Abstract
Background
NADPH-cytochrome-P450 oxidoreductase (CPR) is a ubiquitous enzyme that belongs to a family of diflavin oxidoreductases and is required for activity of the microsomal cytochrome-P450 monooxygenase system. CPR gene-disruption experiments have demonstrated that absence of this enzyme causes developmental defects both in mouse and insect.
Results
Annotation of the sequenced genome of D. discoideum revealed the presence of three genes (redA, redB and redC) that encode putative members of the diflavin oxidoreductase protein family. redA transcripts are present during growth and early development but then decline, reaching undetectable levels after the mound stage. redB transcripts are present in the same levels during growth and development while redC expression was detected only in vegetative growing cells. We isolated a mutant strain of Dictyostelium discoideum following restriction enzyme-mediated integration (REMI) mutagenesis in which redA was disrupted. This mutant develops only to the mound stage and accumulates a bright yellow pigment. The mound-arrest phenotype is cell-autonomous suggesting that the defect occurs within the cells rather than in intercellular signaling.
Conclusion
The developmental arrest due to disruption of redA implicates CPR in the metabolism of compounds that control cell differentiation.
Background
NADPH-cytochrome-P450 oxidoreductase (CPR, EC 1.6.2.4) is a ubiquitous enzyme that is required for activity of the microsomal cytochrome-P450 (CYP) monooxygenase system [1,2]. This system is involved in the metabolic activation and/or detoxification of numerous foreign compounds as well as in the metabolism of endogenous substrates, such as steroids, alkaloids and fatty acids [3,4]. CPR belongs to a family of diflavin oxidoreductases which also includes the flavoprotein subunit of bacterial sulfite reductase (SiR) as well as a methionine synthase reductase and the cytoplasmic NADPH-dependent diflavin oxidoreductase 1 (NDOR1) identified in eukaryotic cells [5-8]. In addition, the diflavin reductase domain is found in fusion with cytochromes P-450 or with hemoprotein forming complex multidomain enzymes such as the cytochromes P450BM3 and the nitric oxide synthases [6].
CPR is a membrane anchored ~78 kDa enzyme which contains one molecule each of FAD and FMN bound as prosthetic groups that facilitate transfer of electrons of NADPH to the prosthetic heme group of CYP [1,2,9]. CPR is also involved in transferring electrons to other molecules, including heme oxygenase, squalene epoxidase and cytochrome b5 [10-12].
Despite the diversity of CYP isoforms that can be found in a single species [13], CPR in most organisms, except in certain plants and some zygomycetes, is encoded by only one gene [14-23]. Inactivation of the single-copy CPR gene in Saccharomyces cerevisiae results in mutants that accumulate only 25% as much ergosterol as observed in wild-type strains which probably accounts for the increased sensitivity of these mutants to the antifungal drug ketoconazole [16,24]. Moreover, it has been reported that cytochrome b5 gene can suppress the phenotype resulting from disruption of the CPR gene and therefore might function as an alternative electron donor for CYP activity in yeast [25-27]. In the fungus Gibberella fujikuroi loss of CPR leads to a reduced growth rate and has a strong influence on gibberellin biosynthesis [20].
CPR gene-disruption experiments in mouse have demonstrated that absence of this enzyme causes defects leading to mid-gestational lethality [28-30]. In situ hybridization studies have shown high levels of CPR expression in mesenchymal cells of the limbs and developing olfactory neuroepithelia [31]. CPR has been implicated in odorant clearance in insect antennae [17] and in ecdysone 20-hydroxylation during insect embryonic development [32].
Annotation of the sequenced genome of D. discoideum [33] revealed the presence of three genes that encode putative members of the diflavin oxidoreductase protein family. DDB0187719 (redC) on chromosome 5 encodes a polypeptide of 633 amino acids, which is 56% similar to human NADPH-dependent diflavin oxidoreductase 1 (NDOR1), a cytoplasmic enzyme highly expressed in cancer cell lines with as yet unknown functions [8]. Two genes, redA (DDB0215407) on chromosome 6 and redB (DDB0190667) on chromosome 1, show about 50% similarity to CPR proteins in humans, rats, Drosophila and yeast. We found that inactivation of redA in Dictyostelium results in developmental arrest at the mound stage.
Results and Discussion
Identification of the disrupted gene in the REMI mutant redA-
The mutant described in this work was isolated from a REMI-mutagenic library screen for morphological mutants of Dictyostelium discoideum. Strain DG1047 was picked because it forms yellow mounds that fail to make proper fruiting bodies. A portion of the disrupted gene was isolated from this strain by plasmid rescue in E. coli [34]. This fragment was used to screen a cDNA library prepared from vegetative cells. The largest cDNA insert (2094 bp) was sequenced and found to encode a putative protein of 631 amino acids with ~50% similarity to CPRs from human, rat, Drosophila, and yeast (Figure 1). The gene was designated redA as a mnemonic that it is likely to act in a redox reaction.
Figure 1.
Alignment of CPR aminoacid sequences. Comparison of Dictyostelium discoideum (Dd) NADPH cytochrome P450 oxidoreductase (RedA) aminoacid sequence with orthologs from Homo sapiens (Hs, P16435), Rattus norvegicus (Rn, P00388), Drosophila melanogaster (Dm, CAA63639) and Saccharomyces cerevisiae (Sc, P16603) using CLUSTAL W program. Identical and conserved amino acids are denoted with (*) and (:), respectively. Semi-conservative changes are indicated with a single dot. Binding domains for FMN, FAD and NADPH are indicated. Closed circles indicate highly conserved aromatic amino-acid residues that are particularly important in flavin binding. Open triangle points residues involved in NADPH discrimination. The ~56 aminoacid and the ~20 aminoacid hydrophobic segments at the N-terminal are indicated by dashed line in human CPR and dotted line in Dictyostelium CPR, respectively.
