Abstract
Due to their involvement in processes such as DNA replication, repair, and recombination, bacterial single-stranded DNA binding (SSB) proteins are essential for the survival of the bacterial cell. Whereas most bacterial SSB proteins form homotetramers in solution, dimeric SSB proteins were recently discovered in the Thermus/Deinococcus group. In this work we characterize the biophysical properties of the SSB protein from Thermus aquaticus (TaqSSB), which is structurally quite similar to the tetrameric SSB protein from Escherichia coli (EcoSSB). The binding of TaqSSB and EcoSSB to single-stranded nucleic acids was found to be very similar in affinity and kinetics. Mediated by its highly conserved C-terminal region, TaqSSB interacts with the χ-subunit of E. coli DNA polymerase III with an affinity that is similar to that of EcoSSB. Using analytical ultracentrifugation, we show that TaqSSB mutants are able to form tetramers in solution via arginine-mediated hydrogen-bond interactions that we identified in the crystal packing of wild-type TaqSSB. In EcoSSB, we identified a homologous arginine residue involved in the formation of higher aggregates and metastable highly cooperative single-stranded DNA binding under low salt conditions.
INTRODUCTION
One of the main requirements for sustainment of life is to conserve the integrity of the genetic material through a variety of different reactions, mainly replication, recombination, or repair. The single-stranded state of DNA (ssDNA) during these reactions is most vulnerable and needs to be protected and shaped for the reactions to follow. ssDNA binding (SSB) proteins play a key role in this process.
SSB proteins in general bind to ssDNA and have little or no affinity to double-stranded DNA. Though some SSB proteins exhibit sequence specificity to fulfill regulatory functions (1), most of them bind with little or no sequence preference. X-ray crystallographic studies showed that SSB proteins share a common structural motif, the oligonucleotide/oligosaccharide binding fold (OB-fold) (2). Most SSB proteins contain this fold more than once in the structure, indicating that there is more than a single binding site for ssDNA in these proteins.
However, the quaternary structure of SSB proteins can differ considerably. Representatives of monomers (3,4), homodimers (5,6), homotetramers (7–9), and heterotrimers (10) have been described. Most widely distributed among different species are the heterotrimeric SSB proteins that occur in eukaryotes and have analogs in the archaea kingdom and the homotetrameric SSB proteins, which are the essential SSB proteins for bacteria and eukaryotic mitochondria.
Only recently has it been shown that SSB proteins from the Thermus/Deinococcus group of bacteria form homodimers instead of homotetramers, with the polypeptide chain of the protomer being almost twice as long as observed for other bacteria (11–13). Sequence comparison and structural studies showed a large similarity to homotetrameric proteins, and it is now accepted that the Thermus/Deinococcus group SSB proteins result from a gene duplication event and must be grouped together with the other bacterial SSBs. This study focuses on the SSB protein from Thermus aquaticus (TaqSSB).
The structure of bacterial ssDNA binding proteins consists of three parts. The N-terminal part of the sequence contains the OB-fold and forms the tetrameric ssDNA binding interface in which four OB-folds act together. In TaqSSB the N-terminal part is much longer. It contains two OB-folds that form a dimer with a structure similar to that of the EcoSSB tetramer (14,15). This ssDNA binding/oligomerization domain is succeeded by a proline- and glycine-rich stretch of amino acids acting as a spacer to the highly conserved region of the last 10 C-terminal amino acids. It has been shown that these last 10 amino acids are essential for the survival of the bacterial cell (16) and that they are a prime target for other proteins interacting with SSB (17–20). Whereas homotetrameric bacterial SSB proteins have four C-termini, the homodimeric TaqSSB has only two of them; the other two were lost during gene duplication and subsequent evolution.
In DNA metabolism, e.g., replication, long stretches of ssDNA have to be covered by SSB proteins. To mimic this situation in vitro, SSB protein binding to long ssDNA, preferably poly(dT), is investigated. This binding can be described as the binding of a multidentate ligand (SSB) to a long linear polymer (ssDNA) (21,22). In such a model, the isolated binding of a single ligand must be treated differently from the adjacent cooperative binding of a second ligand. For the binding of EcoSSB to poly(dT) under high salt conditions, moderate cooperativity has been reported (9,23). Under low salt conditions, however, the formation of the EcoSSB/ssDNA complex is highly cooperative and the cooperatively bound proteins redistribute extremely slowly to a more random and less cooperative distribution (24). From crystal packing interactions observed in the structure of EcoSSB and an EcoSSB/ssDNA complex, this metastable high cooperativity has been speculated to be the result of an interaction of the L45 loops of two adjacently bound EcoSSB molecules (25,26).
In this study, we characterize the ssDNA and protein binding abilities of the TaqSSB protein and investigate the role of the C-terminus and the proline- and glycine-rich spacer in the interaction of TaqSSB with other proteins. We demonstrate that the C-terminus is an important signal of the functional state of SSB that is recognized by other factors of the DNA replication machinery. Furthermore, we describe the self-association of a mutated TaqSSB protein to form a tetramer and show how this tetramerization is modulated by different modifications of the protein. Sequence and structure comparisons suggest that this interaction is conserved in the tetrameric EcoSSB protein, and we show that mutation of one of the involved residues reduces the metastable high cooperativity in the low salt binding mode of EcoSSB. Using these data, we propose a model for a mechanism of the cooperative binding of SSB proteins to ssDNA.
MATERIALS AND METHODS
Buffers and reagents
Poly(dT) and poly(rU) were purchased from GE Healthcare Life Sciences (Munich, Germany). Polynucleotide concentrations are given in monomer residues throughout the text and were determined spectrophotometrically using absorption coefficients of 8600 M−1 cm−1 for poly(dT) at maximum (27) and of 9200 M−1 cm−1 for poly(rU) at 260 nm (28). Protein concentrations were determined spectrophotometrically using the absorption coefficients at 280 nm calculated from amino acid composition (29) unless stated otherwise: 77,920 M−1 cm−1 for wild-type TaqSSB and all TaqSSB mutants, 113,000 M−1 cm−1 for wild-type EcoSSB (30) and EcoSSB R72A, 89,840 M−1 cm−1 for EcoSSBΔ116-167 and EcoSSBG117*, and 29,400 M−1 cm−1 for the χ-subunit of Escherichia coli DNA polymerase III.
Experiments were carried out in potassium phosphate buffer (KPi), pH 7.4, containing NaCl as a neutral salt, where high salt buffer is defined as 0.3 M NaCl, 20 mM KPi, pH 7.4 and low salt buffer used for analytical ultracentrifugation is defined as 5 mM NaCl, 5 mM KPi, pH 7.4, 0.87 M glycerol. In the case of fluorescence titrations, low salt buffer is defined as 1 mM NaCl, 1 mM KPi, pH 7.4, and 100 ppm Tween 20 to compare to results published earlier for EcoSSB and the SSB protein from Deinococcus radiodurans (DraSSB) (13).
