Abstract
A comprehensive study of the thermodynamic redox behavior of the hemes of the ba3 enzyme from Thermus thermophilus, a B-type heme-copper oxygen reductase, is presented. This enzyme, in contrast to those having a single type of heme, allows the B- and A-type hemes to be monitored separately by visible spectroscopy and the reduction potential of each heme to be determined unequivocally. The relative order of the midpoint reduction potentials of each center changed in the pH range from 6 to 8.4, and both hemes present a significant redox-Bohr effect. For instance, at pH 7, the midpoint reduction potentials of the hemes B and A3 are 213 mV and 285 mV, respectively, whereas at pH 8.4, the order is reversed: 246 mV for heme B and 199 mV for heme A3. The existence of redox anticooperativity was established by introducing a redox interaction parameter in a model of pairwise interacting redox centers.
INTRODUCTION
Oxygen reductases are the terminal enzymatic complexes in the respiratory chains of aerobic organisms. Most of them belong to the heme-copper superfamily, which have in their subunit I a low-spin heme and a binuclear oxygen-reducing center (catalytic center) formed by a high-spin heme and a copper ion (1,2). These enzymes oxidize electron donors such as cytochrome c, quinols, high-potential iron sulfur proteins, or copper proteins (e.g., plastocyanins), reducing molecular oxygen to water. A transmembrane difference of electrochemical potential is generated in this reaction in two ways: 1), charge separation, because electrons and protons come from opposite sides of the cytoplasmic membrane (in prokaryotes) or of the mitochondrial membranes, and 2), proton translocation (“pump”) across the membrane (from the N-side to the P-side). The resulting transmembrane difference of electrochemical potential is subsequently used by ATP synthase, leading to the conversion of ADP and inorganic phosphate to ATP.
The initial proposal to classify heme-copper oxygen reductases in families was based on the conservation of key amino acid residues proposed to be part of the intramolecular proton pathways, the so-called D- and K-channels, identified by sequence analysis, site-directed mutagenesis, and x-ray crystallographic three-dimensional structures for mitochondrial-like oxygen reductases (3,4). On the basis of the sequence signatures of their putative proton transfer pathways, later supported by the comparison of kinetic and ligand-binding properties of the binuclear oxygen reduction center, the superfamily of heme-copper terminal oxidases was divided into three families (A, B, and C), with one of them subdivided into two subfamilies (A1 and A2) (5,6). Proton translocation with a stoichiometry close to 1 H+/e− at neutral pH has been clearly demonstrated for types A1 and A2 enzymes. The data for B- and C-type reductases are less clear, although proton pumping, albeit with a lower stoichiometry, has been reported in some cases (7,8).
In recent decades, many studies addressed unsolved questions regarding the function of these enzymes, namely, the ways by which the mechanism of O2 reduction occurs (catalytic cycle), the way protons are translocated, and finally, how these two processes are coupled (9–12). To date, all the models proposed for both the catalytic cycle and the proton-pumping mechanism(s) are based on experiments performed with “canonical” mitochondrial-like oxygen reductases, of the A1-type family, which may be biased as a result of the limited sampling. Experiments using members of the other families are extremely useful to further understand these mechanisms and establish the key elements for their function, taking advantage of the naturally occurring diversity of these enzymes.
The ba3 cytochrome c:oxygen oxidoreductase is one of the two oxygen reductases present in the respiratory chain of the thermophilic Gram-negative bacterium Thermus (T.) thermophilus. This oxygen reductase has four redox centers: the binuclear CuA center in subunit II, which receives electrons from soluble cytochrome c552, and the low-spin heme B and the binuclear oxygen reduction center in subunit I. This enzyme is a member of the B-type family (5), which does not have the conserved residues of the D-channel, retaining only part of the K-channel residues (13). It is a well-studied enzyme in terms of its biochemistry (14–16) and kinetic behavior (7,17–19), and it is one of the few oxygen reductases with a known three-dimensional structure (13,14). Because it incorporates two different types of heme, which have quite different electronic properties in the visible region, it allows the analysis of the redox properties of each heme separately. All these arguments make the T. thermophilus ba3 enzyme an excellent system not only to investigate its own reduction profile but also as a putative model for other B-type enzymes.
MATERIALS AND METHODS
Enzyme preparation
The enzyme was purified to homogeneity as described by Soulimane et al. (13) as judged by SDS-PAGE and visible spectroscopy.
Spectroscopic characterization
Electronic spectra were obtained on a Shimadzu UV-1603 spectrophotometer at room temperature. The sample was buffered in 50 mM Tris-HCl, pH 7.6, 0.1% n-dodecyl-β-d-maltoside (DM).
