 connecting helices I and II in rhodopsin: a site-directed spin-labeling study. Biochemistry. 38:7945–7949. [DOI] [PubMed] [Google Scholar]
- 29.Altenbach, C., K. Cai, H. G. Khorana, and W. L. Hubbell. 1999. Structural features and light-dependent changes in the sequence 306–322 extending from helix VII to the palmitoylation sites in rhodopsin: a site-directed spin-labeling study. Biochemistry. 38:7931–7937. [DOI] [PubMed] [Google Scholar]
- 30.Schertler, G. F. 2005. Structure of rhodopsin and the metarhodopsin I photointermediate. Curr. Opin. Struct. Biol. 15:408–415. [DOI] [PubMed] [Google Scholar]
- 31.Sakmar, T. P., R. R. Franke, and H. G. Khorana. 1991. The role of the retinylidene Schiff base counterion in rhodopsin in determining wavelength absorbance and Schiff base pKa. Proc. Natl. Acad. Sci. USA. 88:3079–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ridge, K. D., N. G. Abdulaev, M. Sousa, and K. Palczewski. 2003. Phototransduction: crystal clear. Trends Biochem. Sci. 28:479–487. [DOI] [PubMed] [Google Scholar]
- 33.Arnis, S., K. Fahmy, K. P. Hofmann, and T. P. Sakmar. 1994. A conserved carboxylic acid group mediates light-dependent proton uptake and signaling by rhodopsin. J. Biol. Chem. 269:23879–23881. [PubMed] [Google Scholar]
- 34.Choi, G., J. Landin, J. F. Galan, R. R. Birge, A. D. Albert, and P. L. Yeagle. 2002. Structural studies of metarhodopsin II, the activated form of the G-protein coupled receptor, rhodopsin. Biochemistry. 41:7318–7324. [DOI] [PubMed] [Google Scholar]
- 35.Salom, D., D. T. Lodowski, R. E. Stenkamp, I. Le Trong, M. Golczak, B. Jastrzebska, T. Harris, J. A. Ballesteros, and K. Palczewski. 2006. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc. Natl. Acad. Sci. USA. 103:16123–16128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gouldson, P. R., N. J. Kidley, R. P. Bywater, G. Psaroudakis, H. D. Brooks, C. Diaz, D. Shire, and C. A. Reyenolds. 2004. Toward the active conformations of rhodopsin and the β2-adrenergic receptor. Proteins. 56:67–84. [DOI] [PubMed] [Google Scholar]
- 37.Nikiforovich, G. V., and G. R. Marshall. 2006. 3D Modeling of the activated states of constitutively active mutants of rhodopsin. Biochem. Biophys. Res. Commun. 345:430–437. [DOI] [PubMed] [Google Scholar]
- 38.Urban, J. D., W. P. Clarke, M. von Zastrow, D. E. Nichols, B. K. Kobilka, H. Weinstein, J. A. Javitch, B. L. Roth, A. Christopoulos, P. M. Sexton, K. J. Miller, M. Spedding, and R. B. Mailman. 2007. Functional selectivity and classical concepts of quantitative pharmacology. J. Pharmacol. Exp. Ther. 320:1–13. [DOI] [PubMed] [Google Scholar]
- 39.Lacapère, J.-J., E. Pebay-Peyroula, J.-M. Neumann, and C. Etchebest. 2003. Determining membrane protein structures: still a challenge! Trends Biochem. Sci. 32:259–270. [DOI] [PubMed] [Google Scholar]
- 40.Crozier, P. S., M. J. Stevens, L. R. Forrest, and T. B. Woolf. 2003. Molecular dynamics simulation of dark-adapted rhodopsin in an explicit membrane bilayer: coupling between local retinal and larger scale conformational change. J. Mol. Biol. 333:493–514. [DOI] [PubMed] [Google Scholar]
- 41.Spijker, P., N. Vaidehi, P. L. Freddolino, P. A. J. Hilbers, and W. A. Goddard 3rd. 2006. Dynamic behavior of fully solvated β2-adrenergic receptor, embedded in the membrane with bound agonist or antagonist. Proc. Natl. Acad. Sci. USA. 103:4882–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu, W., J. Shen, X. Luo, W. Zhu, J. Cheng, K. Yu, J. M. Briggs, G. Jin, K. Chen, and H. Jiang. 2007. Dopamine D1 receptor agonist and D2 receptor antagonist effects of the natural product (−)-stepholidine (SPD): molecular modeling and dynamics simulations. Biophys. J. 93:1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Canutescu, A. A., A. A. Shelenkov, and R. L. Dunbrack, Jr. 2003. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 12:2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zamanakos, G. 2001. A fast and accurate analytical method for the computation of solvent effects in molecular simulations. PhD thesis. Caltech, Pasadena, CA.
