Abstract
Background
Alternative splicing of mutually exclusive exons is an important mechanism for increasing protein diversity in eukaryotes. The insect Mhc (myosin heavy chain) gene produces all different muscle myosins as a result of alternative splicing in contrast to most other organisms of the Metazoa lineage, that have a family of muscle genes with each gene coding for a protein specialized for a functional niche.
Results
The muscle myosin heavy chain genes of 22 species of the Arthropoda ranging from the waterflea to wasp and Drosophila have been annotated. The analysis of the gene structures allowed the reconstruction of an ancient muscle myosin heavy chain gene and showed that during evolution of the arthropods introns have mainly been lost in these genes although intron gain might have happened in a few cases. Surprisingly, the genome of Aedes aegypti contains another and that of Culex pipiens quinquefasciatus two further muscle myosin heavy chain genes, called Mhc3 and Mhc4, that contain only one variant of the corresponding alternative exons of the Mhc1 gene. Mhc3 transcription in Aedes aegypti is documented by EST data. Mhc3 and Mhc4 inserted in the Aedes and Culex genomes either by gene duplication followed by the loss of all but one variant of the alternative exons, or by incorporation of a transcript of which all other variants have been spliced out retaining the exon-intron structure. The second and more likely possibility represents a new type of a 'partially' processed pseudogene.
Conclusion
Based on the comparative genomic analysis of the alternatively spliced arthropod muscle myosin heavy chain genes we propose that the splicing process operates sequentially on the transcript. The process consists of the splicing of the mutually exclusive exons until one exon out of the cluster remains while retaining surrounding intronic sequence. In a second step splicing of introns takes place. A related mechanism could be responsible for the splicing of other genes containing mutually exclusive exons.
Background
Alternative splicing is an important and widespread mechanism that is used by higher organisms to express molecularly distinct mRNAs in response to developmental and cellular contexts [1,2]. Mutually exclusive splicing, in which only one exon is chosen out of a cluster of alternative exons arranged in a tandem array, is a very frequent alternative splicing event on a genome-wide level [3,4]. Several mechanisms have been proposed that explain why only one of the two or more variants is included in the mature mRNA [5-7]. Mostly, Metazoa contain mutually exclusive exons only in pairs. Extreme cases for mutually exclusive splicing are the insects Dscam genes that have arrays of up to 52 variants as observed in the Drosophila Dscam gene [8]. A less dramatic example is the mutually exclusive spliced Drosophila muscle myosin heavy chain gene that can potentially produce 480 different mRNAs [9].
Myosins comprise a large superfamily of actin-based motors that fulfill a variety of cellular functions from cell division, cellular locomotion, and vesicle transport to muscle contraction [10,11]. 35 classes of myosins have been identified to date with each class being responsible for a different function [12-14]. The first myosin was identified in skeletal muscle tissue over hundred years ago (for a review about the history of muscle myosin see [15]) and, since different myosins turned up, it has been referred to as conventional myosin or class-II myosin. Class-II myosins comprise the largest and most extensively studied class not only because the muscle myosin genes and muscles have been in the focus of biophysical and biochemical studies for decades and because the metazoan species are the most studied organisms but also because this class contains the most isoforms per organism [12].
Drosophila melanogaster contains two class-II myosin genes, one encoding the muscle isoforms (Mhc) and one the nonmuscle isoform (zipper) [16]. The Mhc gene produces all different muscle myosins as a result of alternative RNA splicing [9]. This is in contrast to the organisms of most other taxa of the Metazoa lineage, that have a family of muscle myosin heavy chain genes with each gene coding for a protein specialized for a functional niche. For example, the nematode Caenorhabditis elegans expresses six muscle myosins [13], while the ascidian Ciona intestinalis genome contains five muscle myosin heavy chain genes [17] and vertebrate genomes encode up to 22 muscle myosin heavy chain isoforms [12].
The Drosophila Mhc gene consists of 30 exons including five clusters of alternatively spliced exons and one differentially included penultimate exon. Thus, 480 combinations of alternative exons are possible. The four clusters of alternative exons in the motor domain part of the gene code for 120 different variations of the motor domain. In contrast to the muscle myosins of the other metazoa species, changes modulating myosin function are thus limited to four regions in the head domain. These discrete regions of sequence variation have been shown to produce physiological differences among the various muscle types [18]. Although many variations are possible and all alternative exons get expressed at some point in Drosophila's life, only a limited number of combinations seem to be employed. For example, during Drosophila embryogenesis only seven Mhc transcripts have been found to be expressed [18].
The genome of Drosophila melanogaster was the third eukaryotic genome to be completely sequenced [19]. Since then, the number of sequenced organisms has increased rapidly. Of the phylum Arthropoda, the genomes of the mosquitos Anopheles gambiae [20] and Aedes aegypti [21] and the silkworm Bombyx mori [22] have been published, and 17 further insect genomes have been finished of which eleven belong to the Drosophila species group [23,24].
Originally, pseudogenes have been defined as DNA sequences that are derived from functional genes, but acquired such degenerative features as premature stop codons and frameshift mutations, which make them unable to produce functional proteins [25-27]. Non-processed pseudogenes are thought to result from tandem duplications of genes with subsequent accumulation of disabling mutations. Processed pseudogenes lack introns and their original upstream gene regulatory resions and presumably arise by retrotransposition of a mature messenger RNA (mRNA). While non-processed pseudogenes are commonly found near the functional original gene, processed pseudogenes are randomly inserted into the genome. Also, partially processed pseudogenes have been reported that sometimes contain the complete coding region [28,29]. Recent studies have shown, that pseudogenes are not just "Junk" DNA but often exhibit functional roles (for a review see [26]).
Here, we report the comparative genomic analysis of the muscle myosin heavy chain genes of all arthropod species that have completely been sequenced so far. On this basis we propose that the splicing process operates sequentially on the transcript involving the splicing of all unwanted alternative versions of an exon while retaining intronic sequence around the remaining variant.