The D. discoideum CPR encoded by redA shows considerable conservation in the regions proposed to be involved in binding FMN, FAD and NADPH [2,9,18,35-37]. It is worth mentioning that the NADPH-2 region pointed out in Figure 1 contains the three residues Ser-596, Arg-597 and Lys-602 (positions numbered according human CPR) involved in the binding of the enzyme to the 2' phosphate of NADPH [2,38]. In addition, the conserved carboxi terminal motif G/K/N-R-Y-x-x-D-V/T-W is present in D. discoideum CPR. It has been demonstrated that the tryptophan in this motif plays a major role in discrimination of NADPH [36].
Analysis of the predicted RedA amino acid sequence by Signal P and TMHMM programs [39,40] revealed a N-terminal hydrophobic segment of ~20–25 amino acids (Figure 1) that should be sufficient for its anchorage to a membrane. Despite the fact that the hydrophobic N-terminal of mammalian CPRs is approximately 56 amino acids long [2], in plants and in fungi a shorter hydrophobic N-terminal is sufficient for membrane interaction [24,41-43]. Moreover, it has been proposed that interaction of human CPR to membranes and to CYPs is likely to involve additional hydrophobic patches on CPR surface [2,44].
Southern blot analysis of D. discoideum genomic DNA cleaved with a variety of restriction enzymes showed that RedA is encoded by a single copy gene (data not shown). Comparison of redA cDNA sequence with the Dictyostelium genome sequence [33] confirmed this result and showed that redA is an intronless gene located on chromosome 6. In most organisms analyzed, such as humans, mouse, Drosophila, S. cerevisiae and filamentous fungi, the CPR gene is present as a single copy and in the fruit fly two alternative splicing isoforms have been identified [14-18,20,45]. On the other hand, plants and certain fungi often have multiple copies of CPR gene [21,22,41,43,46,47]. We found two CPR genes in the Dictyostelium genome, redA and redB. Even though the amino acid sequences of redA and redB are 52% similar to each other, their nucleotide sequences are highly diverged and the genes appear to have evolved independently for a long time. D. discoideum genome has a third gene (redC) that encodes an additional member of diflavin oxidoreductases family which conserves sequences defined as binding domains for FMN, FAD and NADPH but lacks the N-terminal hydrophobic region found in RedA and RedB.
Expression of redA during growth and development
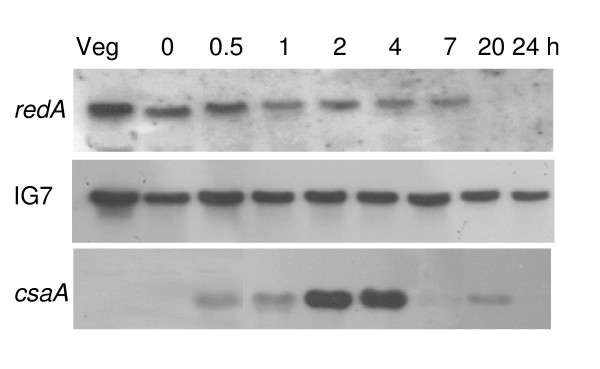
The expression of redA was monitored by Northern blot using a redA cDNA fragment as probe. As shown in Figure 2, a single mRNA species of 2.3 kbp was present in growing cells and decreased in abundance upon starvation of the cells on filter pads. No redA mRNA could be detected late in development (Figure 2 and 3A) in agreement with the redA expression profile determined on microarrays [48]. As a control, we probed for the csaA mRNA encoding the cell adhesion protein gp80 which is highly expressed during early aggregation [49]. This mRNA accumulated rapidly to reach peak levels by 2 hours and decreased after 4 hours of development (Figure 2).
Figure 2.
Expression of redA during growth and development of wild type AX4 cells. Exponentially growing AX4 cells (Veg) were starved on filter pads and harvested at the indicated times (h) after starvation. Identical Northern blots of total RNA samples were probed with redA, IG7 and csaA cDNAs as indicated. IG7 transcript is expressed at similar levels throughout the D. discoideum development [59].
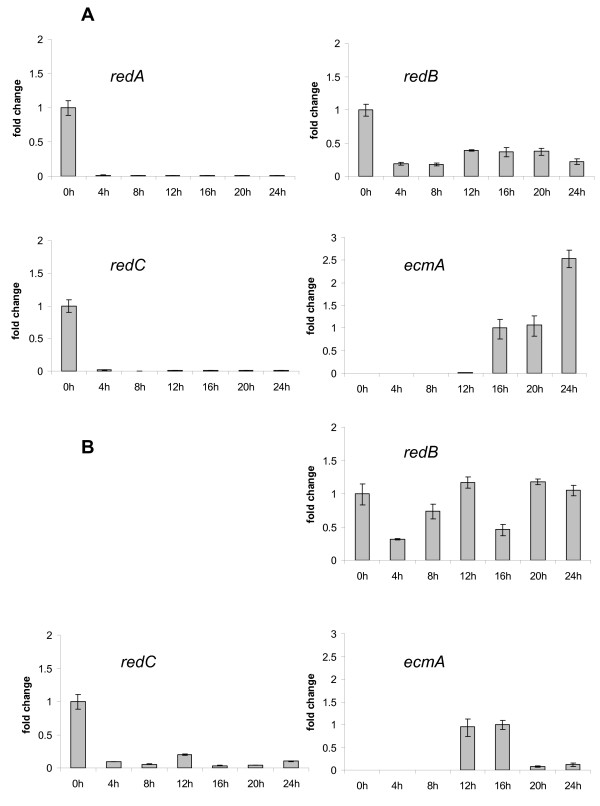
Figure 3.