Redox titrations
Anaerobic potentiometric titrations were monitored by visible spectroscopy in the pH range of 6 to 8.4, at 25°C, and with the following redox mediators, each at a final concentration of 20 μM: N,N-dimethyl-p-phenylene-diamine, p-benzoquinone, 1,2-naphthoquinone-4-sulfonic acid, 1,2-naphthoquinone, trimethylhydroquinone, phenazine methosulfate, 1,4-naphthoquinone, and duroquinone. The protein, at a concentration of 3.75 μM, was buffered in 50 mM Mes-bistrispropane, 0.1% DM at the desired pH value. The pH of the solution was measured at the beginning and at the end of each titration. Sodium dithionite and potassium ferricyanide were used for the reductive or oxidative titrations, respectively. For each potential measured, the solution was equilibrated for at least 5 min after which a visible spectrum from 380 to 700 nm was measured. A combined silver/silver chloride electrode was used, calibrated with a quinhydrone solution at pH 7 before each titration. Redox potentials are quoted in relation to the standard hydrogen electrode.
Data analysis
For each titration, the complete spectral data set, collected from 380 to 700 nm at each redox potential value, was analyzed with the MATLAB software (The MathWorks, Natick, MA). The heme reduction profiles were followed at the maxima of both the α and the Soret bands, at which each heme has distinct fingerprints, 612 nm and 443 nm for heme A3 and 560 nm and 428 nm for heme B, respectively. To characterize quantitatively the heme reduction profile of this enzyme, the midpoint reduction potentials of each heme were determined. This parameter corresponds to the potential at which half of the center is reduced, and it depends on the intrinsic heme potential (i.e., the reduction potential that the redox center would have if there were no other redox centers) and its possible interactions with other redox or protonable centers. The midpoint reduction potentials were determined directly from the changes in absorbance at both maxima of the two main absorption bands of the two hemes as a function of the solution reduction potential. The values obtained are independent of any theoretical model and represent the reduction potential at which the center most easily exchanges electrons from the thermodynamic point of view. To provide a visual guide to the pH dependence of the midpoint reduction potentials of the heme centers, polynomial functions (without physical meaning) were fitted to the data (see Fig. 5 in Results and Discussion section).
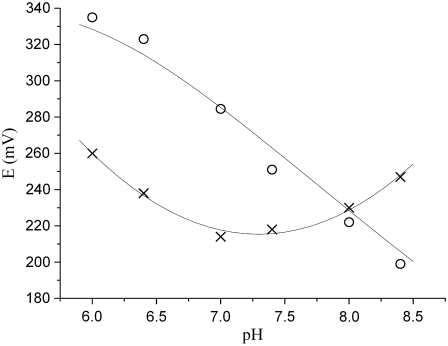
FIGURE 5.
pH dependence of the midpoint potentials of hemes A3 (○) and B (×) for ba3 oxygen reductase from T. thermophilus. The trendlines depicted in the figure are only for a better visualization of the pH effect in each heme (see Materials and Methods section).
Because the titration curves of both hemes do not follow a one-electron Nernst curve behavior, as would occur for isolated one-electron redox centers, electrostatic interactions occur between them. Therefore, the data collected were fitted in the framework of a model of interacting centers. The general theoretical framework governing multiple binding is well known (20–23) and can be sketched as follows. A system with N redox centers has 2N different redox microstates, each of which can be represented by a vector a=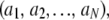 where
where 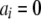 or 1 depending on whether the center i is reduced or oxidized. When the microstate populations are compared, it is convenient to define a reference microstate, which will be taken here as the fully reduced microstate 0 = (0, 0, … , 0). The reduction reaction a→0 involves the capture of
or 1 depending on whether the center i is reduced or oxidized. When the microstate populations are compared, it is convenient to define a reference microstate, which will be taken here as the fully reduced microstate 0 = (0, 0, … , 0). The reduction reaction a→0 involves the capture of 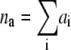 electrons, having an associated reduction potential given by the Nernst equation
electrons, having an associated reduction potential given by the Nernst equation
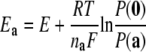 |
(1) |
where  (a) is the probability of microstate a. Rearranging terms we get
(a) is the probability of microstate a. Rearranging terms we get
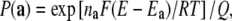 |
(2) |
where the condition 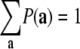 (where the sum extends over all the 2N microstates) is taken into account by introducing the normalization constant
(where the sum extends over all the 2N microstates) is taken into account by introducing the normalization constant
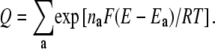 |
(3) |
Given the probabilities 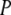 (a), a typical macroscopically measurable redox property
(a), a typical macroscopically measurable redox property 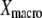 can be expressed as the average of its microscopic (molecular) counterpart
can be expressed as the average of its microscopic (molecular) counterpart 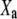 :
:
 |
(4) |
For example, the oxidation probability of the whole system is
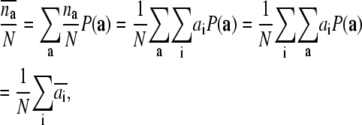 |
(5) |
where
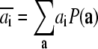 |
(6) |
is the oxidation probability of center i (i.e., the probability that center i is oxidized).