Results
Identification and annotation of the muscle myosin heavy chains
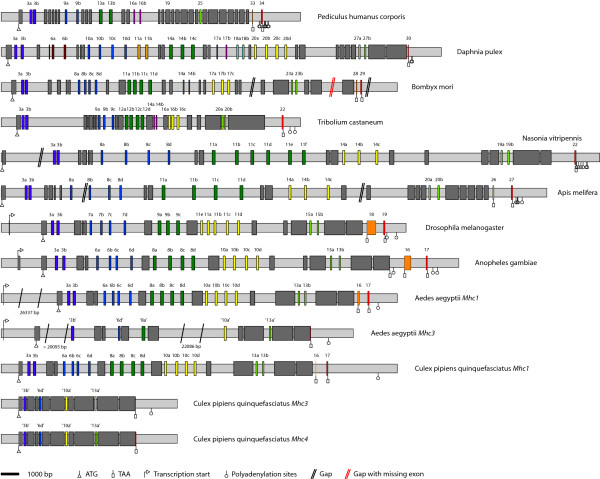
The arthropod muscle myosin heavy chain genes were identified by TBLASTN searches against the corresponding genome data of the different species using the Drosophila melanogaster protein as query (Figure 1, see Additional file 1). The species analysed were the mosquitos Aedes aegypti, Culex pipiens quinquefasciatus and Anopheles gambiae, the silkworm Bombyx mori, the honeybee Apis mellifera, the jewel wasp Nasonia vitripennis, the waterflea Daphnia pulex, the rust-red flour beetle Tribolium castaneum, the body louse Pediculus humanus corporis, and thirteen Drosophila species (Table 1). According to the general nomenclature for myosin sequences [12] the alternatively spliced muscle myosin heavy chain genes are named Mhc1, and the non-muscle myosin heavy chain genes are denoted Mhc2. The sequences were assigned by manual inspection of the genomic DNA sequences. Exons have been confirmed by the identification of flanking consensus intron-exon splice junction donor and acceptor sequences (Figure 1) [30]. Because of the five to nine clusters of mutually exclusive exons and the included or excluded penultimate exon, automatic identification of all exons failed. The genomic sequences of Apis mellifera and Bombyx mori contain several gaps that at least in one case must have contained missing exons. The expression of the myosin genes including the transcription of some of the mutually exclusive exons has been confirmed by analysis of corresponding EST data.
Figure 1.
Diagram of the arthropod Mhc1 genes with exon-intron structure. The gene structures of the arthropod muscle myosins genes are shown using the following color code: light-gray: intron sequences; dark-gray: common exons; colored: alternatively spliced exons. The Drosophila melanogaster Mhc1 gene is shown as representative for all Drosophila sp. Mhc1 genes, because their gene structures only differ in the length of the introns. The transcriptional and translational start sites, the stop codons and polyadenylation sites are shown if they have been determined. Some genes are spread on several contigs. The corresponding gap positions are shown in black, if further exons are not expected, and in red, if exons are definitively missing. The genes are drawn to scale except for the Aedes aegypti genes where the extremely long introns have been shortened. Gaps have been filled with 100 bp although their exact length is unknown.
Table 1.
Nucleotide ID's and number of combinations of alternative exons for the motor domains and the full-length proteins.
| Species | Species Abbr. | Nucleotide ID's GenBank: | Motor domain | Full-length protein |
| Daphnia pulex | Dap | 1536 | > 3072 | |
| Bombyx mori str. Dazao | Bm |
AADK01001734, BAAB01137479 BAAB01017092, AV404226 AADK01040535, AADK01049792 |
192 | 768 |
| Tribolium castaneum str. Georgia GA2 | Tic | AAJJ01000118 | 192 | > 384 |
| Nasonia vitripennis str. SymAX | Nav | AAZX01008059, AAZX01007288 | 144 | > 288 |
| Apis mellifera str. DH4 | Am |
AADG05005753, AADG05005754 AADG05005757 |
96 | 384 |
| Drosophila ananassae TSC#14024-0371.13 | Da | AAPP01015693 | 120 | 480 |
| Drosophila erecta TSC#14021-0224.01 | Der | AAPQ01007075 | 120 | 480 |
| Drosophila grimshawi TSC#15287-2541.00 | Dg | AAPT01021775 | 120 | 480 |
| Drosophila hydei | Dh | X77570 | 120 | 480 |
| Drosophila melanogaster | Dm | NM_165190 | 120 | 480 |
| Drosophila mojavensis TSC#15081-1352.22 | Dmo | AAPU01010481 | 120 | 480 |
| Drosophila persimilis MSH-3 | Drp |
AAIZ01000908, AAIZ01000907 AAIZ01000906, AAIZ01000905 AAIZ01000904, AAIZ01024863 AAIZ01000903 |
120 | 480 |
| Drosophila pseudoobscura MV2-25 | Dp | AAFS01000199 | 120 | 480 |
| Drosophila sechellia Rob3c | Dse | AAKO01001629 | 120 | 480 |
| Drosophila simulans str. white501 | Dss | 120 | 480 | |
| Drosophila virilis TSC#15010-1051.87 | Dv | AANI01016210, AANI01016211 | 120 | 480 |
| Drosophila yakuba Tai18E2 | Dy |
AAEU01002444, AAEU01002445 AAEU01002446 |
120 | 480 |
| Drosophila willistoni TSC#14030-0811.24 | Dw | AAQB01006734 | 120 | 480 |
| Anopheles gambiae str. PEST | Ang | AAAB01008980 | 128 | 768 |
| Aedes aegypti str. Liverpool Mhc1 | Aea | AAGE02009209 | 128 | 512 |
| Aedes aegypti str. Liverpool Mhc3 | Aea | AAGE02009019, AAGE02009018 | 1 | 1 |
| Pediculus humanus corporis str. USDA | Pdc | AAZO01001178 | 16 | 32 |
| Culex pipiens quinquefasciatus JHB Mhc1 | Cpq | AAWU01000999 | 128 | 512 |
| Culex pipiens quinquefasciatus JHB Mhc3 | Cpq | AAWU01000999 | 1 | 1 |
| Culex pipiens quinquefasciatus JHB Mhc4 | Cpq | AAWU01000999 | 1 | 1 |
The untranslated first exons of the genes have been assigned by analysing EST data, if possible. Because untranslated 5' exons were found for all those species for which EST data covering the amino-termini of the genes is available, it is expected that the other arthropod myosin genes also contain untranslated first exons. Accordingly, the unambiguously identified exons have been numbered starting with exon two. Duplicated exons were named in alphabetical order according to the direction of transcription, the exception being the alternatively spliced exon 11 of the Drosophila Mhc1 of which the first of the mutually exclusive exons was named 11e for historical reasons [9]. The differentially included penultimate exons of the Drosophila species have been predicted based on their similarity at the DNA level. Although this exon mainly consists of untranslated bases and its identity between the Drosophila species is almost as low as that found in intron regions, the exon borders are conserved enough to be recognised. The carboxy-terminal exons of the other arthropod Mhc1 genes have been confirmed by analysing EST data, if possible. For TicMhc1 and DapMhc1 only one carboxy-terminal exon could be confirmed by EST data. However, given the exon conservation between all arthropod Mhc1 genes it is expected that both genes contain another carboxy-terminal exon. For Nasonia, EST data is not available. The carboxy-terminal exon of the NavMhc1 gene was identified based on its homology to the other Mhc1 exons. An exon corresponding to the penultimate exon of the other genes could not be identified.