Transcriptional profile of redA, redB, redC and ecmA. Exponentially growing AX4 (A) and redA- (B) cells were starved on filter pads and harvested at the indicated times (h) after starvation. Transcript levels for redA, redB and redC genes are relative to 0 h cells. Fold change for ecmA are relative to transcript levels detected at 16 h. Error bars represent the standard deviation from two independent experiments where qPCR assays were performed in triplicate.
In contrast, we found that redB is constitutively expressed throughout development of AX4 cells, while redC is expressed at detectable levels only in vegetative growing cells being repressed upon cell starvation. As a late development marker we monitored expression of ecmA [50], a prestalk-specific gene (Figure 3A).
CPR mRNA has distinct expression patterns during the development of several tissues and organs in mice, and this expression is not coordinated with expression of CYP genes [29,31,51,52]. The CPR gene is expressed in the early stages of embryonic development, suggesting that CPR-dependent processes may be important at this stage of the embryogenesis [28,31,32,53]. In Drosophila melanogaster the CPR gene shows high levels of expression in various embryonic tissues as well as in the antenna of adults [17]. In parsley and Arabidopsis there are two CPR genes, one of which is constitutively expressed while the other is induced by biotic and abiotic stresses [21,47].
Phenotype of the redA minus mutants
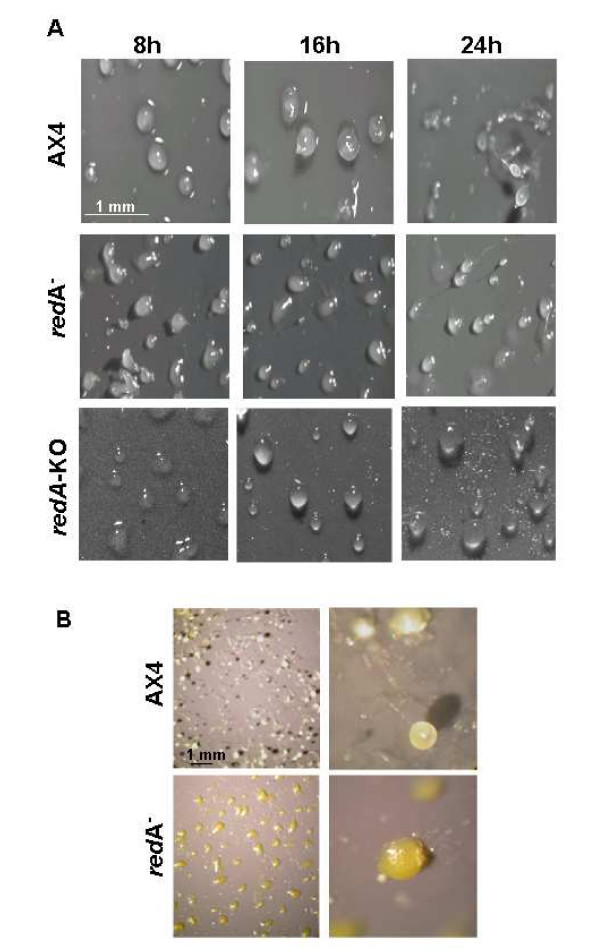
To confirm that the redA- phenotype is due to the disruption of redA, we generated new mutant strains by homologous recombination with the original plasmid pRED isolated from redA REMI mutant. Effective redA disruption was checked by Southern blot analyses of genomic DNA from blasticidin-resistant clones (data not shown) and six independent knockout clones were isolated which showed the same mound-arrest phenotype. One strain (redA-KO) was selected for further analyses. As shown in Figure 4A, when compared to wild type AX4 strain, the majority of cells of redA- and redA-KO mutants failed to make mature fruiting bodies after 24 h development on filters, and was arrested at the mound stage where they accumulated a yellow pigment (Figure 4B). It should be pointed out that the redA-KO mutants form a few tipped aggregates in the mound population after 48 hours starvation (data not shown). On the other hand, the original REMI and the recapitulated mutants did not show any significant differences in their growth curves when compared to the wild type AX4 (data not shown).
Figure 4.
Disruption of redA impairs development at mound stage. (A) Exponentially growing AX4 wild type cells and the mutants redA- and redA-KO were starved on filter pads and photographed at the indicated times (h) after starvation. (B) AX4 fruiting bodies and redA- yellow mounds after 48 hours starvation on filter pads are shown at lower (left) and at 5× higher magnification (right).
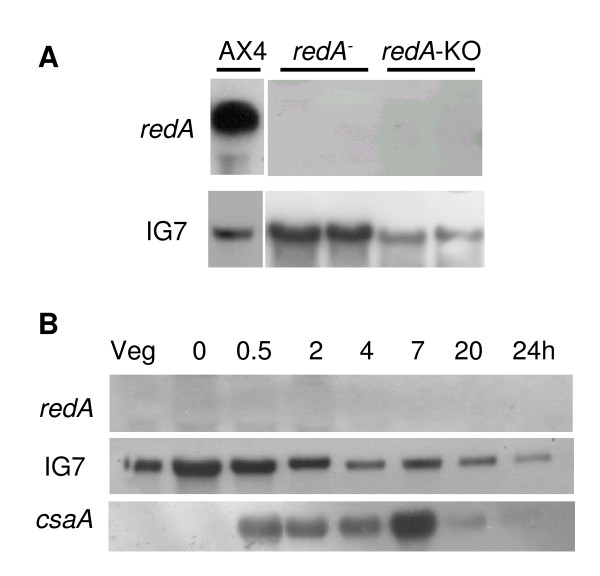
As expected, redA- and redA-KO mutants did not express redA mRNA (Figure 5A). Despite their developmental defect, redA- cells expressed csaA during development on filter pads (Figure 5B). Moreover, redC transcriptional profile is reasonably similar in wild type AX4 and in redA-cells (Figure 3) as its transcript levels strongly decrease upon starvation. On the other hand despite being expressed throughout development both in AX4 and redA- cells, redB transcript accumulates at higher levels in the latter (Figure 3). Also the peak of expression of ecmA was found to be advanced by four hours in the redA- strain as compared to the wild type (Figure 3).