Construction of mutant SSB proteins
The wild-type TaqSSB protein was cloned as described previously (14). Site-directed mutagenesis was performed using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). Mutant SSB proteins are designated by the amino acid exchange, e.g., EcoSSB R72A is an EcoSSB mutant in which the arginine residue at position 72 was replaced by an alanine.
The TaqSSB deletion mutant TaqSSBΔ228-252, in which amino acids 1–227 of TaqSSB are followed by the last 12 amino acids of TaqSSB, was constructed as described previously (14). The mutant TaqSSBΔ228-252 R190A was obtained by site-directed mutagenesis of pETTaqSSBΔ228-252 (14). TaqSSB R229* results from introducing a double-stranded DNA cassette into NdeI/NotI-digested pETTaqSSBentry (14), replacing codon 229 by a stop codon. TaqSSB L262A P263A F264A and TaqSSB E259Q E260Q D261Q were constructed by site-directed mutagenesis of pETTaqSSB (14). EcoSSB R72A was obtained by site-directed mutagenesis of the plasmid pSF1 bearing the ssb gene of E. coli (31).
Expression and purification of proteins
Wild-type TaqSSB, TaqSSBΔ228-252, and TaqSSBΔ228-252 R190A were expressed and purified as described earlier (14). TaqSSB R229* was purified the same way wild-type TaqSSB was, omitting the polyethylenimine precipitation and ResourceQ column, yielding a protein preparation ∼95% pure, as judged from Coomassie brilliant blue stained sodium dodecyl sulfate (SDS) gels. TaqSSB L262A P263A F264A was purified the same way wild-type protein was except for the use of 30% (w/v) (NH4)2SO4 for precipitation. TaqSSB E259Q E260Q D261Q was purified the same way wild-type was except for omission of the TGE400 wash step and the use of 30% (w/v) (NH4)2SO4 for precipitation.
The expression of EcoSSB and EcoSSB R72A was performed using plasmids carrying the E. coli ssb gene under the control of the λ PL promoter in E. coli strain BT317 (32), which allows induction of expression by temperature shift from 30°C to 42°C by denaturing the thermosensitive λ-repressor present in BT317. Wild-type EcoSSB and EcoSSB R72A were prepared as described for wild-type EcoSSB (33). EcoSSBΔ116-167 and EcoSSB G117* were expressed and prepared as described earlier (16). DraSSB and the χ-subunit of the E. coli DNA polymerase III were expressed and purified as described (13).
Fluorescence measurements
Fluorescence titrations were carried out as described earlier (34) in a Jasco FP-6500 fluorimeter (Tokyo, Japan). Excitation wavelength was 295 nm, and the intrinsic tryptophan fluorescence was detected at an emission wavelength of 350 nm. Concentrations of protein and ssDNA did not exceed a total absorption of 0.05 at excitation wavelength throughout the titration to avoid inner filter effects. All titrations were carried out at 22°C in buffers containing 100 ppm Tween 20 to stabilize the proteins against denaturation (35). After each addition, the solution was allowed to reequilibrate for 60–300 s.
Binding curves were analyzed using the model of Schwarz and Watanabe (22) for the binding of a multidentate ligand to a long linear polymer. In this model the concentration of free SSB protein cA is calculated as follows:
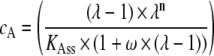 |
with
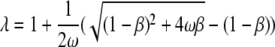 |
and
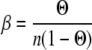 |
where Θ is the degree of saturation, n the binding site size, KAss the binding affinity, and ω the cooperativity. The algorithm is implemented in the BPCfit program package (36), which allows the nonlinear least squares fit of n, KAss, and ω, with the fluorescence of free (Ffree) and nucleic acid bound protein (Fbound) fitted by linear regression. The fluorescence quench is calculated as 1 − Fbound/Ffree.
Stopped-flow kinetics
The determination of the association rate constant of TaqSSB and poly(dT) was investigated using a fluorescence stopped-flow system (Hi-Tech Scientific, Bradford on Avon, UK) measuring tryptophan fluorescence excited at 295 nm. A series of measurements carried out at the same concentrations was accumulated to reduce noise. As described previously (13), data sets from different concentrations were analyzed by global fitting, using the model of Urbanke and Schaper (27) provided by the BPCfit software package (36).
Analytical ultracentrifugation
Analytical ultracentrifugation experiments were performed in a Beckman/Coulter XL-A analytical ultracentrifuge (Beckman/Coulter, Palo Alto, CA) using An50Ti or An60Ti rotors at 20°C. Concentration profiles were measured with the ultraviolet (UV)-absorption scanning optics of the centrifuge. Beckman/Coulter XL-A software was run on a computer attached to the centrifuge to program the centrifuge and record the data.
Sedimentation velocity experiments were carried out with 400 μL samples in double-sector centerpieces at the indicated rotor speeds. To analyze protein-protein interactions and visualize distributions of s-values, the measured concentration profiles A(x,t) (A: measured absorption, x: distance from center of the rotor, t: time of measurement) were evaluated with the program package SEDFIT (37). This program provides a model for diffusion-corrected differential sedimentation coefficient distributions (c(s) distributions). As the areas under the separate peaks in c(s) distributions are a measure of the absorbance of the species represented by the peaks (38), this information can be used to determine binding isotherms (20,39). For highly heterogeneously aggregating samples, the measured sedimentation profiles were converted to cumulative sedimentation coefficient distributions A(s) by
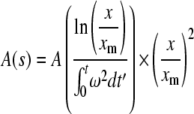 |
as described earlier (20,40), where xm is the radial position of the meniscus, ω is the angular velocity, and s the sedimentation coefficient.
When sedimentation coefficients and molar masses of single species were of interest, these were determined using the BPCfit program package (36), which analyzes the concentration profiles by fitting a numerical solution of Lamm's differential equation (41) to the measured data. For hydrodynamic analysis, measured s-values were corrected to s20°C,W using the partial specific volumes calculated from amino acid composition (42).
Sedimentation equilibrium experiments were carried out with 120 μL samples in six-channel centerpieces in an An50Ti rotor running at 20°C and analyzed as described earlier (13).
RESULTS AND DISCUSSION
Solution properties of TaqSSB
The SSB protein from T. aquaticus (TaqSSB) was expressed in E. coli and purified to homogeneity as judged by SDS gel electrophoresis (43). Sedimentation equilibrium in the analytical ultracentrifuge yielded a molar mass of (59.6 ± 1) kg/mol for TaqSSB (Supplementary Material Fig. S1) and showed no dependence on protein concentration. Since the molar mass of the covalent protein chain of TaqSSB is 30.0 kg/mol, it can be concluded that TaqSSB forms stable dimers in solution. Sedimentation velocity experiments analyzed by least-squares fitting of the Lamm equation (36,41) to the sedimentation profiles resulted in a sedimentation coefficient s20°C,W of 4.04 S and a molar mass of 56.7 kg/mol (data not shown). Conformational heterogeneities of the protein would lead to a broadening of the sedimentation profiles and thus to a drastic underestimation of the molar mass in Lamm equation fitting analysis (44). The good agreement between the molar masses of TaqSSB determined in sedimentation velocity and sedimentation equilibrium experiments shows that the protein is conformationally homogeneous.
