Abstract
Background
Anthrax and plague are diseases caused by Bacillus anthracis and Yersinia pestis respectively. These bacteria are etiological agents for worldwide zoonotic diseases and are considered among the most feared potential bioterror agents. Strain differentiation is difficult for these microorganisms because of their high intraspecies genome homogeneity. Moreover, fast strain identification and comparison with known genotypes may be crucial for naturally occurring outbreaks versus bioterrorist events discrimination.
Results
Thirty-nine B. anthracis and ten Y. pestis strains, representative of the species genetic diversity, were genotyped by Agilent 2100 Bioanalyzer using previously described Multiple Locus VNTR Analysis assays (MLVA). Results were compared to previous data obtained by standard genotyping system (capillary electrophoresis on automatic sequencer) and, when necessary, direct amplicon sequencing. A reference comparison table containing actual fragment sizes, sequencer sizes and Agilent sizes was produced.
Conclusion
In this report an automated DNA electrophoresis apparatus which provides a cheaper alternative compared to capillary electrophoresis approaches was applied for genotyping of B. anthracis and Y. pestis. This equipment, uses pre-cast gels and provides easy transportation, low maintenance and overall general logistic requirements and costs, is easy to set up and provides rapid analysis. This platform is a candidate for on-site MLVA genotyping of biothreat agents as well as other bacterial pathogens. It is an alternative to the more expensive and demanding capillary electrophoresis methods, and to the less expensive but more time-consuming classical gel electrophoresis approach.
Background
Bacillus anthracis is a Gram-positive spore-forming bacillus that causes anthrax [1,2]. This bacterium is commonly found in soil and is responsible for diseases of herbivores and other mammals including humans. Anthrax is still endemic in many countries, Middle East, Africa, North, Central and South America, as well as other areas of the world [3]. The site of entry determines different forms of anthrax, cutaneous, gastrointestinal, and inhalation; the latter form is highly fatal, with a mortality rate of up to 80% in the absence of an adequate antimicrobial therapy.
Yersinia pestis is a Gram-negative bacterium, etiological agent of plague. The bacterium is transmitted by fleas or aerosols, causing different forms of plague: bubonic, septicemic or pneumonic [4,5]. Y.pestis is often associated with the wellknown Black Death plague of the Middle Ages, a pandemic that had killed a third of European population in the 14th and 15th centuries, but approximately 2,000 human cases still occur worldwide each year [5]. Primary pneumonic plague is rapidly progressive and virulent, and, as inhalation anthrax, with a mortality rate close to 100% in the absence of a timely treatment. Y. pestis and B. anthracis are both considered serious threats and potential bioterrorism agents [6] because of their evaluation as bioweapons by Soviet Union and United States laboratories during the past decades. Above all, B. anthracis gained renewed attention in 2001, when letters containing anthrax spores were mailed causing the death of five persons and infecting 17 others, while probably hundreds of people were exposed to infectious spores [7]. Both agents are classified by the US Centre for Disease Control and Prevention in the Bioterrorism Disease Agent List as Category A microrganism, the most dangerous ones, because of easy dissemination and transmission, high mortality and impact to public health.
B. anthracis and Y. pestis both show very low intraspecies genetic diversity [8-10], making very challenging the rapid and accurate differentiation among individual biovars and strains. Nevertheless, finding a way to differenziate the strains by molecular genotyping, remains essential for discrimination between naturally occurring versus intentional outbreaks. The importance of forensic microbiology, as this field is know called, was demonstrated during the 2001 events, and previously by Tokyo [11] and Sverdlovsk [12] incidents. Finally genetic characterization of isolates allows to increase information about worldwide bacterial distribution and epidemiology.
Standard genotyping methods require either highly discriminative but heavy, and relatively expensive devices such as automated capillary electrophoresis devices, or cheaper, easy to use but more time consuming and with lower resolution power such as agarose gels (for a review of bacterial MLVA genotyping see [13]). A new miniaturized platform for quantification and separation of nucleic acid molecules, Agilent 2100 Bioanalyzer, has shown accuracy, precision and high feasibility along with speed and moderate cost reagents. This platform is based on microfluidic technology and allows to analyze 12 DNA samples in 30 minutes. The device, also called "Lab on a Chip", integrates multiple functions onto a single apparatus, so that sample dispensing, separation, detection and analysis are performed on the same support (a 5 × 5 cm chipper-cast gel). Along with limited weight and size (10 kg, 162 × 412 × 290 mm), the above features make the instrument ideal for field use and other on-site investigations. Agilent 2100 can also be easily used by low-expertise staff. A similar system was previously employed to study the genetic variability of bclA gene for strain discrimination within the Bacillus cereus group [14]. In this paper we evaluate this approach for genotyping analysis of the two major biothreat agents.