Figure 5.
Disruption of redA results in cells lacking redA transcript. (A) Total RNA was prepared from exponentially growing AX4 cells (AX4) and from two independent clones of the mutants redA- and redA-KO. Identical Northern blots were probed with redA and IG7 cDNAs as indicated. (B) Exponentially growing redA- cells (Veg) were starved on filter pads and harvested at the indicated times (h) after starvation. Identical Northern blots of total RNA samples were probed with redA, IG7 and csaA cDNAs as indicated.
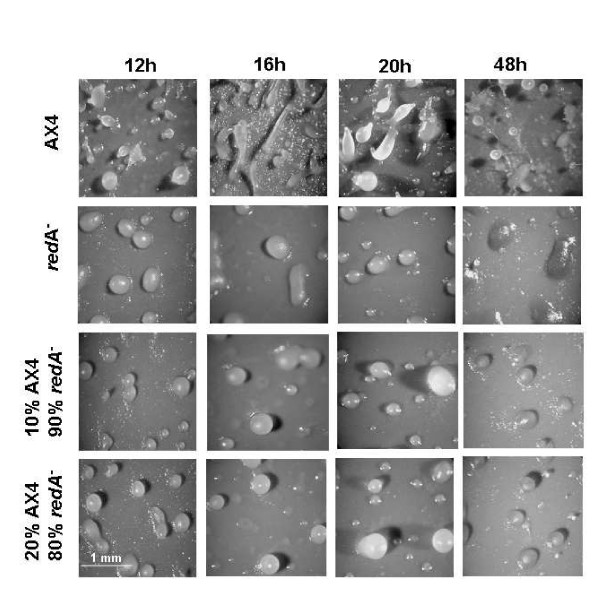
The developmental defect of redA- mutant is not rescued by mixing with AX4 wild type cells. As shown in Figure 6, mixing 10% or 20% of AX4 with redA- mutant did not overcome the mutant mound arrest indicating autonomy of the mutant phenotype. Moreover, the mutant cells did not inhibit wild type cells from forming fruiting bodies when they were developed together in equal numbers (not shown).
Figure 6.
AX4 cells do not rescue redA- phenotype. Exponentially growing redA- and AX4 wild type cells were starved on filter pads mixed at the indicated proportions. At the indicated times (h) after starvation cells were photographed.
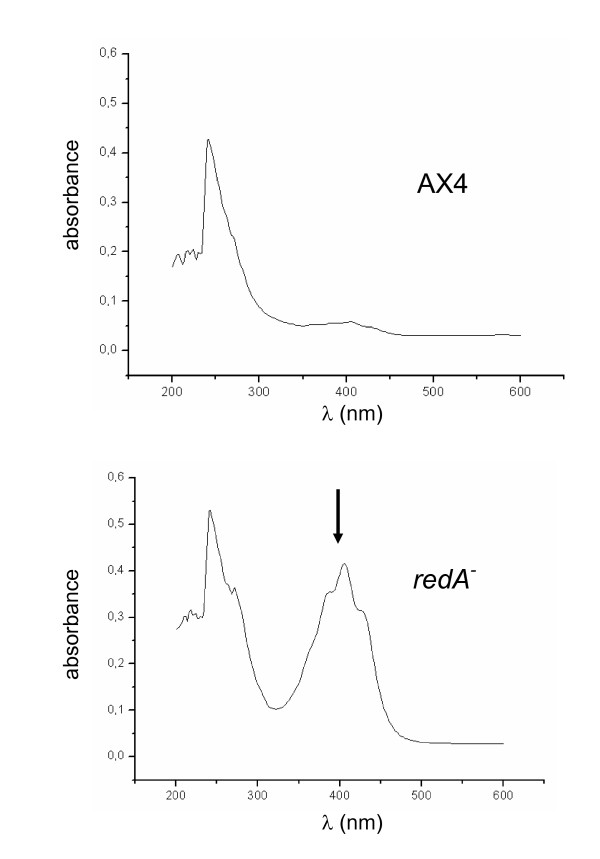
As mentioned above both redA- and redA-KO mutants form yellow mounds upon starvation (Figure 4B). We have observed that after 48 hours filter starvation the mounds and even the filter turn a strong yellow colour. This does not reflect premature spore formation since the yellow mounds do not contain any viable spores (data not shown). Chloroform extracts of redA- mounds collected after 48 hours starvation show an absorption peak at 400 nm which is not observed in AX4 cells (Figure 7). We did not succeed in characterizing the metabolites that accumulate in redA-mutants despite many attempts.
Figure 7.
UV and visible spectra of the chloroform extracts from AX4 and redA-. Chloroform extracts of AX4 and redA- cells collected after 48 hours starvation were analyzed using a UV/Visible spectrophotometer. The arrow points the absorption peak at 400 nm observed in the redA- cell extract.