The calculated molar mass of the dimer and the measured sedimentation coefficient gave a frictional ratio of 1.38. Since hydrated spherical proteins are expected to show a frictional ratio of 1.1–1.2 (44), TaqSSB seems to be either strongly asymmetric or globular with some protrusions. Similar results have been described for DraSSB, another SSB protein from the Thermus/Deinococcus group (13).
Binding of TaqSSB to single-stranded nucleic acids
When binding to single-stranded nucleic acids, the intrinsic tryptophan fluorescence of TaqSSB decreases up to 70%. Fluorescence titrations of TaqSSB with poly(dT) under high salt (0.3 M NaCl, 20 mM KPi) and low salt (1 mM NaCl, 1 mM KPi) conditions are shown in Fig. 1. Whereas the fluorescence quench depends on the salt concentration, ∼54 nucleotides can be bound to a TaqSSB dimer independent of the salt concentration. The cooperative DNA binding constant is estimated to be in the range of 107–108 M−1. Similar results were obtained for the related DraSSB protein (13,45). The constant binding stoichiometry is in contrast to EcoSSB, where at low salt the binding site size is reduced from 65 to 35 nucleotides (9). For DraSSB no distinct differences in binding stoichiometry could be observed, but notable differences in fluorescence quench between low and high salt conditions were seen (13). Thus, it appears that in SSB proteins from the Thermus/Deinococcus group, ssDNA binding under low salt conditions is accompanied by a decrease in fluorescence quench but not in an altered binding site size. Nevertheless, this change in intrinsic tryptophan fluorescence quench points to differences in the interaction of these SSB proteins with ssDNA at high and low salt concentrations. Modeling a TaqSSB-ssDNA complex by docking the DNA structure from an EcoSSB-ssDNA complex at high salt conditions (25) to TaqSSB revealed that ssDNA can bind to TaqSSB in a similar way as it does to EcoSSB, with most of the stacking interactions preserved (14).
FIGURE 1.
Fluorescence titrations of 0.38 μM TaqSSB with poly(dT) in (▪) 300 mM NaCl, 20 mM KPi, pH 7.4, 100 ppm Tween 20 and in (▴) 1 mM NaCl, 1 mM KPi, pH 7.4, 100 ppm Tween 20 at 22°C. Intrinsic protein fluorescence was observed with excitation at 295 nm and emission at 350 nm. The solid lines represent binding isotherms calculated with (▪) KAssω = 6.4 × 107 M−1, n = 54, Qf = 0.72 and (▴) KAssω = 3.1 × 107 M−1, n = 53, Qf = 0.55 (see Materials and Methods).
To estimate the relative affinities of different SSB proteins to single-stranded nucleic acids, it is more adequate to use poly(rU) as a weakly binding substrate (46) than to use the strongly binding poly(dT). Fluorescence titrations at 0.2 M NaCl, 20 mM KPi, 100 ppm Tween 20 showed no measurable difference in the binding affinities of TaqSSB, DraSSB, and EcoSSB (data not shown).
Under high salt conditions, stopped flow analysis of the kinetics of association of TaqSSB to poly(dT) in a concentration range from 10 nM to 0.6 μM protein was performed. The data were compatible with a model derived for EcoSSB (27), where in the rate-determining step only a few nucleotides of ssDNA need to interact with a part of the SSB molecule. The association rate constant was determined to be 2.1 × 108 M−1s−1 (data not shown), a result similar to data obtained for the SSB proteins from E. coli (27), D. radiodurans (13), and human mitochondria (7).
Interaction of TaqSSB and the χ-subunit of DNA polymerase III
For EcoSSB it has been shown that the region of the last 10 amino acids is essential for the interaction with other proteins involved in DNA replication, repair, and recombination (17–20,47). One of the protein-protein interactions mediated by the C-terminus of EcoSSB is the binding to the χ-subunit of the DNA polymerase III holoenzyme. Since TaqSSB also carries the highly conserved C-terminal region, it is likely that a similar interaction between DNA polymerase III χ-subunit and TaqSSB exists. However, for T. aquaticus no gene has been discovered yet to encode a protein homologous to χ. We therefore decided to test the ability of TaqSSB to interact with other proteins via its C-terminus by using E. coli χ as a representative binding partner.
If in analytical sedimentation velocity centrifugation with two different proteins, the resulting sedimentation constant is larger than that of any of the constituents, this is a proof for an interaction between the two proteins. In Fig. 2 a, sedimentation coefficient distributions c(s) of TaqSSB in the absence and presence of χ under high salt conditions are shown. Whereas free TaqSSB shows a sedimentation constant s20°C,W of 4.0 S, in the presence of an excess of χ, s20°C,W increases to 4.4 S; this is a clear indication for complex formation. The same kind of experiment was repeated using different ratios of TaqSSB and χ. Free χ-protein (molar mass 16.6 kg/mol) sedimented with s20°C,W = 2.0 S, independent of the amount of TaqSSB present. TaqSSB and all TaqSSB/χ complexes in the presence of an excess of χ sedimented in a single boundary, showing the reaction to be fast compared to the timescale of sedimentation (48). The sedimentation coefficient of this reaction boundary increased with increasing amounts of bound χ. Since the binding of χ to TaqSSB is weak (see below) and the TaqSSB concentration used was below the inverse association constant, the concentration of free χ sedimenting in the reaction boundary is expected to be rather low (48). Additionally, simulation of the concentration profiles using the BPCfit program package (36) showed that even in the case of the lowest χ-concentration used, <6% of the free χ sedimented in the reaction boundary (data not shown). Therefore, integration of the two c(s) peaks can be used to determine the concentrations of free and bound χ-protein, respectively (38). A binding isotherm (Fig. 2 b) was fitted to the data using a model for noncooperative binding of n χ-molecules to one SSB molecule (20). From this binding isotherm it can be deduced that a TaqSSB dimer can bind up to two χ-proteins with an affinity (KAss) of ∼6 × 104 M−1. Nearly identical results have been obtained for the related DraSSB (13). The tetrameric SSB protein of E. coli binds up to four χ-proteins with an affinity of 2 × 105 M−1 (20). Despite the large evolutionary distance between TaqSSB and EcoSSB, the affinity to the E. coli χ-subunit is nearly unchanged, with the binding stoichiometry correlating directly with the number of C-termini in the native proteins. To prove that this interaction of TaqSSB is actually mediated by its highly conserved C-terminal region, we constructed two mutants, one lacking the last 36 amino acids (TaqSSB R229*) and the second containing the last 10 amino acids but missing the glycine-rich region from amino acids 228–252 (TaqSSBΔ228-252). Whereas TaqSSB R229* is no longer able to bind to the χ-protein, the deletion mutant TaqSSBΔ228-252 binds to χ with the same affinity and stoichiometry as wild-type TaqSSB does (Fig. 2 b). Therefore, the disordered glycine-rich region of TaqSSB (14) does not influence the binding of the χ-protein; however, the highly conserved region of the last 10 amino acids is essential for χ binding.