Results
Experimental design
In order to validate this platform, we compared the data produced by the Agilent 2100 Bioanalyzer to agarose gel based data (B. anthracis, Y. pestis) and capillary gel based data (B. anthracis). Ten Y. pestis and thirty-nine B. anthracis strains were genotyped using previously described sets of 25 VNTR loci [15,16]. These previously genotyped strains [15,16] were chosen to be representative of allelic variability within the two species. Briefly, these genotypes are from strains with the largest genetic distance, encompassing most of the observed allele variation (fig. 1 and 2). The 25 VNTR markers amplified for bacterial genotyping, were arranged into 12 PCR multiplex reactions either for B. anthracis or Y. pestis. This allowed us to use a single DNA 1000 chip for each strain.
Figure 1.
UPGMA dendrogram showing clustering and linkage distances of B. anthracis strains used to validate the Bioanalyzer 25 MLVA genotyping. The results obtained by previously described method [16] and Bioanalyzer genotyping are identical.
Figure 2.
UPGMA dendrogram showing clustering and linkage distances of Y. pestis strains used to validate Agilent Bioanalyzer 2100 based genotyping. The results obtained by previously described method [15] and Bioanalyzer genotyping are identical.
After PCR amplification 1 μl of each reaction was loaded into 12-wells chip (DNA 1000 LabChip Kit). Primer concentrations were experimentally adjusted to obtain a balanced amplification of different fragments. This optimization was facilitated using the quantification data supplied by the 2100 Bioanalyzer (peak area and fluorescence peak level). The vast majority of data produced by Agilent 2100 were not accurate. The discrepancies between observed and expected fragment size (offset in Table 1 and 2) are, presumably, the result of abnormal migration patterns of some repetitive sequences into electrophoretic matrix. In order to check if the offset was reproducible, we tested interchip and intrachip variability in markers containing the smallest repeat units, for which incorrect allelic calling is more probable. The samples were run in triplicate on the same and on different chips, and size data compared. In general, it was observed a low level of inter-intrachip variability (less than 25 % of the repeat unit length). For example, for B. anthracis CG3 marker (UL = 5, so 25% UL = 1,25), we observed 168 ± 1 for allele 1 and 173 ± 1 for allele 2. To correctly convert the Agilent DNA fragment size estimates into repeat copy numbers, it was necessary to establish conversion tables (Table 1 and 2) containing 2100 Bioanalyzer fragment sizes as well as actual sizes corresponding to repeats unit numbers, as previously reported [16].
Table 1.
Comparison between B. anthracis product sizes inferred by Agilent 2100 Bioanalyzer software (OBSERVED SIZE) and actual sizes obtained by direct sequencing of the PCR product or data available in Genbank (EXPECTED SIZE). UL BPS (unit length size).
| MULTIPLEX | MARKER (final primer concentration, μM) | UL BPS | ALLELE | EXPECTED SIZE | OBSERVED SIZE | OFFSET |
| 1 | CG3 (1.1) | 5 | 1 | 153 | 168 | -15 |
| 2 | 158 | 173 | -15 | |||
| BAMS44 (0.8) | 39 | 6 | 339 | 350 | -11 | |
| 8 | 417 | 431 | -14 | |||
| BAMS3 (0.6) | 15 | 21 | 474 | 455 | 19 | |