CPR is a key enzyme in many metabolic processes, as a consequence of its close interactions with cytochrome P450 heme oxygenases. In particular, its participation in synthesis and/or degradation of important cellular compounds, such as retinoic acid, cholesterol and steroid hormones [2,29,30,35,36,54] may be related to abnormalities observed in development of organisms where CPR expression is abolished [28,32,55]. Development of homozygous mouse embryos carrying inactivating mutations in both alleles of the CPR gene is severely impaired, but lethality is only observed 10 to 13 days after zygote formation [28,29]. These findings indicate the importance of CPR in early animal development. Our results point to a role for a D. discoideum CPR in the metabolism of factors which control its cellular differentiation.
Conclusion
The D. discoideum genome encodes three genes (redA, redB and redC) for enzymes of diflavin oxidoreductases family. Disruption of redA led to mutant cells that form yellow mounds that fail to make proper fruiting bodies. The developmental arrest shown by this mutant implicates redA-encoded P450 oxidoreductase in the metabolism of compounds that control cell differentiation.
Methods
Cells and culture conditions
Dictyostelium discoideum strain AX4 and derived transformants were grown in axenic medium (HL-5) or in SM agar plates on lawns of Klebsiella aerogenes [56]. Complete and synchronous developmental program was undertaken by washing cells with 20 mM phosphate buffer (pH 6.4) and depositing them at 5 × 107 on nitrocellulose filters supported on buffer-saturated pads as previously described [56]. Strain DG1047 (redA-) was selected from a HindIII REMI-mutagenised library of strain AX4 selected for integration of the pBSR3 vector which carries the blasticidin S resistance cassette [34,57]. Morphological mutants were recognized by the structures formed within plaques generated by the blasticidin-resistant cells grown on SM agar in association with K. aerogenes.
Cloning of Dictyostelium redA cDNA
Regions flanking the plasmid insertion site in the REMI-mutant were isolated by plasmid rescue as described [34]. Genomic DNA from redA- strain was digested with HindIII ligated and electroporated into Escherichia coli SURE cells (Stratagene). The rescue plasmid pRED was isolated from the ampicillin-resistant bacterial transformants and sequenced. A 2190 bp sequence partially encoding redA gene has been deposited in GenBank (access number AF012946). HindIII-linearized pRED was used to recapitulate redA mutation by homologous recombination as described [34].
A 1.8 kbp genomic fragment obtained from pRED by digestion with HindIII and SmaI was used as probe to screen a Lambda-ZAP (Stratagene) cDNA library derived from AX4 D. discoideum vegetative cells (kindly provided by Dr. Hudson Freeze, The Burnham Institute, La Jolla, USA). Screening of 200,000 plaques under high stringency conditions yielded twenty positives clones that were subjected to in vivo excision from a phagemid by transformation of Escherichia coli XL-1 blue MRF' (Stratagene). The pBluescript SK clone (2B) with the largest cDNA insert was completely sequenced on both strands and the sequence was deposited in GenBank (access number DQ344637).
Molecular cloning procedures were essentially as described [58], unless otherwise noted. DNA sequencing was performed on an ABI 377 automated sequencer (Perkin-Elmer).
Northern Blots
Total RNA was isolated from 5 × 107 D. discoideum cells at various developmental stages by using the Trizol (Invitrogen). Formaldehyde-agarose gel electrophoresis of RNA (20 μg) and transfers to nylon membranes (Amersham), were performed as described [58]. Probes were prepared with gel-purified DNA fragments radiolabeled with [α-32P]dATP and [α-32P]dCTP by the random hexanucleotide priming method (Random Primers DNA Labeling System, Invitrogen).
RT-qPCR
Reverse transcription was carried out with 5 μg of D. discoideum total RNA primed with a mixture of oligo dT and random hexamers using SuperScript First-Strand Synthesis System (Invitrogen). A 20 μg amount of the resulting cDNA were subjected to quantitative PCR using Platinum SYBR Green qPCR SuperMix UDG (Invitrogen) on a GeneAmp 5700 System (Applied Biosystems) using the default thermocycler program for all genes. Threshold values were normalized according to Ct of D. discoideum mitochondrial large subunit rRNA (IG7), which is expressed at similar levels throughout the D. discoideum development [59]. The fold change of each gene was calculated using the 2-ΔCt method [60]. qPCR assays were performed in triplicate with the following gene-specific primer pairs: redA (5'-CCTATGGTGATGGTGTTCCACCAAC-3' and 5'-CCCCACTAAATTGAATATGTGAAAGATTTAAACGA-3'), redB (5'-GCAACCGAAGAAGCAAACGAAGAATACAAT-3' and 5'-CAAAGGTTGAAGACCTGGGAAAGATTCTAA-3'), redC (5'-AGGTGGAGTCTTTGAAAGATGTTGTAAAAATCC-3' and 5'-GGTCCAGGTACTGGTGTTGCAC-3'), ecmA (5'-AGCTGATAGTTGCGATTCCA-3' and 5'-TACCTCCTGTACCACCACCA-3') and rnlA (IG7) (5'-GTGGTTCGGCACCTCGAT-3' and 5'-CACCCCAACCCTTGGAAACT-3').
Chloroform extraction
5 × 108 AX4 and redA- cells developed on filters for 48 hours were extracted with 1 ml chloroform and the organic phase was collected by centrifugation at 3000 × g for 5 min at 4°C. UV/Visible spectra of the chloroform extracts were obtained in UV-2401PC Shimadzu spectrophotometer.