The Drosophila sp. Mhc1 genes, the AeaMhc1 and the CpqMhc1 gene contain consensus polyadenylation signals AATAAA, while the Mhc1 genes of Ang, Am, Dap, Nav, Pdc, and Tic contain polyadenylation signals of type AAAAAA. For the DmMhc1 gene it has been shown that the use of either polyadenylation site is not regulated [31,32] and the same might be true for the two or multiple polyadenylation sites of the other arthropod genes.
Identification of further muscle myosin heavy chain genes in Aedes aegypti and Culex pipiens quinquefasciatus
Surprisingly, a second muscle myosin heavy chain gene has been identified in Aedes aegypti (Figure 1) and named Mhc3. The Mhc3 gene contains the same exon organisation as Mhc1 except that it does not have any cluster of alternatively spliced exons and misses the two carboxy-terminal exons (Figure 1). Many EST clones provide supporting evidence for the deduced carboxy-terminus, the amino-terminal untranslated exon1, and other parts of the gene. The exons related to the alternatively spliced exons of Mhc1 are either identical ("exon3b") or very similar to one of the Mhc1 exons. The protein sequence of Mhc3 has an overall sequence identity of 91.4% to Mhc1. Besides the different carboxy-termini, the largest differences are in loop-1, which is three residues shorter in Mhc3, and in loop-2, which has only six instead of ten glycines and might therefore be structurally more restricted. The Culex pipiens quinquefasciatus genome encodes another two muscle myosin heavy chain genes that are very similar to each other and have been named Mhc3 and Mhc4 (Figure 1). Both have the same exon organisation as the CpqMhc1 gene except that they do not have any cluster of alternatively spliced exons and miss the two carboxy-terminal exons. Another difference is that alternative exons 8 are fused to the following constitutive exons in the Mhc3 and Mhc4 genes. The protein sequence identity between CpqMhc3 and CpqMhc4 is 92.0%, the identity to CpqMhc1 is 84.4% and 90.4%, respectively. Surprisingly, AeaMhc3, CpqMhc3 and CpqMhc4 retained identical variants of the alternatively spliced exons of the corresponding Mhc1 genes.
The BmMhc1, TicMhc1, PdcMhc1 and DapMhc1 genes contain further clusters of alternatively spliced exons
The analysis of the BmMhc1, TicMhc1, PdcMhc1, and DapMhc1 genes revealed further clusters of alternatively spliced exons compared to the DmMhc1 gene. All further sets of alternative exons encode for sequence that is part of the motor domain. The additional alternative exon of Bm, Pdc and Tic is conserved between these three organisms, and is also encoded within the Dap Mhc1 gene. It is located between the alternatively spliced exons 11 and 17 (Bm), alternative exon 13 and constitutive exon 19 (Pdc), and alternative exons 12 and 16 (Tic), respectively, and separated from the neighbouring alternatively spliced exons by constitutively expressed exons (Figure 1). In contrast to the other alternatively spliced exons, these alternatively spliced exons are different in length and amino acid conservation (see Additional file 2, figure S6A). The first part of the exon encodes part of loop-2 (see below), that is a very flexible loop involved in actin-binding. In the arthropod genes it mainly consists of glycines, arginines, and lysines. Thus, the alternatively spliced exons of Bm, Tic, Pdc, and Dap encode different numbers and compositions of these residues. The second part of the alternatively spliced exon is part of the following alpha-helix and hence completely conserved in length and strongly conserved in composition. In addition to this cluster of alternatively spliced exons, the DapMhc1 gene contains three further sets of alternatively spliced exons extending its number of clusters of alternatively spliced exons to nine (compared to five in Drosophila). Alternative exon 6 encodes an alternative P-loop to loop-1 sequence, alternative exon 11 directly follows the alternative exon encoding a structural part near the ATP-binding site, and alternative exon 18 encodes an alternative version of the sequence after loop-2 (Figure 1).
The PdcMhc1 gene encodes a strongly reduced set of possible transcripts
The Pediculus humanus corporis Mhc1 gene contains the most reduced set of alternative exons (Figure 1). It has four sets of alternative exons each comprising two variants. However, the sequence encoding part of the converter domain, which is encoded by sets of three to five alternative exons in the other arthropod genes, has been fused to the following exon forming one constitutive exon in the PdcMhc1 gene (exon 19, Figure 1). Also, the part in the tail domain encoded by a set of two alternative exons in all other arthropod genes is represented by only one exon in the PdcMhc1 gene (exon 25). Altogether, the alternative exons encode for 16 different versions of the motor domain and 32 different mRNAs of the PdcMhc1 gene, compared to potentially 120 different combinations of alternative exons for only the motor domain of the Drosophila Mhc1 gene.