| 24 | 519 | 499 | 20 | |||
| 26 | 549 | 529 | 20 | |||
| 27 | 564 | 546 | 18 | |||
| 28 | 579 | 562 | 17 | |||
| 30 | 609 | 586 | 23 | |||
| 2 | vrrB2 (0.15) | 9 | 4 | 135 | 141 | -6 |
| 6 | 153 | 159 | -6 | |||
| 7 | 162 | 168 | -6 | |||
| 8 | 171 | 177 | -6 | |||
| BAMS5 (0.28) | 39 | 3 | 229 | 230 | -1 | |
| 5 | 307 | 297 | 10 | |||
| 6 | 346 | 333 | 13 | |||
| 7 | 385 | 370 | 15 | |||
| 8 | 424 | 411 | 13 | |||
| BAMS15 (0.6) | 9 | 24 | 418 | 427 | -9 | |
| 27 | 445 | 448 | -3 | |||
| 41 | 571 | 581 | -10 | |||
| 42 | 580 | 595 | -15 | |||
| 44 | 598 | 613 | -15 | |||
| 45 | 607 | 619 | -12 | |||
| 46 | 616 | 624 | -8 | |||
| 3 | BAMS1 (0.6) | 21 | 11 | 380 | 353 | 27 |
| 12 | 401 | 371 | 30 | |||
| 13 | 422 | 387 | 35 | |||
| 14 | 443 | 404 | 39 | |||
| 16 | 485 | 444 | 41 | |||
| vrrC1 (0.28) | 9 | 46 | 517 | 501 | 16 | |
| 48 | 535 | 520 | 15 | |||
| 53 | 583 | 564 | 19 | |||
| 57 | 616 | 595 | 21 | |||
| 4 | BAMS13 (0.28) | 9 | 17 | 337 | 334 | 3 |
| 20 | 364 | 362 | 2 | |||
| 21 | 373 | 370 | 3 | |||
| 24 | 391 | 386 | 5 | |||
| 27 | 427 | 421 | 6 | |||
| 30 | 454 | 452 | 2 | |||
| 31 | 463 | 459 | 4 | |||
| 47 | 607 | 597 | 10 | |||
| 76 | 868 | 807 | 61 | |||
| 5 | vrrB1 (0.15) | 9 | 12 | 193 | 198 | -5 |
| 15 | 220 | 229 | -9 | |||
| 16 | 229 | 237 | -8 | |||
| 19 | 256 | 264 | -8 | |||
| BAMS28 (0.6) | 24 | 10 | 397 | 394 | 3 | |
| 13 | 469 | 468 | 1 | |||
| 14 | 493 | 492 | 1 | |||
| vrrC2 (0.6) | 18 | 17 | 532 | 515 | 17 | |
| 19 | 568 | 549 | 19 | |||
| 21 | 604 | 583 | 21 | |||
| 6 | BAMS53 (1.1) | 12 | 6 | 212 | 224 | -12 |
| 8 | 236 | 249 | -13 | |||
| BAMS31 (0.6) | 9 | 15 | 331 | 332 | -1 | |
| 49 | 637 | 638 | -1 | |||
| 55 | 691 | 674 | 17 | |||
| 56 | 700 | 681 | 19 | |||
| 57 | 709 | 684 | 25 | |||
| 64 | 772 | 733 | 39 | |||
| 65 | 781 | 742 | 39 | |||
| 84 | 952 | 935 | 17 | |||
| 7 | vrrA (0.1) | 12 | 8 | 290 | 293 | -3 |
| 9 | 302 | 305 | -3 | |||
| 10 | 314 | 320 | -6 | |||
| 11 | 326 | 333 | -7 | |||
| BAMS25 (1.1) | 15 | 12 | 376 | 370 | 6 | |
| 13 | 391 | 381 | 10 | |||
| BAMS21 (1.1) | 45 | 9 | 631 | 599 | 32 | |
| 10 | 676 | 629 | 47 | |||
| 8 | BAMS34 (0.4) | 39 | 8 | 425 | 399 | 26 |
| 9 | 503 | 477 | 26 | |||
| 11 | 581 | 550 | 31 | |||
| 9 | BAMS24 (0.4) | 42 | 9 | 511 | 526 | -15 |
| 10 | 553 | 571 | -18 | |||
| 11 | 595 | 608 | -13 | |||
| 10 | pXO1 (0.6) | 3 | 6 | 123 | 135 | -12 |
| 7 | 126 | 138 | -12 | |||
| 8 | 129 | 141 | -12 | |||
| 9 | 132 | 145 | -13 | |||
| BAMS51 (0.4) | 45 | 6 | 358 | 338 | 20 | |
| 8 | 448 | 414 | 34 | |||
| 9 | 493 | 468 | 25 | |||
| 10 | 538 | 508 | 30 | |||
| BAMS22 (0.4) | 36 | 11 | 555 | 527 | 28 | |
| 13 | 627 | 581 | 46 | |||
| 14 | 663 | 624 | 39 | |||
| 15 | 699 | 643 | 56 | |||
| 16 | 735 | 659 | 76 | |||
| 11 | BAMS23 (1.1) | 42 | 5 | 399 | 377 | 22 |
| 9 | 567 | 518 | 49 | |||
| 10 | 609 | 557 | 52 | |||
| 11 | 651 | 572 | 79 | |||
| pXO2 (0.28) | 2 | 6 | 135 | 165 | -30 | |
| 7 | 137 | 168 | -31 | |||
| 8 | 139 | 170 | -31 | |||
| 9 | 141 | 172 | -31 | |||
| 12 | BAMS30 (0.8) | 9 | 6 | 268 | 269 | -1 |
| 17 | 367 | 360 | 7 | |||
| 51 | 673 | 670 | 3 | |||
| 57 | 727 | 685 | 42 | |||
| 60 | 754 | 700 | 54 | |||
| 68 | 826 | 770 | 56 | |||
| 69 | 835 | 776 | 59 | |||
| 72 | 862 | 796 | 66 | |||
| 75 | 889 | 817 | 72 | |||
| 78 | 916 | 840 | 76 | |||
Table 2.