Sporulation efficiency assay
Mutants and wild-type cells were allowed to develop on nitrocellulose filters. At 0, 8, 12, 16, 20, 24 and 48 hours the cells were harvested from the filters with 20 mM phosphate buffer (pH 6.4) and sporulation efficiency was determined by detergent and heating treatment of the cells following plating on SM agar in association with K. aerogenes [56]. The number of plaques in the bacterial lawn indicated the number of viable spores. Wild-type AX4 cells submitted to 5-day starvation were used as a positive control for these experiments, to ensure recovery of fruiting bodies with viable spores.
List of abbreviations used
CPR, NADPH-cytochrome P450 oxidoreductase; FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide.
Authors' contributions
DCGK carried out most of the experimental work and drafted the manuscript. LF performed RT-qPCR assays and helped with chloroform extractions. LCF was involved in the initial steps of this work and helped with cDNA cloning and sequence analyses. WFL isolated strain DG1047, participated in the design of the study and in writing the manuscript. AMDS coordinated the study, participated in its design and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Dr. Claudette Klein for suggestions at the beginning of this work. Funded by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) to A.M. da Silva and from The National Institutes of Health to W.F. Loomis. D.C. Gonzalez-Kristeller, L.C. Fiorini and L. Farage were recipients of FAPESP fellowships. D.C. Gonzalez-Kristeller and A.M. da Silva were partially supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Contributor Information
Daniela C Gonzalez-Kristeller, Email: danig@iq.usp.br.
Layla Farage, Email: layla@iq.usp.br.
Leonardo C Fiorini, Email: lcfiorini@hotpop.com.
William F Loomis, Email: wloomis@ucsd.edu.
Aline M da Silva, Email: almsilva@iq.usp.br.
References
- Vermilion JL, Ballou DP, Massey V, Coon MJ. Separate roles for FMN and FAD in catalysis by liver microsomal NADPH-cytochrome P-450 reductase. J Biol Chem. 1981;256:266–77. [PubMed] [Google Scholar]
- Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci USA. 1997;94:8411–6. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14:611–50. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- Hasler JA. Pharmacogenetics of cytochromes P450. Mol Aspects Med. 1999;20:12–24. doi: 10.1016/s0098-2997(99)00005-9. 25–137. [DOI] [PubMed] [Google Scholar]
- Porter TD. An unusual yet strongly conserved flavoprotein reductase in bacteria and mammals. Trends Biochem Sci. 1991;16:154–8. doi: 10.1016/0968-0004(91)90059-5. [DOI] [PubMed] [Google Scholar]
- Murataliev MB, Feyereisen R, Walker FA. Electron transfer by diflavin reductases. Biochim Biophys Acta. 2004;1698:1–26. doi: 10.1016/j.bbapap.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Olteanu H, Banerjee R. Human methionine synthase reductase, a soluble P-450 reductase-like dual flavoprotein, is sufficient for NADPH-dependent methionine synthase activation. J Biol Chem. 2001;276:35558–63. doi: 10.1074/jbc.M103707200. [DOI] [PubMed] [Google Scholar]
- Paine MJ, Garner AP, Powell D, Sibbald J, Sales M, Pratt N, Smith T, Tew DG, Wolf CR. Cloning and characterization of a novel human dual flavin reductase. J Biol Chem. 2000;275:1471–8. doi: 10.1074/jbc.275.2.1471. [DOI] [PubMed] [Google Scholar]
- Porter TD, Kasper CB. NADPH-cytochrome P-450 oxidoreductase: flavin mononucleotide and flavin adenine dinucleotide domains evolved from different flavoproteins. Biochemistry. 1986;25:1682–7. doi: 10.1021/bi00355a036. [DOI] [PubMed] [Google Scholar]
- Schacter BA, Nelson EB, Marver HS, Masters BS. Immunochemical evidence for an association of heme oxygenase with the microsomal electron transport system. J Biol Chem. 1972;247:3601–7. [PubMed] [Google Scholar]
- Enoch HG, Strittmatter P. Cytochrome b5 reduction by NADPH-cytochrome P-450 reductase. J Biol Chem. 1979;254:8976–81. [PubMed] [Google Scholar]
- Ono T, Ozasa S, Hasegawa F, Imai Y. Involvement of NADPH-cytochrome c reductase in the rat liver squalene epoxidase system. Biochim Biophys Acta. 1977;486:401–7. [PubMed] [Google Scholar]
- Nelson DR. Cytochrome P450 and the individuality of species. Arch Biochem Biophys. 1999;369:1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- Simmons DL, Lalley PA, Kasper CB. Chromosomal assignments of genes coding for components of the mixed-function oxidase system in mice. Genetic localization of the cytochrome P-450PCN and P-450PB gene families and the NADPH-cytochrome P-450 oxidoreductase and epoxide hydratase genes. J Biol Chem. 1985;260:515–21. [PubMed] [Google Scholar]
- Shephard EA, Phillips IR, Santisteban I, West LF, Palmer CN, Ashworth A, Povey S. Isolation of a human cytochrome P-450 reductase cDNA clone and localization of the corresponding gene to chromosome 7q11.2. Ann Hum Genet. 1989;53:291–301. doi: 10.1111/j.1469-1809.1989.tb01798.x. [DOI] [PubMed] [Google Scholar]
- Sutter TR, Loper JC. Disruption of the Saccharomyces cerevisiae gene for NADPH-cytochrome P450 reductase causes increased sensitivity to ketoconazole. Biochem Biophys Res Commun. 1989;160:1257–66. doi: 10.1016/S0006-291X(89)80139-1. [DOI] [PubMed] [Google Scholar]