Conservation of alternatively spliced exons
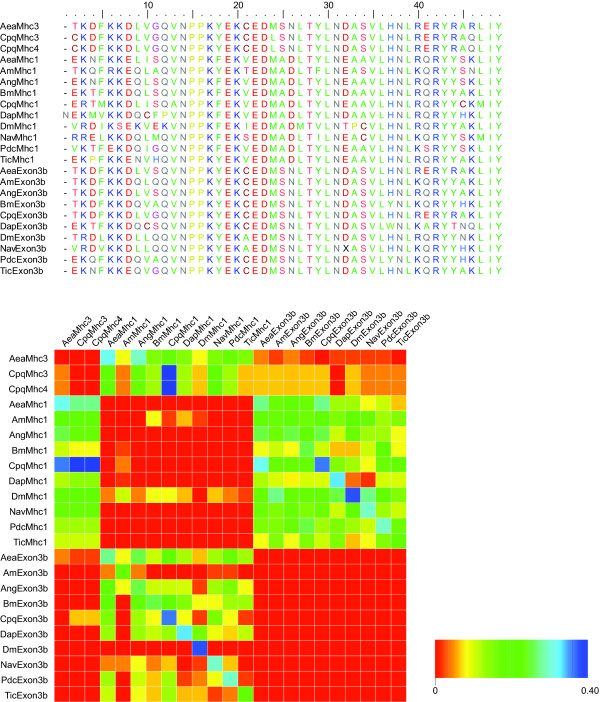
The number of variants differs between the arthropod species for many of the alternatively spliced exons (Figures 1 and 2). For the first set of alternatively spliced exons two variants have been found in all Mhc1 genes. Both differ by two absolutely conserved residues, namely the amino acids alanine and aspartate at positions 25 and 26 in the "a" variants of the exon that are substituted by serine and asparagine in the "b" variants (Figure 3). A slightly less conserved marker for the "b" variants is a cysteine at position 21. Variant 3a of the DapMhc1 is an exception as it has an additional residue at the N-terminus compared to the other Mhc1 variant "a" exons. The DapMhc1 gene encodes three clusters of alternatively spliced exons not found in the other arthropod Mhc1 genes. For all three clusters exons variant "b" is more homologous to the corresponding amino acid sequences of the other Mhc1 proteins than variant "a" (see Additional file 2, figures S2, S4, and S6B). The alternatively spliced exons of BmMhc1, DapMhc1, PdcMhc1 and TicMhc1 covering loop-2 are different in length and starting position (see Additional file 2, figure S6A). However, the "a" variants are more similar to each other than to the "b" variants and the corresponding amino acid sequences of the other Mhc1 proteins. Thus, the common ancestor of Bm, Dap, and Tic had in all probability already contained an "a" and a "b" variant. Completely conserved residues characterizing the "a" variant are a serine at the end of loop-2, a glutamate at position 3, and a leucine at position 8 of the following helix ([G/K/R 8-9]S [G/A]F [Q/M]TVS [S/A]LYR). Except for PdcMhc1, all arthropod Mhc1 genes have two variants of the mutually exclusively spliced exon in the tail (Figure 2; see also Additional file 2, figure S8). The most conserved differences between the two variants are an aspartate at position 14 in variant "b" (either an asparagine or a glutamine in variant "a") and an asparagine at position 24 (an arginine in variant "a"). In addition, at position 15 the "b" variants have a large hydrophobic residue (leucine, methionine, or phenylalanine) while the "a" variants have a small polar residue (serine or threonine). In contrast to the other Mhc1 genes, the "a" variant of DapMhc1 is closer related to the "b" variants than to the other "a" variants.
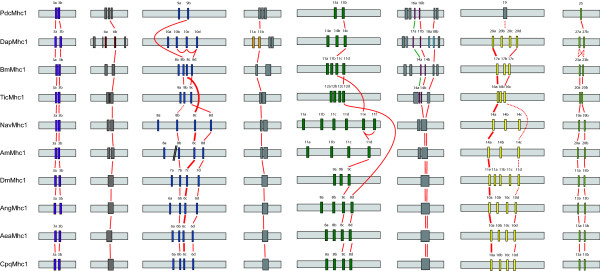
Figure 2.
Relationships between alternatively spliced exon. Sections of the Mhc1 genes of Figure 1 have been aligned showing the relationship between the exon-intron structures of the regions containing alternatively spliced exons. Continuous lines connect variants that are almost identical and thus expected to be derived from a common ancestor. Bold lines connecting alternative exons in regions containing multiple variants per Mhc1 gene highlight particularly conserved exons in these sets. Dotted lines represent putative connections between certain variants although their identity is not very strong on the protein level.
Figure 3.
Sequence conservation in the first set of the alternatively spliced exons. On top, the protein sequence alignment of the alternative exons is shown. The upper sequences, termed Mhc1, Mhc3, and Mhc4, respectively, represent the variant "a" exons. Below, the comparison of the sequence identity between each exon and variant "a" and "b" of every other Mhc1 protein is shown. The graphic has to be read in columns. The higher identity between an exon listed on top and variant "a" or "b" of a certain Mhc1 protein listed on the left side has been set to 1 (red color) while the difference of the lower identity to the value of the higher identity is plotted for the other combination of exons. Thus, in every column the higher identity of the named exon to one of the variants of the other Mhc1 proteins is visualized.
The situation is more complex for the remaining clusters of mutually exclusive exons that contain three to six variants. The exon encoding a loop-helix motif adjacent to the ATP-binding site (blue color in Figure 1) is not as conserved as the other alternatively spliced exons (Figure 2; see also Additional file 2, figure S3). Therefore, it is difficult to identify characteristic residues/motifs for the respective variants. Except for the PdcMhc1 and TicMhc1 genes all genes contain four variants. The variant with the most characteristic residues is variant "c". It is characterized by a positively charged residue at position 8 (arginine or histidine), a conserved arginine at position 21, and a conserved asparagine at position 26. None of these residues appear in any of the other variants at the respective positions. The TicMhc1, PdcMhc1, and DapMhc1 genes have lost this variant. The only strong characteristic of variant "d" is a conserved isoleucine or valine at position 20 that is found in all Mhc1 genes. Variants "a" and "b" do not contain any distinguishing residues.