A pairwise interaction model may be introduced by assuming that all the 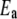 reduction potentials can be written as
reduction potentials can be written as
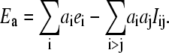 |
(7) |
If we consider a vector with 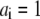 and all
and all 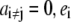 will be the reduction potential of center i when all other centers remain in the reference reduced state; thus, ei may be called the reference reduction potential of site i. The term
will be the reduction potential of center i when all other centers remain in the reference reduced state; thus, ei may be called the reference reduction potential of site i. The term 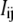 reflects the interaction between centers i and j, with a positive or negative sign indicating a cooperative or anticooperative effect between the centers, respectively. This term does not represent a mere Coulombic electrostatic interaction because it also reflects the energies involving all the properties of the system not explicitly included in the model, such as conformational changes, changes in the oxidation state of other redox groups, and also protonation/deprotonation reactions of protolytic groups. It should be noted that a different reference state could be chosen (e.g., the fully oxidized state), but this will lead to a completely equivalent pairwise model, changing accordingly the physical meaning of
reflects the interaction between centers i and j, with a positive or negative sign indicating a cooperative or anticooperative effect between the centers, respectively. This term does not represent a mere Coulombic electrostatic interaction because it also reflects the energies involving all the properties of the system not explicitly included in the model, such as conformational changes, changes in the oxidation state of other redox groups, and also protonation/deprotonation reactions of protolytic groups. It should be noted that a different reference state could be chosen (e.g., the fully oxidized state), but this will lead to a completely equivalent pairwise model, changing accordingly the physical meaning of 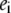 and
and 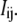
Given some experimentally measured quantities 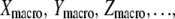 the set of
the set of 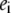 and
and 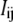 values that gives the optimal fit between those quantities and the computed
values that gives the optimal fit between those quantities and the computed 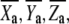 … can de determined. That set corresponds to the pairwise model that best describes the experimental data. In particular, the experimental quantities may be the values defining one or several E-dependent properties (e.g., they may be the points of one or several reduction curves).
… can de determined. That set corresponds to the pairwise model that best describes the experimental data. In particular, the experimental quantities may be the values defining one or several E-dependent properties (e.g., they may be the points of one or several reduction curves).
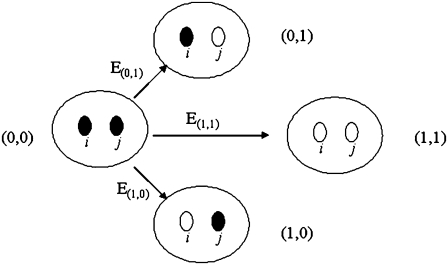
Fig. 1 is a schematic representation of the transitions between the reference (fully reduced) microstate and each of the other partially or fully oxidized microstates, as for the case of T. thermophilus ba3. The parameters obtained in the analysis are determined from the fit (ei and Iij) and are referred to as reference reduction parameters because they are model dependent. The best fit was obtained using the nonlinear least-squares Marquardt-Levenberg algorithm (as implemented in the gnuplot program), by simultaneously considering the absorbance data from the α and Soret absorption bands characteristic of each heme.
FIGURE 1.
Schematic representation of the transitions between the reference (fully reduced) microstate and each of the other partially or fully oxidized microstates. The reduction potentials shown are the ones following a Nernst equation (Eq. 1).
Once the parameters are obtained, it is possible to calculate the populations of all microstates as a function of the solution reduction potential. From these populations the equilibrium constant that describes the electron transfer between two centers can be determined by
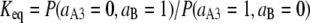 |
(8) |
RESULTS AND DISCUSSION
Redox titrations of the hemes of ba3 oxygen reductase from T. thermophilus were carried out at different pH values from 6 to 8.4. Taking advantage of the different electronic proprieties of each heme in the visible region, we monitored their individual behavior at the corresponding maxima of their α and Soret bands, 612 nm and 443 nm for heme A3 and 560 nm and 428 nm for heme B, respectively (Fig. 2). Following the heme reduction profiles at two distinct wavelengths related to the same electronic transition minimizes possible spectral interferences and validates the method. The titrations were fully reversible, as exemplified in Fig. 3, and several reduction titrations are presented in Fig. 4.
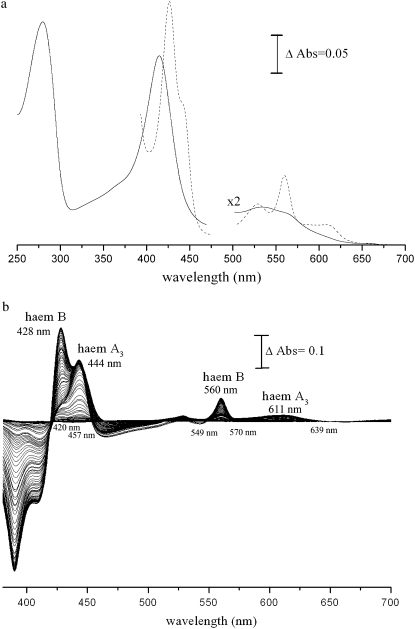
FIGURE 2.