- Hovemann BT, Sehlmeyer F, Malz J. Drosophila melanogaster NADPH-cytochrome P450 oxidoreductase: pronounced expression in antennae may be related to odorant clearance. Gene. 1997;189:213–9. doi: 10.1016/S0378-1119(96)00851-7. [DOI] [PubMed] [Google Scholar]
- Yadav JS, Loper JC. Cloning and characterization of the cytochrome P450 oxidoreductase gene from the zygomycete fungus Cunninghamella. Biochem Biophys Res Commun. 2000;268:345–53. doi: 10.1006/bbrc.2000.2124. [DOI] [PubMed] [Google Scholar]
- Porter TD, Kasper CB. Coding nucleotide sequence of rat NADPH-cytochrome P-450 oxidoreductase cDNA and identification of flavin-binding domains. Proc Natl Acad Sci USA. 1985;82:973–7. doi: 10.1073/pnas.82.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malonek S, Rojas MC, Hedden P, Gaskin P, Hopkins P, Tudzynski B. The NADPH-cytochrome P450 reductase gene from Gibberella fujikuroi is essential for gibberellin biosynthesis. J Biol Chem. 2004;279:25075–84. doi: 10.1074/jbc.M308517200. [DOI] [PubMed] [Google Scholar]
- Mizutani M, Ohta D. Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol. 1998;116:357–67. doi: 10.1104/pp.116.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunic B, Truan G, Breskvar K, Pompon D. Functional cloning, based on azole resistance in Saccharomyces cerevisiae, and characterization of Rhizopus nigricans redox carriers that are differentially involved in the P450-dependent response to progesterone stress. Mol Genet Genomics. 2001;265:930–40. doi: 10.1007/s004380100492. [DOI] [PubMed] [Google Scholar]
- Van Bogaert IN, Develter D, Soetaert W, Vandamme EJ. Cloning and characterization of the NADPH cytochrome P450 reductase gene (CPR) from Candida bombicola. FEMS Yeast Res. 2007 doi: 10.1111/j.1567-1364.2007.00262.x. [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K, Lamb DC, Kelly DE, Manning NJ, Kelly SL. The N-terminal membrane domain of yeast NADPH-cytochrome P450 (CYP) oxidoreductase is not required for catalytic activity in sterol biosynthesis or in reconstitution of CYP activity. J Biol Chem. 1998;273:4492–6. doi: 10.1074/jbc.273.8.4492. [DOI] [PubMed] [Google Scholar]
- Truan G, Epinat JC, Rougeulle C, Cullin C, Pompon D. Cloning and characterization of a yeast cytochrome b5-encoding gene which suppresses ketoconazole hypersensitivity in a NADPH-P-450 reductase-deficient strain. Gene. 1994;142:123–7. doi: 10.1016/0378-1119(94)90366-2. [DOI] [PubMed] [Google Scholar]
- Lamb DC, Kelly DE, Manning NJ, Kaderbhai MA, Kelly SL. Biodiversity of the P450 catalytic cycle: yeast cytochrome b5/NADH cytochrome b5 reductase complex efficiently drives the entire sterol 14-demethylation (CYP51) reaction. FEBS Lett. 1999;462:283–8. doi: 10.1016/S0014-5793(99)01548-3. [DOI] [PubMed] [Google Scholar]
- Rogers KM, Pierson CA, Culbertson NT, Mo C, Sturm AM, Eckstein J, Barbuch R, Lees ND, Bard M. Disruption of the Candida albicans CYB5 gene results in increased azole sensitivity. Antimicrob Agents Chemother. 2004;48:3425–35. doi: 10.1128/AAC.48.9.3425-3435.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen AL, O'Leary KA, Kasper CB. Association of multiple developmental defects and embryonic lethality with loss of microsomal NADPH-cytochrome P450 oxidoreductase. J Biol Chem. 2002;277:6536–41. doi: 10.1074/jbc.M111408200. [DOI] [PubMed] [Google Scholar]
- Otto DM, Henderson CJ, Carrie D, Davey M, Gundersen TE, Blomhoff R, Adams RH, Tickle C, Wolf CR. Identification of novel roles of the cytochrome p450 system in early embryogenesis: effects on vasculogenesis and retinoic acid homeostasis. Mol Cell Biol. 2003;23:6103–16. doi: 10.1128/MCB.23.17.6103-6116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes V, Otto DM, Dickmann L, Schmidt K, Schuhbaur B, Henderson C, Blomhoff R, Wolf CR, Tickle C, Dolle P. Rescue of cytochrome P450 oxidoreductase (Por) mouse mutants reveals functions in vasculogenesis, brain and limb patterning linked to retinoic acid homeostasis. Dev Biol. 2007;303:66–81. doi: 10.1016/j.ydbio.2006.10.032. [DOI] [PubMed] [Google Scholar]
- Keeney DS, Waterman MR. Two novel sites of expression of NADPH cytochrome P450 reductase during murine embryogenesis: limb mesenchyme and developing olfactory neuroepithelia. Dev Dyn. 1999;216:511–7. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<511::AID-DVDY19>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Horike N, Takemori H, Nonaka Y, Sonobe H, Okamoto M. Molecular cloning of NADPH-cytochrome P450 oxidoreductase from silkworm eggs. Its involvement in 20-hydroxyecdysone biosynthesis during embryonic development. Eur J Biochem. 2000;267:6914–20. doi: 10.1046/j.1432-1033.2000.01796.x. [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A, Loomis WF. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci USA. 1992;89:8803–7. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GC, Tew DG, Wolf CR. Dissection of NADPH-cytochrome P450 oxidoreductase into distinct functional domains. Proc Natl Acad Sci USA. 1994;91:8710–4. doi: 10.1073/pnas.91.18.8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohr O, Paine MJ, Friedberg T, Roberts GC, Wolf CR. Engineering of a functional human NADH-dependent cytochrome P450 system. Proc Natl Acad Sci USA. 2001;98:81–6. doi: 10.1073/pnas.98.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen AL, Kasper CB. Differential contributions of NADPH-cytochrome P450 oxidoreductase FAD binding site residues to flavin binding and catalysis. J Biol Chem. 2000;275:41087–91. doi: 10.1074/jbc.M008380200. [DOI] [PubMed] [Google Scholar]