FIGURE 2.
Interaction of TaqSSB with E. coli DNA polymerase III χ-subunit at 20°C and 300 mM NaCl, 20 mM KPi, pH 7.4. (a) c(s) distributions of sedimentation velocity experiments in the analytical ultracentrifuge for 2.65 μM wild-type TaqSSB in the absence (dashed curve) or presence (solid curve) of 31.8 μM χ-protein. (b) Binding isotherm from sedimentation velocity experiments (see Materials and Methods) for the binding of χ-protein to wild-type TaqSSB (▴), TaqSSBΔ228-252 (▾), and TaqSSB R229* (▪). The solid line is a theoretical binding isotherm calculated for the binding of two χ-molecules to a TaqSSB dimer with an affinity KAss of 6.5 × 104 M−1.
Higher assembly of TaqSSB molecules in the crystal structure
In their interaction with long ssDNA, bacterial SSB proteins show positive binding cooperativity (49). This cooperative binding most probably is the result of a protein-protein interaction connecting SSB proteins adjacently bound on ssDNA. Crystal packing represents a subset of possible protein-protein interactions, some of which may have biological meaning. In the crystal structure of TaqSSB that we recently solved (Protein Data Bank (PDB) code: 2ihe) (14), there is an interaction that relates the TaqSSB molecules in the neighboring unit cells by rotation around the twofold axis. This rotation enables hydrogen-bond interactions between residues R190 and D193 of one molecule and D193 and R190 of the other molecule, respectively. Together with the two R226 residues of both molecules, the contact area resembles a short zipper-like structure of four arginine and two aspartate residues (Fig. 3 a). It involves not only the hydrogen bonds described but also more than 100 van der Waals contacts. In the crystal this contact area buries a surface of 448 Å2, and the resulting TaqSSB oligomer is compatible with the ssDNA binding model obtained by docking the ssDNA structure from an EcoSSB/ssDNA complex to TaqSSB (14), whereas other crystal packing interactions that we have observed are not.
FIGURE 3.
Arginine-mediated hydrogen-bond interaction of neighboring TaqSSB molecules as observed in x-ray analysis shown for the modeled TaqSSB-ssDNA and EcoSSB-ssDNA higher assembly complexes. (a) ssDNA is shown in gray metallic color and TaqSSB molecules (PDB code: 2ihe) involved in hydrogen bonding are in red and blue. The atoms making hydrogen bonds or van der Waals contacts are shown in Corey, Pauling, and Koltun representation. Note that the glycine-rich C-terminal region of TaqSSB in conformation C1 overlaps with the bound ssDNA, whereas conformation C2 is fully compatible with ssDNA binding. The close-up (right-hand) shows that the arginine-mediated interaction does not interfere with the binding of ssDNA. (b) Superposition of the interacting structures connecting neighboring TaqSSB and EcoSSB molecules. Parts of the TaqSSB molecules joined by the arginine-mediated interaction are shown in green. Superimposed are parts of the EcoSSB molecules (PDB code: 1eyg) shown in blue. The areas for the different molecules are indicated by different background colors. Dotted lines indicate distances (in Å) between TaqSSB interacting residues, and the dashed lines indicate the distances for EcoSSB. Although the aspartate 193 residue is not conserved in EcoSSB, the described arginine-mediated interaction can be realized through hydrogen bonds between arginine 72 and serine 75 residues.
The residues arginine 190 and aspartate 193 forming the arginine-mediated interaction motif are highly conserved in SSB proteins from the Thermus/Deinococcus group, which may imply an important function. Furthermore, superposition of the DraSSB structure (PDB code: 1se8 (50)) onto TaqSSB molecules interacting through these residues shows that this mode of interaction can be formed in DraSSB if one allows some flexibility in the glycine-rich region. Sequence alignments of the SSB proteins from E. coli and T. aquaticus reveal that the arginine residue 72 of EcoSSB is homologous to arginine 190 of TaqSSB. The aspartate residue 193 of TaqSSB, which makes the hydrogen bond to arginine 190, is replaced by serine in EcoSSB. Superposition of the EcoSSB structure to the crystal packing of TaqSSB reveals that arginine 72 can form a hydrogen bond to serine 75 (Fig. 3 b). Thus, a protein-protein interaction via these hydrogen bonds should also be possible in the case of the tetrameric EcoSSB protein. This makes the arginine-mediated interaction motif a good candidate for a general element of protein-protein interactions of SSB proteins, as found in the cooperative binding of SSB to ssDNA.
In the crystal packing of DraSSB, neighboring dimers are held together by interactions involving the β-hairpin connector that links the two OB-folds in the DraSSB monomer (50). It was suggested that this interaction may be present in all Deinococcus/Thermus SSB proteins and that it may help these organisms survive in hostile environments (50). In our crystal structure of wild-type TaqSSB, however, the region connecting the two OB-folds is disordered, implying a high degree of conformational flexibility (14). The same holds true for the TaqSSBΔ228-252 R190A mutant (see below), which formed a completely different crystal packing (PDB code: 2ihf) (14), and for the structure of TaqSSB published by Jedrzejczak et al. (15).
Formation of TaqSSB tetramers in solution
To test whether the interactions between adjacent TaqSSB molecules observed in the crystal packing of wild-type TaqSSB can also lead to the formation of higher oligomers of TaqSSB in solution, we used sedimentation velocity (Fig. 2 a) and sedimentation equilibrium (Supplementary Material Fig. S1) studies of wild-type TaqSSB under low salt conditions. We could not find any indication for the formation of higher oligomers. Even at a TaqSSB concentration of 26 μM, the c(s) distribution shows only a single peak at 4.0 S (data not shown). For the deletion mutant TaqSSBΔ228-252, however, in which the DNA binding domain is directly fused to the highly conserved region of the last 10 amino acids, an equilibrium between dimers and tetramers can be observed (Fig. 4 a). This equilibrium depends on the protein concentration, and at 15.4 μM TaqSSBΔ228-252 the major fraction of the protein is tetrameric (Fig. 4 a). Determination of the apparent molar mass of this mutant in sedimentation equilibrium experiments as a function of protein concentration (data not shown) resulted in a saturation curve reaching a maximum of 110 kg/mol. This corresponds to the molar mass of tetrameric TaqSSBΔ228-252, which can be calculated from the amino acid composition to be 110.5 kg/mol. The mutant TaqSSB R229* lacking the complete C-terminal region, including the negatively charged part of the last 10 amino acids, however, is insoluble at low salt conditions.
FIGURE 4.