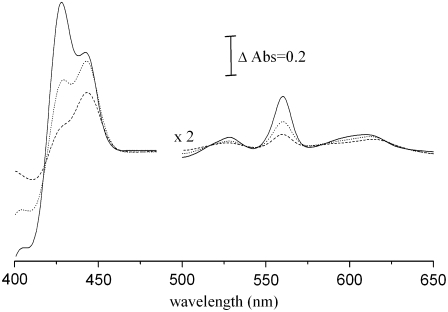
(a) UV/visible spectra of the oxidized (solid curve) and dithionite-reduced (dashed curve) ba3 oxygen reductase from T. thermophilus (enzyme concentration 1.2 μM). (b) Difference of the spectra of the reduced ba3 oxygen reductase on successive additions of dithionite minus the spectrum of the fully oxidized enzyme. The difference spectra were obtained during the redox titration of the enzyme at pH 7.
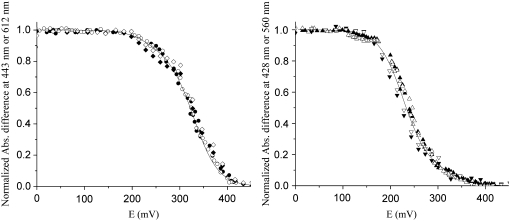
FIGURE 3.
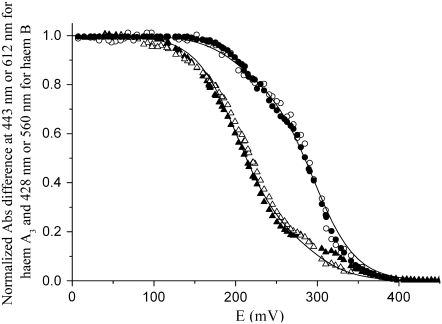
Oxidative and reductive redox titrations at pH of the ba3 oxygen reductase from T. thermophilus monitored by visible spectroscopy. (Left) Data obtained for the high-spin heme. (Right) Data for the low-spin heme. Data were collected at 443 (solid circle, oxidative; solid diamond, reductive) and 612 nm (open circle, oxidative; open diamond, reductive) for heme A3 and at 428 (▴, oxidative; ▾, reductive) and 560 nm (Δ, oxidative; ▿, reductive) for heme B.
FIGURE 4.
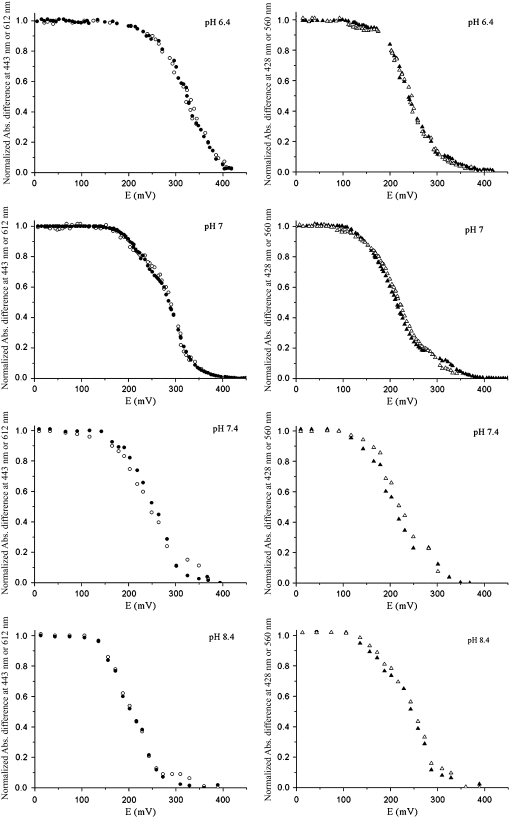
Reductive redox titrations at pH 6.4, 7, 7.4, and 8.4 of the ba3 oxygen reductase from T. thermophilus monitored by visible spectroscopy. On the left are data obtained for the high-spin heme, and on the right for the low-spin heme. Data were collected at 443 (•) and 612 nm (○) for heme A3 and at 428 (▴) and 560 nm (Δ) for heme B.