- Sem DS, Kasper CB. Interaction with arginine 597 of NADPH-cytochrome P-450 oxidoreductase is a primary source of the uniform binding energy used to discriminate between NADPH and NADH. Biochemistry. 1993;32:11548–58. doi: 10.1021/bi00094a011. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst. 1997;8:581–99. doi: 10.1142/S0129065797000537. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Ro DK, Ehlting J, Douglas CJ. Cloning, functional expression, and subcellular localization of multiple NADPH-cytochrome P450 reductases from hybrid poplar. Plant Physiol. 2002;130:1837–51. doi: 10.1104/pp.008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranbarger TJ, Forward BS, Misra S. Regulation of NADPH-cytochrome P450 reductase expressed during Douglas-fir germination and seedling development. Plant Mol Biol. 2000;44:141–53. doi: 10.1023/A:1006425025702. [DOI] [PubMed] [Google Scholar]
- He F, Chen YT. Cloning and heterologous expression of the NADPH cytochrome P450 oxidoreductase genes from an industrial dicarboxylic acid-producing Candida tropicalis. Yeast. 2005;22:481–91. doi: 10.1002/yea.1227. [DOI] [PubMed] [Google Scholar]
- Backes WL, Kelley RW. Organization of multiple cytochrome P450s with NADPH-cytochrome P450 reductase in membranes. Pharmacol Ther. 2003;98:221–33. doi: 10.1016/S0163-7258(03)00031-7. [DOI] [PubMed] [Google Scholar]
- van den Brink HJ, van Zeijl CM, Brons JF, van den Hondel CA, van Gorcom RF. Cloning and characterization of the NADPH cytochrome P450 oxidoreductase gene from the filamentous fungus Aspergillus niger. DNA Cell Biol. 1995;14:719–29. doi: 10.1089/dna.1995.14.719. [DOI] [PubMed] [Google Scholar]
- Benveniste I, Lesot A, Hasenfratz MP, Kochs G, Durst F. Multiple forms of NADPH-cytochrome P450 reductase in higher plants. Biochem Biophys Res Commun. 1991;177:105–12. doi: 10.1016/0006-291X(91)91954-B. [DOI] [PubMed] [Google Scholar]
- Koopmann E, Hahlbrock K. Differentially regulated NADPH:cytochrome P450 oxidoreductases in parsley. Proc Natl Acad Sci USA. 1997;94:14954–9. doi: 10.1073/pnas.94.26.14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranfar N, Fuller D, Loomis WF. Genome-wide expression analyses of gene regulation during early development of Dictyostelium discoideum. Eukaryot Cell. 2003;2:664–70. doi: 10.1128/EC.2.4.664-670.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma PC, Siu CH. A pharmacologically distinct cyclic AMP receptor is responsible for the regulation of gp80 expression in Dictyostelium discoideum. Mol Cell Biol. 1990;10:3297–306. doi: 10.1128/mcb.10.7.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuckolls GH, Osherov N, Loomis WF, Spudich JA. The Dictyostelium dual-specificity kinase splA is essential for spore differentiation. Development. 1996;122:3295–305. doi: 10.1242/dev.122.10.3295. [DOI] [PubMed] [Google Scholar]
- O'Leary KA, Kasper CB. Molecular basis for cell-specific regulation of the NADPH-cytochrome P450 oxidoreductase gene. Arch Biochem Biophys. 2000;379:97–108. doi: 10.1006/abbi.2000.1862. [DOI] [PubMed] [Google Scholar]
- Simmons DL, Kasper CB. Quantitation of mRNAs specific for the mixed-function oxidase system in rat liver and extrahepatic tissues during development. Arch Biochem Biophys. 1989;271:10–20. doi: 10.1016/0003-9861(89)90250-6. [DOI] [PubMed] [Google Scholar]
- Stromstedt M, Keeney DS, Waterman MR, Paria BC, Conley AJ, Dey SK. Preimplantation mouse blastocysts fail to express CYP genes required for estrogen biosynthesis. Mol Reprod Dev. 1996;43:428–36. doi: 10.1002/(SICI)1098-2795(199604)43:4<428::AID-MRD4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Sevanian A, Nordenbrand K, Kim E, Ernster L, Hochstein P. Microsomal lipid peroxidation: the role of NADPH – cytochrome P450 reductase and cytochrome P450. Free Radic Biol Med. 1990;8:145–52. doi: 10.1016/0891-5849(90)90087-Y. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–6. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Sussman M. Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 1987;28:9–29. doi: 10.1016/s0091-679x(08)61635-0. [DOI] [PubMed] [Google Scholar]
- Shaulsky G, Kuspa A, Loomis WF. A multidrug resistance transporter/serine protease gene is required for prestalk specialization in Dictyostelium. Genes Dev. 1995;9:1111–22. doi: 10.1101/gad.9.9.1111. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons, Inc; 1995. [Google Scholar]
- Early AE, Williams JG. A Dictyostelium prespore-specific gene is transcriptionally repressed by DIF in vitro. Development. 1988;103:519–24. doi: 10.1242/dev.103.3.519. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]