Tetramerization of mutated TaqSSB proteins at 20°C monitored by sedimentation velocity experiments in the analytical ultracentrifuge at 50,000 rpm. (a) 15.4 μM TaqSSBΔ228-252 (solid curve) and 15.4 μM TaqSSBΔ228-252 R190A (dashed curve) in 5 mM NaCl, 5 mM KPi, pH 7.4, 0.87 M glycerol and (b) 7.7 μM TaqSSBΔ228-252 in 5 mM (solid curve), 25 mM (shaded curve), and 50 mM (dashed curve) NaCl with 5 mM KPi, pH 7.4, 0.87 M glycerol each.
To test whether the TaqSSBΔ228-252 tetramers are held together by the arginine-mediated interactions that we identified in the crystal packing of wild-type TaqSSB, we replaced arginine 190 by alanine in the deletion mutant TaqSSBΔ228-252, yielding TaqSSBΔ228-252 R190A. The c(s) distribution clearly shows a drastically reduced ability of this mutant to form tetramers in solution (Fig. 4 a), proving that arginine 190 plays a key role in tetramerization of TaqSSBΔ228-252. However, further residues must be involved in this interaction, since the formation of tetramers is not completely prevented by the exchange of arginine 190. To demonstrate that the tetramerization of TaqSSBΔ228-252 occurs via an interaction having a strong electrostatic component, Fig. 4 b shows c(s) distributions for sedimentation velocity experiments at different salt concentrations. At a protein concentration of 7.6 μM TaqSSBΔ228-252 50 mM NaCl almost completely prevents the formation of tetramers.
There are at least two explanations for why the deletion mutant TaqSSBΔ228-252 is able to form tetramers in solution whereas the wild-type protein is not. On one hand, the bulky glycine-rich region of the wild-type protein can occlude the region that must be accessible for the formation of the R190-D193 hydrogen bonds. On the other hand, the C-terminus could compete with the interaction motif, and the removal of the flexible, glycine-rich region could prevent the C-terminus to reach this region. To test the latter possibility we constructed two C-terminal triple mutants and investigated their tetramerization behavior in sedimentation velocity experiments. In the TaqSSB E259Q E260Q D261Q mutant, we replaced the negatively charged acidic amino acids by neutral ones. In the mutant TaqSSB L262A P263A F264A, we exchanged the ultimate three hydrophobic amino acids by alanine. For EcoSSB it has been shown that the exchange of the penultimate proline residue by serine in vivo results in a UV- and temperature-sensitive replication deficient E. coli strain (51). However, both triple mutants showed no tendency to form tetramers in solution and therefore behaved like wild-type TaqSSB (data not shown). A specific interaction of the highly conserved C-terminus of TaqSSB with the region forming the R190-D193 interactions to prevent tetramer formation thus can be ruled out. It is therefore most likely that the bulky glycine-rich region of wild-type TaqSSB prevents formation of the tetramer.
Formation of higher aggregates of EcoSSB
To experimentally test whether tetrameric SSB proteins are able to form higher aggregates under low salt conditions using the described arginine-mediated interaction, we examined a 33 μM solution of EcoSSB in a sedimentation velocity experiment (Fig. 5). In this experiment EcoSSB showed a broad distribution of sedimentation coefficients, where half of the material sedimented with more than 9 S and only a small fraction showed sedimentation coefficients in the range of 4–5 S expected for the native EcoSSB tetramer (7). This is a clear indication of protein aggregation. We also investigated the sedimentation behavior of EcoSSBΔ116-167, a deletion mutant in which the DNA-binding domain is directly fused to the highly conserved C-terminus and thus corresponds to the TaqSSBΔ228-252 mutant. This mutant is able to form even larger aggregates than wild-type EcoSSB, resulting in sedimentation coefficients higher than 100 S (data not shown). Even the EcoSSB mutant G117*, which lacks the negatively charged C-terminal region of EcoSSB, is even insoluble under low salt conditions, indicating extensive aggregation. The single exchange of arginine 72 of wild-type EcoSSB to alanine, however, results in the complete disappearance of the tendency to form higher aggregates (Fig. 5). The c(s) analysis of the sedimentation of EcoSSB R72A (not shown) reveals the protein to be exclusively tetrameric. Thus, in the case of EcoSSB, arginine 72 is able to promote interactions between tetramers in solution, resulting in the formation of large aggregates.
FIGURE 5.
Aggregation of EcoSSB and EcoSSB R72A at low salt. Sedimentation velocity experiments in the analytical ultracentrifuge of 33 μM wild-type EcoSSB (dashed curve) and EcoSSB R72A (solid curve) at 20°C and 16,000 and 50,000 rpm, in 5 mM NaCl, 5 mM KPi, pH 7.4, 0.87 M glycerol. Sedimentation profiles at both speeds were converted into a cumulative distribution of absorption over sedimentation coefficient (see Materials and Methods) and combined.
The difference in the oligomerization behavior of TaqSSB and EcoSSB presumably is based on the fact that the native TaqSSB protein is a dimer, whereas the native EcoSSB protein is a tetramer. Each EcoSSB tetramer can use its four arginine residues at position 72 to interact with other EcoSSB tetramers, resulting in a whole network of EcoSSB proteins. In the case of the dimeric TaqSSB protein, only two interactions can be made by each molecule. The fact that the TaqSSB R229* mutant is insoluble under low salt conditions indicates that the negative net charge of TaqSSBΔ228-252 prevents the formation of protein fibers, as observed in crystal packing.
Cooperativity of ssDNA binding
Under low salt conditions EcoSSB and long ssDNA form highly cooperative complexes, which have been shown to redistribute extremely slowly to a random less cooperative complex (24). Therefore, incubation of EcoSSB with an excess of ssDNA results in a nonrandom distribution of EcoSSB molecules on the ssDNA strands, which can be detected using electrophoretic mobility shift assays or analytical ultracentrifugation experiments (13,24). ssDNA highly saturated with SSB as well as ssDNA with few SSB molecules bound can be found, leading to a bifurcation of the SSB distribution. This ssDNA binding cooperativity is most probably the result of protein-protein interactions connecting SSB molecules bound adjacently on ssDNA.
To test whether the described arginine-mediated interaction identified in the crystal structure of TaqSSB is involved in such a cooperative ssDNA binding, we examined complexes of EcoSSB R72A with poly(dT) (length ∼1400 nucleotides) under low salt conditions. Analytical ultracentrifugation of a mixture of EcoSSB R72A and poly(dT) shortly (1 h) after mixing gave two different complexes—a complex with few SSB molecules bound sedimented with ∼11 S and a highly SSB-saturated poly(dT) complex with 15 S (Fig. 6 b)—indicating high binding cooperativity. For wild-type EcoSSB the two types of complexes sedimented with ∼9 S and 16 S, respectively (Fig. 6 a). This larger difference indicates an even higher cooperativity of binding. However, the bifurcation of complex size is metastable, and reshuffling the protein to form a random distribution sedimenting in a single boundary is completed after 4 days for EcoSSB R72A and is not completed for wild-type EcoSSB even after 14 days (Fig. 6). Thus, the metastable highly cooperative binding is diminished by the R72A mutation. This indicates that the arginine-mediated interaction motif is one of the components of the protein-protein interactions leading to the metastable highly cooperative binding. However, further interactions, such as the loop L45 interaction discovered in the crystal packing of EcoSSB and an EcoSSB/ssDNA complex (25,26), must be involved in the metastable high cooperativity.