The variation of the heme midpoint reduction potentials as a function of pH is shown in Fig. 5. As mentioned in the Materials and Methods section, the trendlines presented in the figure are shown only to make the illustration clearer. In the pH range from 6 to 7.4, the midpoint reduction potential of the high-spin heme is always higher than that of the low-spin heme. By analysis of difference spectra (reduced minus oxidized) of samples at the midpoint reduction potentials of each heme center, it is unequivocally observed that, at pH < 8, the reduction of the high-spin heme A3 begins at higher potentials than the reduction of the low-spin heme B. Fig. 6 shows the difference of the spectra of the partially reduced enzyme at two different reduction potentials, corresponding to the midpoint reduction potential of each heme and the spectrum of the fully reduced enzyme minus the spectrum of the fully oxidized enzyme, at pH 7. At 285 mV, half of the high-spin heme is reduced, whereas 80% of the low-spin heme is still oxidized. By contrast, at 213 mV, which is the half-reduction potential of the low-spin heme, more than 80% of the high-spin heme is already reduced. It can be concluded that the high-spin heme has a higher reduction potential than the low-spin heme and that the two hemes have quite different redox potentials. This indicates that the equilibrium for electron transfer between the two hemes is shifted toward the state in which the high-spin heme is reduced. A calculation of this equilibrium constant using an equation such as  is not possible because the hemes are not redox-independent entities. The relative order of the heme midpoint reduction potentials and consequently the equilibrium of the electron transfer between them vary with pH (see Table 1). At pH 8 or higher, the heme with the higher midpoint reduction potential is now heme B, so it is the first to become reduced during reductive titrations.
is not possible because the hemes are not redox-independent entities. The relative order of the heme midpoint reduction potentials and consequently the equilibrium of the electron transfer between them vary with pH (see Table 1). At pH 8 or higher, the heme with the higher midpoint reduction potential is now heme B, so it is the first to become reduced during reductive titrations.
FIGURE 6.
Difference of the spectra of partially reduced, at 213 mV (dotted curve) and 285 mV (dashed curve) and of the fully reduced at 0 mV (solid curve) enzyme minus the spectrum of the fully oxidized enzyme. The chosen difference spectra were obtained at solution reduction potentials corresponding to the midpoint potentials of each heme. It can be observed that at the solution reduction potential corresponding to the midpoint potential of heme A (285 mV) heme B is 20% reduced, whereas at the solution reduction potential corresponding to the midpoint potential of heme B (213 mV), heme A3 is more than 80% reduced. This clearly indicates that the hemes have different midpoint potentials, heme A having the higher one.
TABLE 1.
Midpoint reduction potentials obtained for the hemes at different pH values as explained in Materials and Methods
| Experimental midpoint reduction potentials (experimental error ∼10 mV)
|
Model parameters (standard errors between 1 and 2.5 mV)
|
|||||
|---|---|---|---|---|---|---|
| pH | EA3 (mV) | EB (mV) | eA3 (mV) | eB (mV) | IA3B(mV) | Keq |
| 6 | 336 | 264 | 293 | 257 | −50 | 4.06 |
| 6.4 | 323 | 238 | 280 | 235 | −46 | 7.00 |
| 7 | 285 | 213 | 241 | 207 | −49 | 3.76 |
| 7.4 | 251 | 218 | 224 | 208 | −37 | 1.86 |
| 8 | 222 | 231 | 205 | 209 | −39 | 0.86 |
| 8.4 | 199 | 246 | 191 | 215 | −39 | 0.39 |
Reference potentials, reference interaction potentials, and Keq obtained for the hemes at different pH values using the two-interaction-center model; best fit was given by the gnuplot software.
These results indicate that the electron affinities of both centers, measured by their midpoint reduction potentials, are influenced by pH, which is evidence of proton uptake by the enzyme during the reductive part of the catalytic cycle. Because of the complexity of these systems, pKa values can not be determined. Although the equation 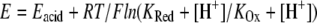 is often used to describe the pH dependence of midpoint reduction potentials, it is valid only in a system of a single redox center in the presence of a single protonatable group. In this case, not only are two redox centers present, but also more than one protonatable group within the protein is expected to interact with them (24–26). Therefore, the Ka values would not have a precise physical meaning and were not determined.
is often used to describe the pH dependence of midpoint reduction potentials, it is valid only in a system of a single redox center in the presence of a single protonatable group. In this case, not only are two redox centers present, but also more than one protonatable group within the protein is expected to interact with them (24–26). Therefore, the Ka values would not have a precise physical meaning and were not determined.
It is also clearly shown in Fig. 5 that the pH dependence is different for each heme, suggesting that, in addition to possible homotropic electrostatic interactions between them, each center may have different electrostatic interactions with protonable centers. This heterotropic cooperativity, or redox-Bohr effect, together with the homotropic cooperativities among the redox centers are proposed to have a fundamental role in the oxygen reductase mechanism.