The alternatively spliced exons spanning the relay helix and the relay loop are the longest and most conserved of the mutually exclusive exons (see Additional file 2, figure S5). The variability ranges from two variants in the Pediculus Mhc1 gene to six variants in the Nasonia gene (Figures 1 and 2). The least conserved part of the exon is the relay loop that is not embedded in the motor domain. In this region, characteristic residues for certain variants are found. Variant "c" is characterized by a conserved glutamine at position 49 and either a glutamine or an asparagine at position 50. A copy of this variant is present in all Mhc1 genes except that of Tic. Another conserved variant is variant "d" characterized by a glutamine at position 49 followed by a proline at position 50. This variant appears in the Mhc1 genes of Aea, Ang, Cpq, Tic, and Bm. Similar to the situation for the alternatively spliced exon at the ATP-binding site, the other variants are not conserved enough to define characteristic residues. It is thus not clear which were present in the ancient arthropod gene and which arose through exon duplication in the individual genes. Again, the DapMhc1 is the exception because its first two variants, characterized by two conserved methionines at positions 42 and 55, differ from all other variants.
The variants of the cluster of alternative exons encoding part of the converter domain also show a high degree of variability (Figure 2; see also Additional file 2, figure S7). Two of the variants have characteristic features. Variant "a" is the most conserved of the variants at the protein level having a conserved methionine at position 9 and a conserved cysteine at position 26. These residues do not appear in any of the other variants of this cluster. Variant "a" of this cluster is conserved in the Mhc1 genes of all species and therefore must have been present in their common ancestor. The last of the variants has a characteristic feature at the DNA level. The intron following the last variant always has a GT 5' splice site. This is in contrast to all other variants of this exon whose following introns have a GC 5' splice site. At the amino acid level this variant is characterized by a lysine at position 2, a cysteine at position 5 and a glutamate at position 20.
Wherever EST and/or cDNA data was available a differentially excluded penultimate exon could be identified. These exons are very short (one to thirteen residues) and not conserved (see Additional file 2, figure S9), and therefore similar exons have not been predicted for the species for which EST data is not available. For Ang three carboxy-termini have been identified. Based on EST data the AngMhc1 transcript may also end with a short extension to the antepenultimate exon. This C-terminus is similar to that found for AeaMhc3 and CpqMhc4 and might be used in a similar combination of the other alternatively spliced exons.
Phylogenetic analysis of the arthropod muscle myosin heavy chain genes
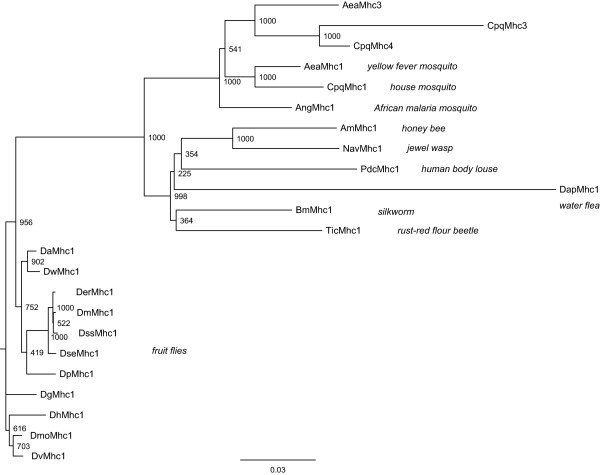
A phylogenetic tree of all arthropod Mhc1 protein sequences, always incorporating the first variant of the clusters of alternatively spliced exons and excluding the differentially included penultimate exon, has been generated (Figure 4). In general, the tree reflects the phylogenetic relationship between the species. The AeaMhc3 sequence is most closely related to the CpqMhc3 and the CpqMhc4 sequence implicating that the last common ancestor of Aedes and Culex already had one of these genes. The phylogeny of the Drosophila species slightly differs compared to other analyses [23]. Thus, the DaMhc1 sequence would have been expected to separate after the divergence of the DpMhc1 sequence. Similarly, the DseMhc1 gene would have been expected to be the closest relative of the DssMhc1 sequence. Overall, the sequence identity is very high. Between DapMhc1 and the other sequences the identity is 70.6 – 77.9%, while it is between 77.0% and 99.7% between the other species.
Figure 4.
Phylogenetic tree of the arthropod muscle myosin heavy chain proteins. The amino acid sequences of the full-length proteins were aligned manually. Because of their incompleteness the sequences of Drosophila persimilis and Drosophila yakuba have been omitted from the tree calculation. Support values for each internal branch were obtained by 1,000 bootstrap steps. The scale bar corresponds to 0.1 estimated amino acid substitutions per site.
Predicting the gene structure of an ancient Mhc1 gene
Whenever intron positions are shared between the genes, the corresponding type of splice site is conserved, with the exception of the shared exon 9 (AmMhc1), exon 10 (TicMhc1), exon 9 (BmMhc1), and the alternatively spliced exon 11 of DapMhc1 (Figure 5). All introns have consensus dinucleotide borders except those downstream of the last variant of the cluster of alternative exons encoding part of the motor domain (homologs of exon 11 in DmMhc1), which have a GC dinucleotide at the 5' donor site instead of the consensus GT. The 3' exons of these alternatively spliced exons again have a consensus GT site. Exon '10a' of AeaMhc3 is almost identical to exon 10a of AeaMhc1 and the following intron also has a GC dinucleotide at the 5' donor site. In contrast to the introns following the exons 9 of AmMhc1, NavMhc1, and BmMhc1, and the intron following exon 10 of PdcMhc1 that have a consensus GT site, exon 10 of TicMhc1 has a GC 5' donor site. The intron following exon 11a of DapMhc1 starts with a consensus GT site, while the intron following exon 11b starts with the absolutely rare GA dinucleotide. Also, all split codons are shared between the genes.
Figure 5.
Diagram of the arthropod Mhc1 proteins. The exon-intron structure of the Mhc1 genes is shown based on the protein sequence. Exons are shown as boxes while introns are represented by spaces. The same colour scheme has been used as in Figure 1. Numbers on alternative exons denote the number of variants. The exons are drawn that the intron positions align between the different Mhc1 genes. Thus, the exon lengths are not drawn to scale (e.g. the exons encoding the variable loop-2 are different in lengths). On the right side, the protein sequence of Drosophila melanogaster Mhc1 is shown as reference. Dotted lines connect amino acids that are derived from split codons.