FIGURE 6.
Metastable cooperativity of wild-type EcoSSB and EcoSSB R72A binding to poly(dT) under low salt conditions (5 mM NaCl, 5 mM KPi, pH 7.4, 0.87 M glycerol). Mixtures of 35 μM poly(dT) (length ∼1400 nucleotides) and 0.5 μM wild-type EcoSSB (a) or EcoSSB R72A (b) were analyzed in sedimentation velocity experiments in the analytical ultracentrifuge (23,000 rpm, 20°C) after 1 h (solid), 2 days (shaded), 4 days (dotted, b), and 14 days (dashed, a) incubation at 22°C. The differential sedimentation coefficient distributions were obtained using the program SEDFIT (37). Whereas the initially nonrandomly distributed EcoSSB R72A protein redistributes within 4 days, the wild-type EcoSSB/poly(dT) complexes are not at equilibrium even after 14 days of incubation.
Under high salt conditions (≥0.2 M NaCl) EcoSSB binds with moderate cooperativity to single-stranded nucleic acids (24). Since the intrinsic tryptophan fluorescence of EcoSSB is strongly quenched by binding to single-stranded nucleic acids, fluorescence titrations can be used to characterize the binding parameters. Fluorescence titrations using poly(dT) as a substrate reveal that binding site size and fluorescence quench of wild-type EcoSSB and the EcoSSB R72A mutant are virtually identical (Supplementary Material Fig. S2a). However, fluorescence titrations with poly(dT) cannot be used to determine binding affinity and cooperativity of this interaction, since the titrations are stoichiometric, even at 5 M NaCl (24). Therefore, we used the weaker binding substrate poly(rU) to get information about the cooperative binding affinities. Fluorescence titrations were performed at 0.2 M NaCl and 0.25 M NaCl using protein concentrations in the range of 60–350 nM (examples are shown in Supplementary Material Fig. S2b). Our data reveal the cooperative affinity (KAssω) of the EcoSSB R72A mutant to be reduced by a factor of 2–3 compared to the wild-type protein. Whether this difference in cooperative affinity is a consequence of a difference in the isolated binding affinity (KAss) or the cooperativity (ω) cannot be deduced from our data. A difference in isolated binding affinity should involve a direct interaction of the R72 residue with DNA. Therefore, we examined the crystal structure of an EcoSSB/ssDNA complex (PDB code: 1eyg (25)). In this structure the R72 side chain is disordered. However, the closest possible distance is between the region where the positive charge of R72 must be located and the phosphate backbone is larger than 6.6 Å, making a direct interaction unlikely.
CONCLUSIONS
The SSB protein of T. aquaticus, TaqSSB, is a dimeric bacterial SSB protein, which is very similar in three-dimensional structure (14,15) and biophysical properties to the tetrameric EcoSSB. The cooperative binding affinities of both proteins for single-stranded nucleic acids are nearly identical, and the ssDNA binding kinetics can be described by the same model and yield similar association rate constants. However, TaqSSB, as well as DraSSB (13), shows no reduced binding site size at low salt conditions as described for EcoSSB (9) but does show a markedly reduced fluorescence quench.
As described for EcoSSB (19,20), TaqSSB is able to make protein-protein interactions using the highly conserved region of the last 10 C-terminal amino acids. Since TaqSSB forms dimers in solution, it can bind up to two χ-subunits of DNA polymerase III of E. coli with approximately the same affinity as described for the tetrameric EcoSSB, which can bind up to four χ-proteins (20). The glycine-rich region of TaqSSB is not required for this interaction.
In the x-ray structure of TaqSSB, we observed an interaction between two N-terminal DNA binding/dimerization domains of neighboring TaqSSB dimers mediated by hydrogen bonds between arginine 190 and aspartate 193. Whereas wild-type TaqSSB shows no tendency to form higher oligomers in solution, a deletion mutant, TaqSSBΔ228-252—in which the highly conserved C-terminal region is directly fused to the N-terminal DNA binding/dimerization domain—uses this arginine-mediated interaction motif to form tetramers under low salt conditions. Using site-directed mutagenesis we could show that the absence of higher oligomers in wild-type TaqSSB is not caused by the C-terminal region itself. Therefore, the bulky glycine-rich region prevents the formation of tetramers in wild-type TaqSSB. This is supported by structural data for DraSSB (50) in which the glycine-rich region blocks the site for which we can show that it would otherwise be able to form hydrogen bonds between adjacent molecules.
Sequence alignment of EcoSSB and TaqSSB reveals arginine 72 of EcoSSB to be homologous to arginine 190 of TaqSSB. Structural comparison shows that arginine 72 of EcoSSB can connect two SSB tetramers by an intermolecular hydrogen bond to serine 75 (Fig. 3 b). This hydrogen bond leads to an aggregation of EcoSSB under low salt conditions, which can be prevented by an exchange of arginine 72 by alanine. Thus, an intermolecular interaction mediated by the arginine-mediated interaction motif found in TaqSSB seems to be a general feature of SSB proteins. For EcoSSB, we also showed this interaction to be involved in metastable highly cooperative ssDNA binding at low salt conditions.
If the arginine-mediated intermolecular interaction leads to cooperativity in the binding of SSB to long ssDNA, a conformational change involving the glycine-rich region is necessary to make the arginine accessible. Previous studies showed that the binding of EcoSSB to long ssDNA results in a conformational change of the protein, which makes the glycine-rich region more easily accessible to the action of proteases (52) and exposes the C-terminal region to the binding of other proteins involved in DNA metabolism, e.g., the χ-subunit of DNA polymerase III (20). In the crystal structure of wild-type TaqSSB, we observed two conformations of the glycine-rich region (C1 and C2) accompanied by a proline cis-trans isomerization (14). Docking the ssDNA from the EcoSSB-ssDNA complex onto the TaqSSB structure was only possible when the glycine-rich region was in conformation C2. Conformation C1, however, is incompatible with ssDNA binding (Fig. 3 a). Therefore, in the case of TaqSSB a conformational change induced by ssDNA binding seems to be likely, too. In our solution experiments of TaqSSB, we could show that the conformation of the glycine-rich region observed in free SSB and the formation of the R190-D193 hydrogen bonds are mutually exclusive. In Fig. 7, we propose a schematic model of how this interaction could be involved in ssDNA binding cooperativity. The model shows that ssDNA binding induces a conformational change in the flexible glycine-rich region of SSB, thereby facilitating the cooperative interaction of neighboring SSB molecules via the arginine-mediated hydrogen bonds and additional interactions. The conformational change makes the C-terminus more easily accessible to other proteins. For EcoSSB it has been shown recently that the removal of the last 42 amino acids favors the highly cooperative low salt ssDNA binding mode even at higher salt concentrations (53). Therefore, it has been speculated that the binding of the χ-protein to the C-terminus of EcoSSB may also result in a stabilization of the low salt binding mode, inducing high ssDNA binding cooperativity during DNA replication (53).