Comparison between Y. pestis product sizes inferred by Agilent 2100 Bioanalyzer software (OBSERVED SIZE) and actual sizes obtained by direct sequencing of the PCR product or data available in Genbank (EXPECTED SIZE). UL BPS (unit length size).
| MULTIPLEX | MARKER (final primer concentration, μM) | UL BPS | ALLELE | EXPECTED SIZE | OBSERVED SIZE | OFFSET |
| 1 | ypms51 (0.2) | 21 | 1 | 186 | 185 | 1 |
| 2 | 207 | 207 | 0 | |||
| ypms01* (0.2) | 18 | 6 | 283 | 279 | 4 | |
| 7 | 301 | 291 | 10 | |||
| 9 | 337 | 329 | 8 | |||
| 10 | 355 | 344 | 11 | |||
| 2 | ypms04 (0.2) | 17 | 5 | 179 | 187 | -8 |
| 6 | 196 | 205 | -9 | |||
| 8 | 230 | 241 | -11 | |||
| 9 | 247 | 259 | -12 | |||
| ypms62* (0.2) | 9 | 5 | 270 | 258 | 12 | |
| 6 | 279 | 267 | 12 | |||
| 9 | 306 | 285 | 21 | |||
| 10 | 315 | 294 | 21 | |||
| 12 | 333 | 308 | 25 | |||
| 15 | 360 | 331 | 29 | |||
| 20 | 405 | 366 | 39 | |||
| 3 | ypms07 (0.2) | 10 | 7 | 164 | 168 | -4 |
| 8 | 174 | 178 | -4 | |||
| 9 | 184 | 187 | -3 | |||
| ypms05 (0.2) | 17 | 10 | 274 | 285 | -11 | |
| 11 | 291 | 300 | -9 | |||
| 4 | ypms74 (0.2) | 15 | 5 | 180 | 178 | 2 |
| 6 | 195 | 190 | 5 | |||
| 7 | 210 | 203 | 7 | |||
| 8 | 225 | 217 | 8 | |||
| ypms15* (0.2) | 15 | 9 | 275 | 272 | 3 | |
| 10 | 290 | 284 | 6 | |||
| 5 | ypms35 (0.2) | 15 | 8 | 204 | 208 | -4 |
| 9 | 219 | 222 | -3 | |||
| ypms40* (0.2) | 17 | 7 | 253 | 252 | 1 | |
| 8 | 270 | 266 | 4 | |||
| 9 | 287 | 277 | 10 | |||
| 6 | ypms38 (0.2) | 16 | 6 | 201 | 200 | 1 |
| 7 | 217 | 215 | 2 | |||
| 8 | 233 | 230 | 3 | |||
| ypms20* (0.2) | 15 | 8 | 288 | 285 | 3 | |
| 9 | 303 | 300 | 3 | |||
| 7 | ypms41 (0.2) | 17 | 5 | 183 | 188 | -5 |
| 6 | 200 | 207 | -7 | |||
| 7 | 217 | 221 | -4 | |||
| ypms56* (0.2) | 16 | 7 | 258 | 265 | -7 | |
| 9 | 290 | 296 | -6 | |||
| 8 | ypms45 (0.2) | 12 | 6 | 149 | 147 | 2 |
| 7 | 161 | 159 | 2 | |||
| ypms44 (0.2) | 17 | 7 | 233 | 239 | -6 | |
| 8 | 250 | 260 | -10 | |||
| ypms09* (0.2) | 18 | 22 | 732 | 671 | 61 | |
| 32 | 930 | 847 | 83 | |||
| 35 | 984 | 912 | 72 | |||
| 9 | ypms71 (0.2) | 14 | 5 | 157 | 163 | -6 |
| 6 | 171 | 178 | -7 | |||
| ypms46 (0.2) | 7 | 3 | 238 | 243 | -5 | |
| 4 | 245 | 250 | -5 | |||
| 5 | 252 | 256 | -4 | |||
| 6 | 259 | 263 | -4 | |||
| 10 | 308 | 313 | -5 | |||
| 12 | 322 | 326 | -4 | |||
| 10 | ypms70 (0.2) | 9 | 5 | 137 | 145 | -8 |
| 6 | 146 | 155 | -9 | |||
| 8 | 164 | 174 | -10 | |||
| 9 | 173 | 185 | -12 | |||
| 11 | 191 | 205 | -14 | |||
| ypms54 (0.2) | 22 | 6 | 259 | 265 | -6 | |
| 7 | 281 | 289 | -8 | |||
| 11 | ypms69 (0.2) | 16 | 5 | 163 | 164 | -1 |
| 6 | 179 | 177 | 2 | |||
| 7 | 195 | 195 | 0 | |||
| ypms06 (0.2) | 60 | 3 | 306 | 309 | -3 | |
| 4 | 366 | 374 | -8 | |||