In the part encoding the motor and the neck domain, all intron positions are shared by at least two genes (Figure 5). In the coiled-coil tail domain, all genes have lost several introns so that the exons are considerably longer and the intron positions in many cases are not identical. Assuming, that introns have in most cases been lost and were not gained during evolution [33], an ancient arthropod Mhc1 gene can be reconstructed (Figure 5). The ancient Mhc1 gene is expected to contain all intron positions that appear in at least one of the analysed Mhc1 genes. In the motor domain, the proposed ancient Mhc1 gene structure completely resembles the DapMhc1 gene. The exon lengths are between 30 and 210 bp. The exons in the tail domain are considerably longer (up to 480 bp).
Structural implications of the alternatively spliced exons
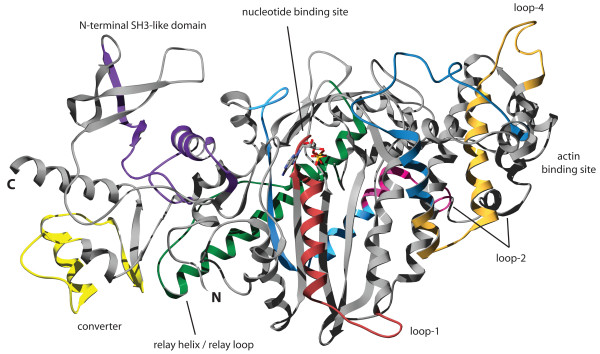
The locations of the alternatively spliced exons of DmMhc1 in the motor domain have been discussed in detail elsewhere [34]. The position of the additional alternatively spliced exons of the BmMhc1, TicMhc1, PdcMhc1, and DapMhc1 genes in the structure of the motor domain are shown in Figure 6. The alternative exons of DapMhc1 encoding the structural part from the P-loop to loop-1 have identical P-loop sequences. The loop-1 sequences are identical in length but differ significantly in composition. Studies have shown that the flexibility of this loop affects the rate of ADP and phosphate release, with greater flexibility leading to an enhancement in the rate of product release [35]. Although the amino acid composition is different between the alternative variants, both contain two glycines and a similar overall charge. The alternative exons of DapMhc1 including loop-4 are similar in length and composition. This region of the motor domain has not been investigated so far and therefore functional consequences of differences in the two variants cannot be drawn. Loop-4 has been postulated to be important for the proper localization of class-I myosins that contain elongated loops that sterically interact with actin-binding proteins [36] but the loop-4 sequences are almost identical between the two DapMhc1 variants and the two variants must therefore modulate a different property of the motor domain. The loop-2 sequence is modulated by alternative exons in the BmMhc1, DapMhc1, PdcMhc1, and TicMhc1 genes. By studies of the Dictyostelium class-2 myosin with its loop-2 replaced with the analogous loop from four other myosins with different enzymatic activities, loop-2 was shown to be involved in the weak and the strong binding interactions with actin [37]. It also plays an important role in the rate-limiting step of Pi release [38,39]. The exon variants of the BmMhc1, DapMhc1, PdcMhc1, and TicMhc1 genes encoding the loop-2 sequence have identical numbers of lysine and arginine residues. The "a" variants are always one residue shorter and have only four instead of five glycines. These differences are, however, very subtle and their influence on actin binding is expected to be very small. The variants of the alternative exon in DapMhc1 following loop-2 are very similar. This part of the motor domain has also not been investigated so far.
Figure 6.
Structure of the myosin motor domain. The structure of the motor domain of the class-II myosin of Dictyostelium discoideum has been used to highlight the regions encoded by alternatively spliced exons in arthropod Mhc1 genes. The color-coding is the same as in Figure 1 allowing the identification of corresponding regions.
Discussion
25 muscle myosin heavy chain genes have been identified in 22 species of the Arthropoda. All sequences share strong homology to the alternatively spliced Mhc1 gene that was first described in Drosophila melanogaster [9]. The genes contain five to nine clusters of mutually exclusive exons and an penultimate exon that might either be included or excluded in the mRNA, and were assigned by manual inspection of the genomic DNA sequences (Figure 1). Because of the many clusters of alternatively spliced exons automatic identification of all exons failed. This is probably also the main reason for the wrong prediction of the exon organisation of the Anopheles Mhc1 gene (supplementary material of [20]).
Altogether, alternative splicing of Mhc1 transcripts could result in several hundred differently spliced mRNAs (Table 1). The Pediculus Mhc1 gene has the least alternatives for its alternatively spliced exons resulting in a theoretical maximum of 32 different mRNAs, while the water flea gene could result in at least 3072 different mRNAs. Thus, except for Pediculus, Nasonia, and Apis mellifera all arthropod Mhc1 genes, for which all exons could be identified, outscore the 480 mRNA possibilities of Drosophila melanogaster. Although the number of possible transcripts seems vast compared to the number of different muscle myosin heavy chain genes in other metazoa species, the regions to modulate the function of the protein are limited to five to nine. In Drosophila melanogaster, all alternative exons are expressed depending on the developmental stage, but only a limited number of combinations seem to be employed [18]. Whether all alternative exons are expressed in the other arthropod species and which combinations are used has yet to be determined.
The phylogenetic analysis of the Mhc1 protein sequences agrees with the expected phylogenetic relationship between the species. There are two notable exceptions in the Drosophila species section of the tree. The DseMhc1 sequence would have been expected to be the closest relative of the DssMhc1 sequence, and the DaMhc1 sequence would have been expected to separate after the split of the DpMhc1 and DrpMhc1 sequences. There are two possible ways to explain this observation. Either, the Mhc1 genes have evolved asynchronously as has been found for many yeast genes [40] or the genes might have incorporated back-mutations. The sequence identities of 96.1 to 99.7% are very high, and thus only a few mutations would lead to a different phylogenetic classification.
The Tribolium castaneum, Pediculus humanus corporis, and Bombyx mori Mhc1 genes contain one additional and the Daphnia pulex Mhc1 gene contains four additional clusters of alternatively spliced exons compared to the Drosophila melanogaster gene (Figure 1, Figure 2). All additional alternatively spliced exons are mutually exclusive and encode parts of the motor domain. The additional exons of the Tic, Pdc, and Bm Mhc1 genes encode alternative versions of the loop-2 sequence while the additional exons of the Dap Mhc1 gene are spread over the entire motor domain. In each case, the 3' variant is more homologous to the corresponding sequences in the other Mhc1 genes than the 5' variant (Figure 2).