The data presented in Fig. 4 also show evidence for the existence of homotropic interactions affecting the heme reduction profiles, as the titration curves of this enzyme do not follow a the single-electron Nernst curve. The three-dimensional crystallographic data for this enzyme (13) (as well as for all determined so far for heme copper reductases (4,3)) show that the heme iron atoms are at a distance of ∼14 Å, which indicates the possibility of electrostatic interactions between them. Therefore, a pairwise-interacting redox centers model, described in the Material and Methods section, was employed to describe the thermodynamic redox behavior of the hemes of the enzyme. It should be stressed that, in addition to the interaction between the two hemes, their interactions with the copper centers (CuA in subunit II and CuB in the binuclear site) can not be disregarded. In the case of CuA, because of its different subunit localization and its distance to the other centers (19.0, 21.8, and 21.6 Å to the heme B Fe, heme A3 Fe, and CuB, respectively (13)), electrostatic interactions are expected to be negligible. As for CuB, which is at 4.4 Å from the iron of heme A3 (13), a distance where electrostatic interactions may have an important role, a three-redox-center model should have been applied. Unfortunately, the reduction profile of the CuB center can not be monitored by visible spectroscopy. With no data available, the introduction of more variables by extending the interaction model to three centers would give rise to a multiple-solution system with too many degrees of freedom. So, in approximation, and only for qualitative purposes, the two pairwise redox centers model was applied.
As an example, Fig. 7 shows the best fit obtained for the redox titration at pH 7. In this case, the reference reduction potentials used to describe both curves were 241 mV for heme A3, 207 mV for heme B, and −49 mV for the interaction term. The interaction parameter that best fits the curves is always negative and equal to or higher than −50 mV (Table 1). Imposing a null interaction always leads to a worse fit. Thus, a redox interaction between the hemes is present. The negative value shows that this interaction is anticooperative; i.e., the oxidation (or reduction) of one heme makes the oxidation (or reduction) of the other one more difficult. With the model parameters determined, the equilibrium constant Keq discussed above, which refers to the electron distribution between the hemes, can be computed. This equilibrium is strongly influenced by pH, as can be observed from the data in Table 1: Keq has values >1 in the pH range where the high-spin heme presents the highest midpoint reduction potential, and values <1 when the order of midpoint reduction potentials is reversed.
FIGURE 7.
Redox titration curve obtained for the ba3 oxygen reductase from T. thermophilus at pH 7. Data were collected at 443 (•) and 612 nm (○) for heme A3, and at 428 (▴) and 560 nm (Δ) for heme B. Solid curves are the best fits obtained by the two-interacting-centers model given by gnuplot software with the following reference potential and interaction parameter: EA3 = 241 mV, EB = 207 mV, and IA3B = −49 mV.
A direct comparison between our findings and results for other oxygen reductases is hampered by differences in the methodologies for analyzing the redox data, which are not even presented in most cases. Furthermore, the analyses of the pH dependencies of the heme redox potentials are not correct, as explained above, because the use of the classical equations is not valid for 1), redox interacting centers, and 2), the pKa values thus determined can not be assigned to specific amino acid residues, as already demonstrated theoretically for multiple enzymes. Regarding the present case of the T. thermophilus ba3 oxygen reductase, as observed before (14), it does not have the behavior usually shown for other oxygen reductases, namely the mitochondrial-like ones, for which it has been assumed that either both hemes have the same reduction potential (28) or the low-spin heme has the highest potential. In the enzyme studied here, not only do the hemes have significantly different redox potentials but also their relative order varies with pH, with the high-spin heme having the higher reduction potential in the pH range from 6 to 7.4. This order of heme electron affinities was already reported for the aa3 enzyme from A. ambivalens, also a B-type enzyme (29), and for the T. thermophilus ba3 enzyme, from a single redox titration at pH 7 (14), although the reduction potentials reported were not equivalent to the ones now obtained. In this last case, the reduction of each individual heme was fitted to Nernst curves described by two independent midpoint potentials. Thus, the reduction of the two redox centers, a two-electron process in total, was described by four independent reduction potentials, which is conceptually not correct. Nevertheless, our results are qualitatively the same; i.e., at pH 7 the high-spin heme has a higher reduction potential than the low-spin heme, which is opposite to the results obtained for the A-type enzymes. The relative reduction potentials of the hemes have also been determined by a kinetic approach using pulse radiolysis. The difference between the reduction potential of the hemes is reported to be −25 mV at pH 7.5 with heme A3 having the higher reduction potential (30). This difference is in close agreement to that reported here.
The order of the heme potentials for the B-type oxidoreductases so far studied is different from that of A1-type enzymes. According to the literature on mitochondrial-like enzymes (11,31), the existence of redox-Bohr effects between groups within the protein that interact with the redox centers of the enzyme creates a complex network of cooperativities that ensure a downhill electron transfer process coupled with proton uptake. Apparently, in the B-type family of heme copper oxygen reductases, this network of cooperativities is different from the one proposed for A1-type oxidoreductases.
The hypothesis that each type of oxygen reductase has a different mechanism to perform the same function cannot be disregarded, but the structural similarities between these enzymes seem to favor the existence of a common mechanism for the coupling of e/H+ transfer. The results so far obtained for the B-type enzymes indicate that at physiological pH the order of the heme reduction potentials does not determine the proper function of oxygen reductases.