| 6 | 486 | 504 | -18 | |||
| 8 | 606 | 625 | -19 | |||
| 9 | 666 | 674 | -8 | |||
| 12 | ypms73 (0.2) | 18 | 5 | 207 | 212 | -5 |
| 6 | 225 | 231 | -6 | |||
| ypms21* (0.2) | 18 | 9 | 329 | 339 | -10 | |
| 10 | 347 | 358 | -11 | |||
| 11 | 365 | 375 | -10 | |||
* modified primer set (increased final fragment size)
Once these reproducible offset values are taken into account, the MLVA assay as run on the Agilent exhibited concordant results with previous typing results for B. anthracis and Y. pestis [15,16]. We were able to resolve closely sized DNA fragments, and the analyzed samples showed concordance either with interchip repetitions or with expected size data. These data, used for UPGMA cluster analysis, generate dendrograms with linkage distance perfectly identical to those previously obtained by standard methodologies (Fig. 1 and 2). For each individual strain the Agilent assay is completed in about 30 minutes after PCR amplification. For a single strain to be genotyped, this time is significantly lower than 3 hours described in previous assay [16].
Genotyping B. anthracis with the 25-loci MLVA
To fit all the 25 loci into a 12-wells chip the B. anthracis loci were combined into 5 triplex, 3 duplex and 4 singleplex (Table 1) PCR amplifications. The arrangement of different loci in the same multiplex is such as to avoid overlapping of VNTR markers size ranges. For four B. anthracis loci multiplexing was impossible, because of the large allele size range.
Primer sequences were as described in [16], loci combination and final primers concentrations are reported in Table 1. Under these conditions, we were able to amplify each expected fragment, and the software (Agilent 2100 Expert version B.02.03.SI307, firmware C.01.055) could identify amplicon size (Fig. 3). The offset values for all loci and alleles were measured and are presented in Tables 1 and 2. For chromosomal markers, reproducibility of the observed data allowed correct assignment of each allele for every locus (<25% of unit length). However, for plasmid markers, since their shorter repeat unit and PCR product size compared to the chromosomal ones, we observed variability not exceeding one single additional base for each allele in repeated runs. Even with this additional base, correct assignment could be reproducibly done for the plasmid loci.
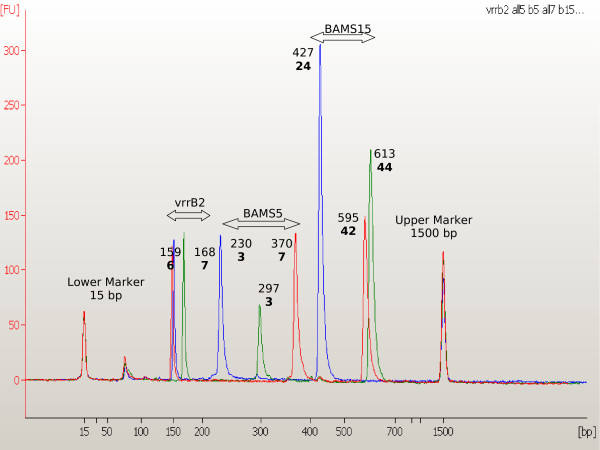
Figure 3.
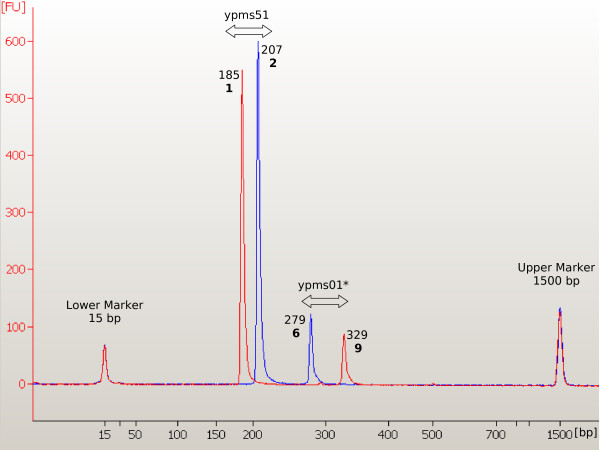
Multiplex n. 2 electropherogram comparison between three different B. anthracis strains (depicted in red, blue and green). For every amplicon the observed size and, in bold, the corresponding allele enumeration of markers vrrB2 BAMS5, and BAMS15 are shown.