FIGURE 7.
Schematic depiction of the proposed model of cooperative binding of SSB to ssDNA. A TaqSSB-like model for SSB is used. The OB-fold containing core and the glycine-rich region are depicted in gray, the arginine-mediated interaction motif as a hatched rod, the C-terminal 10 amino acids as a dark gray rod, and ssDNA as a black line. In the isolated protein the region responsible for cooperative protein-protein interaction is occluded by the glycine-rich region. Only when ssDNA is bound to the SSB molecules will this region become accessible and the glycine-rich region and the C-terminus adopt conformations in which they can be recognized more easily by other proteins.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
We thank Lidia Litz for excellent technical assistance, Dr. Claus Urbanke for help with the analytical ultracentrifuge and valuable discussions, Drs. Joachim Greipel and Dietmar J. Manstein for helpful discussions, Tobias Knust for help with mutagenesis and protein preparation, and Dr. Jan Faix for critically reading the manuscript.
This work was supported by a HiLF grant from Hannover Medical School to U.C. and by grant MA1081/5-3 from the Deutsche Forschungsgemeinschaft.
Gregor Witte's present address is Gene Center and Dept. of Chemistry and Biochemistry, University of Munich, Feodor-Lynen-Str. 25, D-81377 Munich, Germany.
Editor: David P. Millar.
References
- 1.Swamynathan, S. K., A. Nambiar, and R. V. Guntaka. 1998. Role of single-stranded DNA regions and Y-box proteins in transcriptional regulation of viral and cellular genes. FASEB J. 12:515–522. [DOI] [PubMed] [Google Scholar]
- 2.Suck, D. 1997. Common fold, common function, common origin? Nat. Struct. Biol. 4:161–165. [DOI] [PubMed] [Google Scholar]
- 3.Shamoo, Y., A. M. Friedman, M. R. Parsons, W. H. Konigsberg, and T. A. Steitz. 1995. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 376:362–366. [DOI] [PubMed] [Google Scholar]
- 4.Tucker, P. A., D. Tsernoglou, A. D. Tucker, F. E. Coenjaerts, H. Leenders, and P. C. van der Vliet. 1994. Crystal structure of the adenovirus DNA binding protein reveals a hook-on model for cooperative DNA binding. EMBO J. 13:2994–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folkers, P. J. M., M. Nilges, R. H. A. Folmer, R. N. H. Konings, and C. W. Hilbers. 1994. The solution structure of the Tyr41→His mutant of the single- stranded DNA binding protein encoded by gene V of the filamentous bacteriophage M13. J. Mol. Biol. 236:229–246. [DOI] [PubMed] [Google Scholar]
- 6.Skinner, M. M., H. Zhang, D. H. Leschnitzer, Y. Guan, H. Bellamy, R. M. Sweet, C. W. Gray, R. N. H. Konings, A. H.-J. Wang, and T. C. Terwilliger. 1994. Structure of the gene V protein of bacteriophage f1 determined by multiwavelength x-ray diffraction on the selenomethionyl protein. Proc. Natl. Acad. Sci. USA. 91:2071–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curth, U., C. Urbanke, J. Greipel, H. Gerberding, V. Tiranti, and M. Zeviani. 1994. Single-stranded-DNA-binding proteins from human mitochondria and Escherichia coli have analogous physicochemical properties. Eur. J. Biochem. 221:435–443. [DOI] [PubMed] [Google Scholar]
- 8.Greipel, J., C. Urbanke, and G. Maass. 1989. The single-stranded DNA binding protein of Escherichia coli: physicochemical properties and biologic functions. In Protein-Nucleic Acid Interaction. W. Saenger and U. Heinemann, editors. Macmillan, London. 61–86.
- 9.Lohman, T. M., and W. Bujalowski. 1990. E. coli single strand binding protein: multiple single stranded DNA binding modes and cooperativities. In The Biology of Nonspecific Protein DNA Interactions. A. Revzin, editor. CRC Press, Boca Raton, FL. 131–168.
- 10.Iftode, C., Y. Daniely, and J. A. Borowiec. 1999. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34:141–180. [DOI] [PubMed] [Google Scholar]
- 11.Dabrowski, S., M. Olszewski, R. Piatek, A. Brillowska-Dabrowska, G. Konopa, and J. Kur. 2002. Identification and characterization of single-stranded-DNA-binding proteins from Thermus thermophilus and Thermus aquaticus—new arrangement of binding domains. Microbiology. 148:3307–3315. [DOI] [PubMed] [Google Scholar]
- 12.Eggington, J. M., N. Haruta, E. A. Wood, and M. M. Cox. 2004. The single-stranded DNA-binding protein of Deinococcus radiodurans. BMC Microbiol. 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witte, G., C. Urbanke, and U. Curth. 2005. Single-stranded DNA-binding protein of Deinococcus radiodurans: a biophysical characterization. Nucleic Acids Res. 33:1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fedorov, R., G. Witte, C. Urbanke, D. J. Manstein, and U. Curth. 2006. 3D structure of Thermus aquaticus single-stranded DNA-binding protein gives insight into the functioning of SSB proteins. Nucleic Acids Res. 34:6708–6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jedrzejczak, R., M. Dauter, Z. Dauter, M. Olszewski, A. Dlugolecka, and J. Kur. 2006. Structure of the single-stranded DNA-binding protein SSB from Thermus aquaticus. Acta Crystallogr. D: Biol. Crystallogr. 62:1407–1412. [DOI] [PubMed] [Google Scholar]
- 16.Curth, U., J. Genschel, C. Urbanke, and J. Greipel. 1996. In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 24:2706–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cadman, C. J., and P. McGlynn. 2004. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 32:6378–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handa, P., N. Acharya, and U. Varshney. 2001. Chimeras between single-stranded DNA-binding proteins from Escherichia coli and Mycobacterium tuberculosis reveal that their C-terminal domains interact with uracil DNA glycosylases. J. Biol. Chem. 276:16992–16997. [DOI] [PubMed] [Google Scholar]
- 19.Kelman, Z., A. Yuzhakov, J. Andjelkovic, and M. O'Donnell. 1998. Devoted to the lagging strand-the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 17:2436–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Witte, G., C. Urbanke, and U. Curth. 2003. DNA polymerase III chi subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 31:4434–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGhee, J. D., and P. H. Von Hippel. 1974. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 86:469–489. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz, G., and F. Watanabe. 1983. Thermodynamics and kinetics of co-operative protein-nucleic acid binding. I. General aspects of analysis of data. J. Mol. Biol. 163:467–484. [DOI] [PubMed] [Google Scholar]