A similar conservation is found for alternative exons with multiple variants (Figure 2). In almost all cases, the most 3' variant is the most conserved one. Of the cluster of alternative exons encoding part of the motor domain near the ATP-binding site (exon 7 in DmMhc1), the last of the variants is the only variant that is conserved in all species. The other variants are either missing in certain species, or are very similar to each other as well as to those of other species, so that it is not clear whether they have been derived from independent variant duplications or whether they were present in a common ancestor. Thus, all variants except for the most 3' variant evolved after the separation of Daphnia from the other species. The variants encoding the relay-helix and the relay-loop are highly conserved. Conserved differences confine to only one or two residues. The penultimate of the variants seems to be the most conserved, although mutation of one residue might change this. The exon encoding part of the converter domain has two highly conserved variants, the most 5' and the most 3' variants. The most 3' variant is distinguished from all other variants of this set of alternative exons at the DNA level because the following intron starts with a GT donor site. The most 5' exon is the most important, though not the only, determinant for flight capabilities [41,42].
Based on the exon-intron patterns of the 21 Mhc1 genes the gene structure of the ancient arthropod Mhc1 gene can be predicted. The prediction is based on the assumption that it is very unlikely that the different species, distributed over a broad taxonomic range, invented introns at the same positions independently from each other. In the first half of the genes encoding the motor and the neck domain, all intron positions are shared by at least two genes (Figure 5). The exons encoding the coiled-coil tail domain starting at amino acid 850 are considerably longer and the intron positions in almost all genes are not identical. It is highly probable that further sequencing of arthropod Mhc1 genes will reveal different exon-intron patterns in the tail region while intron positions with one or more of the already analysed genes will be shared. Comparing the intron rich DapMhc1 and PdcMhc1 genes with the mosquito and Drosophila Mhc1 genes, it is apparent that intron loss is a major determinant of arthropod Mhc1 gene evolution. Loss of intron events have also been found for many other arthropod genes [33]. However, as long as data from further arthropod species is missing, it cannot be excluded that some of the introns in the tail region, that are not shared between the analyzed arthropods, have been gained during evolution. Very recently, an analysis of eleven Drosophila genomes showed, that a small number of introns have been gained in these species [43]. The ancient Mhc1 gene is expected to contain all intron positions that appear in at least one of the analysed Mhc1 genes. Analysis of Mhc1 genes of further species might add additional intron positions especially in the tail region. The exon lengths of the ancient Mhc1 gene are between 30 and 210 bp in the motor domain and up to 480 bp in the tail region. These short exons (compared to e.g. the Drosophila Mhc1 gene) resemble exon lengths in vertebrates and further comparative analysis with vertebrate muscle myosin heavy chain genes will reveal the gene structure of the ancient Metazoa gene.
In addition to the Mhc1 gene, Aedes aegypti encodes a further muscle myosin heavy chain gene, named Mhc3 that encodes only one variant of each of the alternatively spliced exons of the Mhc1 gene. The presence of this gene is not an artefact from sequencing or the assembly process. Both genes, Mhc1 and Mhc3, are very different at the DNA level, and both are confirmed by several EST clones, although the translated exons show high identities. That also means, that the Mhc3 gene, that does not encode any alternatively spliced exons, is expressed during the life cycle of Aedes aegypti. However, there is not enough data that shows that the Mhc3 gene is expressed in a biological important (e.g. muscle-specific) manner. Note that the combination of alternatively spliced exons does not correspond to any of the tissue-specific combinations found in Drosophila [18]. The Culex pipiens quinquefasciatus genome contains another two muscle myosin heavy chain genes in addition to the Mhc1 gene, named Mhc3 and Mhc4, that, similarly to AeaMhc3, encode only one variant of most of the alternatively spliced exons of the Mhc1 gene. In one case, the intron between the presumed variant of the alternatively spliced exons and the following constitutive exon disappeared. Unfortunately, there is not enough EST data available for Culex pipiens quinquefasciatus to support any of the myosin heavy chain genes. AeaMhc3, CpqMhc3, and CpqMhc4 retained the same variants of the alternative exons of the corresponding Mhc1 genes. The presence of these further muscle myosin heavy chain genes is very surprising because the number of alternatively spliced exons in the Mhc1 genes already allows for the transcription of several hundred different muscle myosin isoforms. How could it happen that the genomes of Aedes aegypti and Culex pipiens quinquefasciatus encode such genes? According to the phylogenetic tree of the myosin heavy chain genes, the Mhc3 and Mhc4 genes obviously appeared in the common ancester of Aedes and Culex after the divergence from Anopheles gambiae. In addition, there is no evidence for a (partial) second muscle myosin heavy chain gene in the Anopheles gambiae genome. Also, the carboxy-terminal ends of AeaMhc3 and CpqMhc4, that are 3' elongations of the last constitutive exon, do not exist in the AeaMhc1 and CpqMhc1 genes but have an identical counterpart in the AngMhc1 gene that is also supported by several EST clones. It is unlikely that these three organisms have developed such a carboxy-terminal end of the myosin gene independently from each other. Instead, it is more probable that the ancient AeaMhc1 and CpqMhc1 genes have lost this specific carboxy-terminus after incorporation of the Mhc3 and Mhc4 genes into the genome. This would mean that this carboxy-terminus is only used in the specific combination of alternatively spliced exons as found in the AeaMhc3 and CpqMhc4 genes. Whether this is also true for the AngMhc1 gene has to be verified. Based on their identity in sequence and gene structure it is most probable that CpqMhc3 has been derived by gene duplication of CpqMhc4 or CpqMhc4 is a duplication of CpqMhc3.