In conclusion, we present a detailed study of the thermodynamic redox behavior of a member of the B -type heme copper oxygen reductase family, between pH 6 and 8.4, being unequivocally shown that, at physiological pH, the midpoint reduction potential of the high-spin heme can be significantly higher than the reduction potential of the low-spin heme. Furthermore, we observed that, in addition to homotropic interactions between the hemes, heterocooperativies also affect the heme reduction behaviors, promoting an inversion of their order along the pH range studied. This result suggests that the coupling between proton and electron transfer does not depend on the relative order of the heme potentials and that it can be related instead with the reduction of dioxygen to water at the binuclear site. A way to address the study of the functional mechanism of oxygen reductases must involve the cross-link between experimental data obtained for different enzymes, belonging to different heme-copper oxygen reductases families, acquired by different techniques, with theoretical models and calculations, all analyzed without premature generalizations. Finding the common denominator may be a step for understanding how these enzymes operate.
Acknowledgments
Ricardo Louro, David Turner, and João Vicente are acknowledged for the critical reading of the manuscript.
Filipa L. Sousa and Andreia F. Veríssimo are recipients of grants from Fundação para a Ciência e a Tecnologia, SFRH XXI/BD/27972/2006 and SFRH XXI/BD/14388/2003, respectively. This work was supported by Fundação para a Ciência e a Tecnologia (POCTI/BME/45122/2002 to MP; POCI/BIA-PRO/58608/2004 to M.T.).
Editor: Michael Edidin.
References
- 1.Ferguson-Miller, S., and G. T. Babcock. 1996. Heme/copper terminal oxidases. Chem. Rev. 7:2889–2907. [DOI] [PubMed] [Google Scholar]
- 2.Michel, H., J. Behr, A. Harrenga, and A. Kannt. 1998. Cytochrome c oxidase: structure and spectroscopy. Annu. Rev. Biophys. Biomol. Struct. 27:329–356. [DOI] [PubMed] [Google Scholar]
- 3.Iwata, S., C. Ostermeier, B. Ludwig, and H. Michel. 1995. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 376:660–669. [DOI] [PubMed] [Google Scholar]
- 4.Tsukihara, T., H. Aoyama, E. Yamashita, T. Tomizaki, H. Yamaguchi, K. Shinzawa-Itoh, R. Nakashima, R. Yaono, and S. Yoshikawa. 1996. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 272:1136–1144. [DOI] [PubMed] [Google Scholar]
- 5.Pereira, M. M., M. Santana, and M. Teixeira. 2001. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim. Biophys. Acta. 1505:185–208. [DOI] [PubMed] [Google Scholar]
- 6.Pereira, M. M., and M. Teixeira. 2004. Proton pathways, ligand binding and dynamics of the catalytic site in haem-copper oxygen reductases: a comparison between the three families. Biochim. Biophys. Acta. 1655:340–346. [DOI] [PubMed] [Google Scholar]
- 7.Kannt, A., T. Soulimane, G. Buse, A. Becker, E. Bamberg, and H. Michel. 1998. Electrical current generation and proton pumping catalyzed by the ba3- type cytochrome c oxidase from Thermus thermophilus. FEBS Lett. 434:17–22. [DOI] [PubMed] [Google Scholar]
- 8.Arslan, E., A. Kannt, L. Thony-Meyer, and H. Hennecke. 2000. The symbiotically essential cbb(3)-type oxidase of Bradyrhizobium japonicum is a proton pump. FEBS Lett. 470:7–10. [DOI] [PubMed] [Google Scholar]
- 9.Michel, H. 1999. Cytochrome c oxidase: catalytic cycle and mechanisms of proton pumping–a discussion. Biochemistry. 38:15129–15140. [DOI] [PubMed] [Google Scholar]
- 10.Wikstrom, M. 2004. Cytochrome c oxidase: 25 years of the elusive proton pump. Biochim. Biophys. Acta. 1655:241–247. [DOI] [PubMed] [Google Scholar]
- 11.Xavier, A. V. 2004. Thermodynamic and choreographic constraints for energy transduction by cytochrome c oxidase. Biochim. Biophys. Acta. 1658:23–30. [DOI] [PubMed] [Google Scholar]
- 12.Capitanio, N., T. V. Vygodina, G. Capitanio, A. A. Konstantinov, P. Nicholls, and S. Papa. 1997. Redox-linked protolytic reactions in soluble cytochrome-c oxidase from beef-heart mitochondria: redox Bohr effects. Biochim. Biophys. Acta. 1318:255–265. [DOI] [PubMed] [Google Scholar]