- 23.Greipel, J., G. Maass, and F. Mayer. 1987. Complexes of the single-stranded DNA-binding protein from Escherichia coli (Eco SSB) with poly(dT). An investigation of their structure and internal dynamics by means of electron microscopy and NMR. Biophys. Chem. 26:149–161. [DOI] [PubMed] [Google Scholar]
- 24.Lohman, T. M., L. B. Overman, and S. Datta. 1986. Salt-dependent changes in the DNA binding co-operativity of Escherichia coli single strand binding protein. J. Mol. Biol. 187:603–615. [DOI] [PubMed] [Google Scholar]
- 25.Raghunathan, S., A. G. Kozlov, T. M. Lohman, and G. Waksman. 2000. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 7:648–652. [DOI] [PubMed] [Google Scholar]
- 26.Raghunathan, S., C. S. Ricard, T. M. Lohman, and G. Waksman. 1997. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-Å resolution. Proc. Natl. Acad. Sci. USA. 94:6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urbanke, C., and A. Schaper. 1990. Kinetics of binding of single-stranded DNA binding protein from Escherichia coli to single-stranded nucleic acids. Biochemistry. 29:1744–1749. [DOI] [PubMed] [Google Scholar]
- 28.Kowalczykowski, S. C., N. Lonberg, J. W. Newport, and P. H. VonHippel. 1981. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. J. Mol. Biol. 145:75–104. [DOI] [PubMed] [Google Scholar]
- 29.Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohman, T. M., and L. B. Overman. 1985. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem. 260:3594–3603. [PubMed] [Google Scholar]
- 31.Bayer, I., A. Fliess, J. Greipel, C. Urbanke, and G. Maass. 1989. Modulation of the affinity of the single-stranded DNA-binding protein of Escherichia coli (E. coli SSB) to poly(dT) by site-directed mutagenesis. Eur. J. Biochem. 179:399–404. [DOI] [PubMed] [Google Scholar]
- 32.Landwehr, M., U. Curth, and C. Urbanke. 2002. A dimeric mutant of the homotetrameric single-stranded DNA binding protein from Escherichia coli. Biol. Chem. 383:1325–1333. [DOI] [PubMed] [Google Scholar]
- 33.Lohman, T. M., J. M. Green, and R. S. Beyer. 1986. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry. 25:21–25. [DOI] [PubMed] [Google Scholar]
- 34.Curth, U., J. Greipel, C. Urbanke, and G. Maass. 1993. Multiple binding modes of the single-stranded DNA binding protein from Escherichia coli as detected by tryptophan fluorescence and site-directed mutagenesis. Biochemistry. 32:2585–2591. [DOI] [PubMed] [Google Scholar]
- 35.Curth, U., I. Bayer, J. Greipel, F. Mayer, C. Urbanke, and G. Maass. 1991. Amino acid 55 plays a central role in tetramerization and function of Escherichia coli single-stranded DNA binding protein. Eur. J. Biochem. 196:87–93. [DOI] [PubMed] [Google Scholar]
- 36.Kindler, B. 1997. AKKUPROG: Evaluation of chemical reaction kinetics and analysis of biopolymers using ultracentrifugation. Application to protein-DNA interactions. PhD thesis. Universität Hannover, Hanover, Germany.
- 37.Schuck, P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78:1606–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dam, J., and P. Schuck. 2005. Sedimentation velocity analysis of heterogeneous protein-protein interactions: sedimentation coefficient distributions c(s) and asymptotic boundary profiles from Gilbert-Jenkins theory. Biophys. J. 89:651–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urbanke, C., G. Witte, and U. Curth. 2005. Sedimentation velocity method in the analytical ultracentrifuge for the study of protein-protein interactions. Methods Mol. Biol. 305:101–114. [DOI] [PubMed] [Google Scholar]
- 40.Gralén, N., and G. Lagermalm. 1952. A contribution to the knowledge of some physico-chemical properties of polystyrene. J. Phys. Chem. 56:514–523. [Google Scholar]
- 41.Lamm, O. 1929. Die Differentialgleichung der Ultrazentrifugierung. Arkiv Matematik Astronomi Fysik. 21B:1–4. [Google Scholar]
- 42.Durchschlag, H. 1986. Specific volumes of biological macromolecules and some other molecules of biological interest. In Thermodynamic Data for Biochemistry and Biotechnology. H. Hinz, editor. Springer, Berlin/Heidelberg. 45–128.
- 43.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. [DOI] [PubMed] [Google Scholar]
- 44.Lebowitz, J., M. S. Lewis, and P. Schuck. 2002. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 11:2067–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eggington, J. M., A. G. Kozlov, M. M. Cox, and T. M. Lohman. 2006. Polar destabilization of DNA duplexes with single-stranded overhangs by the Deinococcus radiodurans SSB protein. Biochemistry. 45:14490–14502. [DOI] [PubMed] [Google Scholar]
- 46.Bujalowski, W., and T. M. Lohman. 1987. A general method of analysis of ligand-macromolecule equilibria using a spectroscopic signal from the ligand to monitor binding. Application to Escherichia coli single-strand binding protein-nucleic acid interactions. Biochemistry. 26:3099–3106. [DOI] [PubMed] [Google Scholar]
- 47.Genschel, J., U. Curth, and C. Urbanke. 2000. Interaction of E. coli single-stranded DNA binding protein (SSB) with exonuclease I. The carboxy-terminus of SSB is the recognition site for the nuclease. Biol. Chem. 381:183–192. [DOI] [PubMed] [Google Scholar]
- 48.Dam, J., C. A. Velikovsky, R. A. Mariuzza, C. Urbanke, and P. Schuck. 2005. Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s). Biophys. J. 89:619–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lohman, T. M., and M. E. Ferrari. 1994. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 63:527–570. [DOI] [PubMed] [Google Scholar]
- 50.Bernstein, D. A., J. M. Eggington, M. P. Killoran, A. M. Misic, M. M. Cox, and J. L. Keck. 2004. Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc. Natl. Acad. Sci. USA. 101:8575–8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chase, J. W., J. J. L'Italien, J. B. Murphy, E. K. Spicer, and K. R. Williams. 1984. Characterization of the E. coli ssb113 mutant single- stranded DNA binding protein. Cloning of the gene, DNA and protein sequence analysis, HPLC, peptide mapping and DNA binding studies. J. Biol. Chem. 259:805–814. [PubMed] [Google Scholar]
- 52.Williams, K. R., E. K. Spicer, M. B. Lopresti, R. A. Gugenheimer, and J. W. Chase. 1983. Limited proteolysis studies on the E. coli single-stranded DNA binding protein. J. Biol. Chem. 258:3346–3355. [PubMed] [Google Scholar]
- 53.Roy, R., A. G. Kozlov, T. M. Lohman, and T. Ha. 2007. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J. Mol. Biol. 369:1244–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]