- 45.McDonald, I. K., and J. M. Thornton. 1994. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238:777–793. [DOI] [PubMed] [Google Scholar]
- 46.Phillips, J. C., R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kale, and K. Schulten. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim, K.-T., S. Brunett, M. Iotov, R. B. McClurg, N. Vaidehi, S. Dasgupta, S. Taylor, and W. A. Goddard 3rd. 1997. Molecular dynamics for very large systems on massively parallel computers: the MPSim program. J. Comput. Chem. 18:501–521. [Google Scholar]
- 48.Reference deleted in proof.
- 49.Hall, S. E. 2005. Development of a structure prediction method for G-protein coupled receptors. PhD thesis. Caltech, Pasadena, CA.
- 50.Floriano, W. B., N. Vaidehi, and W. A. Goddard 3rd. 2004. Making sense of olfaction through predictions of the 3-D structure and function of olfactory receptor. Chem. Senses. 29:269–290. [DOI] [PubMed] [Google Scholar]
- 51.Vaidehi, N., W. B. Floriano, R. Trabanino, S. Hall, P. Freddolino, E. J. Choi, G. Zamanakos, and W. A. Goddard 3rd. 2002. Structure and function prediction for G-protein coupled receptors. Proc. Natl. Acad. Sci. USA. 99:12622–12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friesner, R. A., R. B. Murphy, M. P. Repasky, L. L. Frye, J. R. Greenwood, T. A. Halgren, P. C. Sanschagrin, and D. T. Mainz. 2006. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 49:6177–6196. [DOI] [PubMed] [Google Scholar]
- 53.Hannawacker, A., C. Krasel, and M. J. Lohse. 2002. Mutation of Asn293 to Asp in transmembrane helix VI abolishes agonist-induced but not constitutive activity of the β2-adrenergic receptor. Mol. Pharmacol. 62:1431–1437. [DOI] [PubMed] [Google Scholar]
- 54.Carmine, R. D., P. Molinari, M. Sbraccia, C. Ambrosio, and T. Costa. 2004. “Induced-fit” mechanism for catecholamine binding to the β2-adrenergic receptor. Mol. Pharmacol. 66:356–363. [DOI] [PubMed] [Google Scholar]
- 55.Wieland, K., H. M. Zuurmond, C. Krasel, A. P. I. Jzerman, and M. J. Lohse. 1996. Involvement of Asn-293 in stereospecific agonist recognition and in activation of the β2-adrenergic receptor. Proc. Natl. Acad. Sci. USA. 93:9276–9281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zongren, W., D. S. Thiriot, and A. E. Ruoho. 2001. Tyr99 in the transmembrane domain 5 of the β2-adrenergic receptor interacts directly with the pharmacophore of a unique fluorenone based antagonist. Biochem. J. 354:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kikkawa, H., H. Kurose, M. Isogaya, Y. Sato, and T. Nagao. 1997. Differential contribution of two serine residues of wild type and constitutively active β2-adrenoceptors to the interaction with β2-selective agonists. Br. J. Pharmacol. 121:1059–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kobilka, B. K., and X. Deupi. 2007. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol. Sci. 28:397–406. [DOI] [PubMed] [Google Scholar]