Genotyping Y. pestis with the 25-loci MLVA
To fit all the 25 loci into 12 chip wells the Y. pestis 25 markers were grouped into 11 duplex and one triplex (Table 2).
The majority of primer sequences were as described in [15] but for 8 Y. pestis markers new primer pairs were redesigned to allow multiplexing (Table 3). Thus, final amplicon size was increased to avoid any possible overlap with other different loci into the new multiplexed reactions (Fig. 4). For Y. pestis markers (for which repeat units are at least 7 bp long [15,17]), we observed variability up to two bases (at the shortest repeat units) and, as previously described for B. anthracis, discrepancies not exceeding 25% of the repeat unit size.
Table 3.
Modified primers for Y. pestis from [15] designed for the new multiplexed reactions.
| YP MODIFIED MARKERS | PRIMER SEQUENCE |
| YPms01 | F: ACTTCGATGATTATTTTGTGCGTA R: TGTTTGGTGCTATTGCCGTA |
| YPms09 | F: TTTTTGCCGCACTGAACATA R: TCTGATGATTGTTGTGGTCGT |
| YPms15 | F: ACAAGCAAGTTGCGCAGATA R: CAAGTTCGCTTTCTGTGTCG |
| YPms20 | F: GCCAGAATAATGGCCGTAAA R: GTGGTTGTTCTTCACGTTGC |
| Ypms21 | F: ACCGGCTTAAAGCAGATTGA R: TTCCCTGTACTCGATTGTTGTG |
| YPms40 | F: TCCTGCTGCTGAGTTCATCTT R: TCATGTGCAATAGGCGTTGT |
| YPms56 | F: GCTCTAACAACGCCGGTAAA R: ATGGCATCAACCGACTGACT |
| YPms62 | F: AAACGGTCGTTAACGGAAGA R: GCTGAACAGCCCCATAAAAC |
Figure 4.
Multiplex n. 1 electropherogram comparison between two different Y. pestis strains (depicted in red and blue). For every amplicon the observed size and, in bold, the corresponding allele enumeration of markers ypms51 and ypms01 are shown. Yms01 is a locus for which modified primers pair is used.
Discussion
The intentional release of anthrax spores by mail in 2001 in the United States caused the death of five persons by inhalation anthrax and publicly demonstrated the bioweapon-associated threat, of which only the community of biodefence experts was previously aware. This increased the demands for a better genetic characterization of bioterrorism agents, in order to distinguish between natural outbreaks and/or intentional release of micro-organisms, and to help trace back the origin of an aggression. We have here evaluated the Agilent 2100 "Lab on a Chip" platform for genotyping of B. anthracis and Y. pestis strains on. This assay runs in 30 minutes one 12 multiplexed PCR reactions chip, genotyping a single isolate at a time. For this reason this system is faster compared to automated sequencing devices, either slab gel or capillary based when a single strain has to be genotyped. Given the possibility to compare different chip runs in different times this device can immediately show identities or diversities between new isolates compared to already characterized ones. Moreover it has shown a high degree of automation either for amplicon separation or for digital output of results. Compared to previous genotyping methods, the Bionalyzer is more effective than standard ethidium bromide slab gel electrophoresis, giving reproducible, precise and more sensible output for the shortest repeat units. Since no fluorescent primers are required the assay is cheaper to maintain than CEQ8000 or other sequencing machines. A rough estimation of the total cost of a 25 loci assay indicates that genotyping on this equipment is at least four to ten times less expensive than a capillary system assay, at low to medium throughput level. The equipment itself is cheaper, and both consumables and equipment can be stored for a long period of time and activated when needed.
Finally, the results are comparable to those obtained by automated sequencers. This platform appears particularly useful when response time is the critical factor. We propose therefore such a system as a method suitable for high resolution identification of biothreat agents. To date this system is the most effective genotyping technology available for on-site investigations. Also, this platform may be used for fast quality-checking of type collection, and DNA preparations for biosecurity and strain accountability purposes. Typing data produced by the MLVA approach can be easily compared using shared internet databases, as illustrated for instance in [18].