- 13.Soulimane, T., G. Buse, G. P. Bourenkov, H. D. Bartunik, R. Huber, and M. E. Than. 2000. Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from Thermus thermophilus. EMBO J. 19:1766–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellwig, P., T. Soulimane, G. Buse, and W. Mantele. 1999. Electrochemical, FTIR, and UV/VIS spectroscopic properties of the ba(3) oxidase from Thermus thermophilus. Biochemistry. 38:9648–9658. [DOI] [PubMed] [Google Scholar]
- 15.Zimmermann, B. H., C. I. Nitsche, J. A. Fee, F. Rusnak, and E. Munck. 1988. Properties of a copper-containing cytochrome ba3: a second terminal oxidase from the extreme thermophile Thermus thermophilus. Proc. Natl. Acad. Sci. USA. 85:5779–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oertling, W. A., K. K. Surerus, O. Einarsdottir, J. A. Fee, R. B. Dyer, and W. H. Woodruff. 1994. Spectroscopic characterization of cytochrome ba3, a terminal oxidase from Thermus thermophilus: comparison of the a3/CuB site to that of bovine cytochrome aa3. Biochemistry. 33:3128–3141. [DOI] [PubMed] [Google Scholar]
- 17.Giuffre, A., E. Forte, G. Antonini, E. D'Itri, M. Brunori, T. Soulimane, and G. Buse. 1999. Kinetic properties of ba3 oxidase from Thermus thermophilus: Effect of temperature. Biochemistry. 38:1057–1065. [DOI] [PubMed] [Google Scholar]
- 18.Koutsoupakis, K., S. Stavrakis, E. Pinakoulaki, T. Soulimane, and C. Varotsis. 2002. Observation of the equilibrium CuB-CO complex and functional implications of the transient heme a3 propionates in cytochrome ba3-CO from Thermus thermophilus. Fourier transform infrared (FTIR) and time-resolved step-scan FTIR studies. J. Biol. Chem. 277:32860–32866. [DOI] [PubMed] [Google Scholar]
- 19.Siletsky, S., T. Soulimane, N. Azarkina, T. V. Vygodina, G. Buse, A. Kaulen, and A. Konstantinov. 1999. Time-resolved generation of membrane potential by ba3 cytochrome c oxidase from Thermus thermophilus. Evidence for reduction-induced opening of the binuclear centre. FEBS Lett. 457:98–102. [DOI] [PubMed] [Google Scholar]
- 20.Ben-Naim, A. 1992. Statistical Thermodynamics for Chemists and Biochemists. Plenum Press, New York.
- 21.Hill, T. 1960. An Introduction to Statistical Thermodynamics. Addison-Wesley, Reading, MA.
- 22.Schellman, J. 1975. Macromolecular binding. Biopolymers. 14:999–1018. [Google Scholar]
- 23.Wyman, J., and S. Gill. 1990. Binding and Linkage. University Science Books, Mill Valley, CA.
- 24.Popovic, D. M., and A. A. Stuchebrukhov. 2004. Electrostatic study of the proton pumping mechanism in bovine heart cytochrome c oxidase. J. Am. Chem. Soc. 126:1858–1871. [DOI] [PubMed] [Google Scholar]
- 25.Kannt, A., C. R. Lancaster, and H. Michel. 1998. The coupling of electron transfer and proton translocation: electrostatic calculations on Paracoccus denitrificans cytochrome c oxidase. Biophys. J. 74:708–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soares, C. M., A. M. Baptista, M. M. Pereira, and M. Teixeira. 2004. Investigation of protonatable residues in Rhodothermus marinus caa3 haem-copper oxygen reductase: comparison with Paracoccus denitrificans aa3 haem-copper oxygen reductase. J. Biol. Inorg. Chem. 9:124–134. [DOI] [PubMed] [Google Scholar]
- 27.Reference deleted in proof.
- 28.Gorbikova, E. A., K. Vuorilehto, M. Wikstrom, and M. I. Verkhovsky. 2006. Redox titration of all electron carriers of cytochrome c oxidase by Fourier transform infrared spectroscopy. Biochemistry. 45:5641–5649. [DOI] [PubMed] [Google Scholar]
- 29.Todorovic, S., M. M. Pereira, T. M. Bandeiras, M. Teixeira, P. Hildebrandt, and D. H. Murgida. 2005. Midpoint potentials of hemes a and a3 in the quinol oxidase from Acidianus ambivalens are inverted. J. Am. Chem. Soc. 127:13561–13566. [DOI] [PubMed] [Google Scholar]
- 30.Farver, O., Y. Chen, J. A. Fee, and I. Pecht. 2006. Electron transfer among the CuA-, heme b- and a3-centers of Thermus thermophilus cytochrome ba3. FEBS Lett. 580:3417–3421. [DOI] [PubMed] [Google Scholar]
- 31.Papa, S. 2005. Role of cooperative H(+)/e(-) linkage (redox bohr effect) at heme a/Cu(A) and heme a(3)/Cu(B) in the proton pump of cytochrome c oxidase. Biochemistry (Mosc.). 70:178–186. [DOI] [PubMed] [Google Scholar]