Conclusion
In this paper we describe a fieldable genotyping method for B. anthracis and Y. pestis. This method is an adaptation on Agilent 2100 Bioanalyzer of previously described 25 loci MLVA. The system was validated by characterizing thirty-nine B. anthracis and ten Y. pestis isolates, demonstrating to be, for a single genotyping, more rapid than traditional methods. The transfer on a new platform maintains reproducibility and precision to unambigously identify alleles. This method is shown to be a valid alternative to standard genotyping techniques for field characterization of important biothreat agents.
Methods
Bacterial strains and isolates
The strains and DNA samples genotyped for this report are from the collection maintained by the French Ministry of Defence at Centre d'Etude du Bouchet (CEB), from Istituto Superiore di Sanità (ISS) Italian collections and from the Italian Reference Center for Anthrax (ISZ, Foggia) [16].
VNTR amplification and analysis
DNA purification was performed as reported elsewhere [15,19]. Up to 30 ng of genomic DNA for each PCR reaction were used for DNA amplification in a final volume of 15 μl containing: 1× PCR reaction buffer (10 mM Tris-HCl, 1,5 mM MgCl2, 50 mM Kcl pH 8.3), 0.2 mM dNTPs, 1 U Taq polymerase and the appropriate concentrations of each primer as reported in Marker columns Table 1 and 2. PCR amplifications were conducted on a Peltier Thermal Cycler DNA Engine DYAD (MJ Research) as follow: initial denaturation step at 96°C for 3 min, 36 cycles of denaturation at 95°C for 20 seconds, annealing at 60°C for 30 seconds and extension at 72°C for 2 min. The reactions were terminated by a final incubation at 72°C for 5 min. Each reaction was loaded into chip wells prepared according to manufacturer recommendations (DNA 1000 LabChip Kit). Each chip contains 16 wells, 12 for the samples, 3 for gel mix (1 labeled black G and 2 grey G) and 1 for the ladder. Briefly, the black G well of DNA LabChip support was filled with 9 μl of a gel containing intercalating dye. The DNA LabChip was pressurized by a syringe in a priming station device and released after 60 sec. Then, two grey G wells were filled with 9 μl of gel-dye mix. After gel preparation, each sample well was loaded with 1 μl of PCR reaction and 5 μl of internal marker (containing two MW size standards of 15 and 1500 bp). Finally 1 μl of DNA ladder was loaded in the ladder well, the chip vortexed for 60 sec and inserted into Agilent 2100 Bioanalyzer. During the run the instrument analyzed sequentially every sample, showing electropherogram, virtual gel image and table data.
Authors' contributions
GF, SG and ACia did the set up of the B. anthracis 25-MLVA assay. VP, SG and ACia did the set up of the Y.pestis 25-MLVA assay. ACia, RDes and SG participated to typing work. FL, ACas and GV did the error checking analysis. VP, ACia, and ACar did various sequence analysis. RD'am and ACas did error checking of overall sequence analysis. GV was in charge of the Bionumerics database and clustering analyses. FL, ACia, RD'am and GV conceived the study. FL and GV wrote the report. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
Work facilitating the accountability of dangerous pathogens is supported by the French "Délégation Générale pour l'Armement" (DGA). This work was part of the European BiodefenceWork on the typing and molecular epidemiology of dangerous pathogens is supported by the French and Italian Ministry of Defence. SG is supported by the "Progetto Antrace" – ISS-Ministero della Salute, within the framework of the "Italy-US Collaboration Program". ACia and RDes are partially supported from the same project.
Contributor Information
Andrea Ciammaruconi, Email: andrea.ciammaruconi@gmail.com.
Saverio Grassi, Email: saverio.grassi@gmail.com.
Riccardo De Santis, Email: riccardo.desantis@gmail.com.
Giovanni Faggioni, Email: giovanni.faggioni@gmail.com.
Valentina Pittiglio, Email: sly.vapi@libero.it.
Raffaele D'Amelio, Email: raffaele.damelio@uniroma1.it.
Alessandra Carattoli, Email: carattoli@iss.it.
Antonio Cassone, Email: cassone@iss.it.
Gilles Vergnaud, Email: gilles.vergnaud@u-psud.fr.
Florigio Lista, Email: florigio.lista@esercito.difesa.it.
References
- Turnbull PC. Introduction: anthrax history, disease and ecology. Curr Top Microbiol Immunol. 2002;271:1–19. doi: 10.1007/978-3-662-05767-4_1. [DOI] [PubMed] [Google Scholar]
- Mock M, Fouet A. Anthrax. Annu Rev Microbiol. 2001;55:647–71. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- Van Ness GB. Ecology of anthrax. Science. 1971;172:303–7. doi: 10.1126/science.172.3990.1303. [DOI] [PubMed] [Google Scholar]
- Perry RD, Fetherston JD. Yersinia pestis – etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins SE, Rollins SM, Ryan ET. Yersinia pestis and the plague. Am J Clin Pathol. 2003;119:S78–85. doi: 10.1309/DQM9-3R8Q-NQWB-FYU8. [DOI] [PubMed] [Google Scholar]
- Bossi P, Garin D, Guihot A, Gay F, Crance JM, Debord T, Autran B, Bricaire F. Bioterrorism: management of major biological agents. Cell Mol Life Sci. 2006;63:2196–212. doi: 10.1007/s00018-006-6308-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush LM, Abrams BH, Beall A, Johnson CC. Index case of fatal inhalational anthrax due to bioterrorism in the United States. N Engl J Med. 2001;345:1607–1610. doi: 10.1056/NEJMoa012948. [DOI] [PubMed] [Google Scholar]
- Price LB, Hugh-Jones M, Jackson PJ, Keim P. Genetic diversity in the protective antigen gene of Bacillus anthracis. J Bacteriol. 1999;181:2358–2362. doi: 10.1128/jb.181.8.2358-2362.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PJ, Hill KK, Laker MT, Ticknor LO, Keim P. Genetic comparison of Bacillus anthracis and its close relatives using amplified fragment length polymorphism and polymerase chain reaction analysis. J Appl Microbiol. 1999;87:263–269. doi: 10.1046/j.1365-2672.1999.00884.x. [DOI] [PubMed] [Google Scholar]
- Adair DM, Worsham PL, Hill KK, Klevytska AM, Jackson PJ, Friedlander AM, Keim P. Diversity in a variable-number tandem repeat from Yersinia pestis. J Clin Microbiol. 2000;38:1516–1519. doi: 10.1128/jcm.38.4.1516-1519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Keim P, Kaufmann AF, Keys C, Smith KL, Taniguchi K, Inouye S, Kurata T. Bacillus anthracis incident, Kameido, Tokyo, 1993. Emerg Infect Dis. 2004;10:117–120. doi: 10.3201/eid1001.030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M, Guillemin J, Hugh-Jones M, Langmuir A, Popova I, Shelokov A, Yampolskaya O. The Sverdlovsk anthrax outbreak of 1979. Science. 1994;266:1202–1208. doi: 10.1126/science.7973702. [DOI] [PubMed] [Google Scholar]
- Vergnaud G, Pourcel C. Multiple Locus VNTR (Variable Number of Tandem Repeat) Analysis (MLVA) In: Stackebrandt E, editor. Molecular Identification, Systematics and Population Structure of Prokaryotes. Springer-Verlag; 2006. pp. 83–104. [Google Scholar]
- Castanha ER, Swiger RR, Senior B, Fox A, Waller LN, Fox K. Strain discrimination among B. anthracis and related organisms by characterization of bclA polymorphisms using PCR coupled with agarose gel or microchannel fluidics electrophoresis. J Microbiol Methods. 2006;64:27–45. doi: 10.1016/j.mimet.2005.04.032. [DOI] [PubMed] [Google Scholar]
- Le Fleche P, Hauck Y, Onteniente L, Prieur A, Denoeud F, Ramisse V, Sylvestre P, Benson G, Ramisse F, Vergnaud G. A tandem repeats database for bacterial genomes: application to the genotyping of Yersinia pestis and Bacillus anthracis. BMC Microbiol. 2001;1:2. doi: 10.1186/1471-2180-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lista F, Faggioni G, Valjevac S, Ciammaruconi A, Vaissaire J, le Doujet C, Gorge O, De Santis R, Carattoli A, Ciervo A, Fasanella A, Orsini F, D'Amelio R, Pourcel C, Cassone A, Vergnaud G. Genotyping of Bacillus anthracis strains based on automated capillary 25-loci multiple locus variable-number tandem repeats analysis. BMC Microbiol. 2006;6:33. doi: 10.1186/1471-2180-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel C, Andre-Mazeaud F, Neubauer H, Ramisse F, Vergnaud G. Tandem repeats analysis for the high resolution phylogenetic analysis of Yersinia pestis. BMC Microbiol. 2004;4:22. doi: 10.1186/1471-2180-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MLVA internet database http://mlva.u-psud.fr/
- Valjevac S, Hilaire V, Lisanti O, Ramisse F, Hernandez E, Cavallo JD, Pourcel C, Vergnaud G. Comparison of minisatellite polymorphisms in the Bacillus cereus complex: a simple assay for large-scale screening and identification of strains most closely related to Bacillus anthracis. Appl Environ Microbiol. 2005;71:6613–23. doi: 10.1128/AEM.71.11.6613-6623.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]