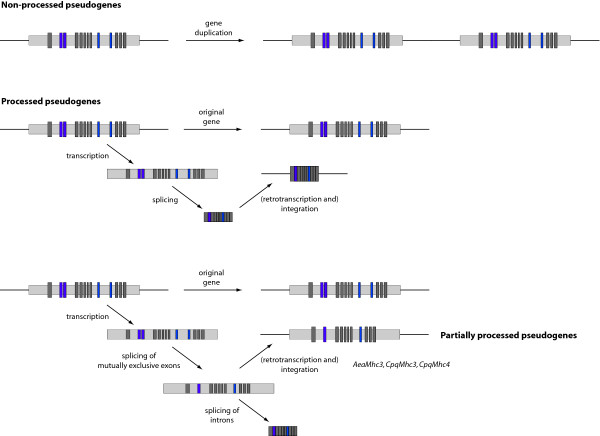
There are two possibilities as to how the Mhc3 and Mhc4 genes could have appeared in the common ancestor of Aedes and Culex. The genes have either been derived from a duplication of the Mhc1 gene as part of a single gene or chromosomal region duplication event. Or, a partially spliced transcript of Mhc1 has been reincorporated into the genome (Figure 7). If the Mhc3 and Mhc4 genes had been derived from duplication, then all variants except one of the alternative exons of only one of the (then) two Mhc genes had to be lost in addition to the loss of both terminal exons in Mhc3. Given the number of possible transcripts of the Mhc1 gene and the possibility to duplicate alternative exons, it is very unlikely that there would be a need for a second gene with the same set of alternative exons. If it were advantageous to keep two almost identical genes, it would be very unlikely that only one of the genes has lost all except one of its alternative exons. In addition, there must have been a very strong evolutionary pressure to keep exactly this special combination of alternative exons. The second possibility would mean that in the first step during the splicing process all alternatively spliced exons, which are not needed, are removed leaving introns between the remaining alternatively spliced and constitutive exons (Figure 7). In the second step, all introns are spliced to yield the mRNA for translation. In the case of the Mhc3 and Mhc4 genes, the transcript containing one combination of alternative exons but all introns would have been integrated into the genome, probably after retrotranscription. How should this type of genes be called? At least the AeaMhc3 gene is completely transcribed, and also CpqMhc3 and CpqMhc4 do not contain any premature stop codons or frameshift mutations. However, compared to the corresponding Mhc1 genes they retained only one variant exon of each of the alternative exons. Thus, they do not belong to the non-processed pseudogenes. We would rather regard them as a new type of partially processed pseudogenes.
Figure 7.
Model for the process of alternative splicing. The model describes the three different origins of pseudogenes. Non-processed pseudogenes are often found adjacent to their paralogous functional gene and retain the same exon-intron structure. Processed pseudogenes are marked by the absence of both 5' promotor sequence and introns, the presence of flanking direct repeats, and are randomly integrated into the genome. In the case of the arthropod Mhc genes, these get in the first step transcribed. In a second step, the alternative exons get spliced resulting in a certain combination of alternative exons and retaining the exon-intron structure. In the case of AeaMhc3, CpqMhc3, and CpqMhc4, these transcripts have been integrated into the genome. Normally in a third step, the introns get spliced revealing the final mRNA ready for translation. Dark grey bars represent constitutive and coloured bars alternatively spliced exons. Light grey bars represent non-coding sequence.
Conclusion
25 arthropod muscle myosin heavy chain genes have been identified and analysed. Compared to the well-studied gene of Drosophila melanogaster other arthropod genes might contain up to four additional alternatively spliced exons encoding part of the motor domain. This considerably extends the possibilities of other arthropod species to fine-tune myosin and thus muscle characteristics. An ancient arthropod muscle myosin heavy chain gene has been reconstructed whose gene structure can best be explained if introns are lost and not gained during evolution of this gene. Aedes aegypti and Culex pipiens quinquefasciatus even encode further muscle myosin heavy chain genes that, however, have lost all except one variant of the alternatively spliced exons. These genes most probably entered the genome by reincorporating a certain processed transcript and not via a gene or genomic region duplication event. If the gene has been derived from a processed transcript then splicing of alternative exons must involve a first step, in which all other variants are spliced out leaving intronic sequence around the variant of choice. In a second step, all introns are spliced.
Methods
Identification and annotation of the arthropod muscle myosin heavy chains
The genes for Aea, Ang, Am, Bm, Cpq, Dm, Drp, Dp, Dse, Dss, Dy, Dw, Pdc, and Tic Mhc1 and Mhc3 have been obtained by TBLASTN searches against the insects section of the NCBI wgs database (Table 1)[44]. The genes for the Da, Der, Dg, Dmo, and Dv Mhc1 have been obtained using the BLAT alignment tool [45] against the UCSC Genome Browser database [46,47]. The DhMhc1 sequence was derived from the NCBI nonredundant database. The DapMhc1 sequence has been obtained by a TBLASTN search against the 9× assembly of the Daphnia pulex genome provided by the DOE Joint Genome Institute [48] and the Daphnia Genomics Consortium [49]. The NavMhc1 gene was derived from version 1.0 of the Nasonia vitripennis assembly provided by the Human Genome Sequencing Center at Baylor College of Medicine [50]. The exons of the genes were predicted by manual inspection of the nucleotide sequences. For the correct prediction of the transcriptional start and the 3' terminal exons, the analysis of cDNA and EST data, that has been obtained from the EST section of NCBI's nucleotide database, was necessary. In particular, the following data has been obtained: For TicMhc1, only a small amount of EST data is available, confirming the prediction of exon2. There is not enough data to exclude a further untranslated 5' exon, as well as further C-terminal exons. For AngMhc1, several EST and cDNA clones support exon1 and the different C-termini. The C-termini of AeaMhc1 are also supported by several EST clones (e.g. GenBank ID DV384821). Exon1 of AeaMhc3 is supported by EST data. Exon1 of AeaMhc3 has been used for the identification of exon1 of AeaMhc1, as there is no direct evidence by EST data. Surprisingly, it is found 26,432 bp before the translation start codon ATG. For AmMhc1, the N-terminus is not supported by EST or cDNA data. Therefore it is not clear whether there might be an additional 5' untranslated exon. The C-termini are supported by several EST and cDNA clones (e.g. GenBank ID CK629939). The C-terminus of DapMhc1 is supported by EST data (e.g. GenBank ID BJ927473), while there is no EST data for the N-terminus. For BmMhc1, exon2 is supported by EST data. However, the corresponding EST clones are not long enough to exclude a further 5' untranslated exon. Both C-termini of BmMhc1 are supported by EST clones (e.g. GenBank ID BP179837). The genomic DNA of the BmMhc1 gene contains a gap in the coiled-coil tail region. The missing amino acid sequence has been derived from EST data. However, the exon/intron structure in the corresponding region remains unresolved.
Analysis of the relationship of the alternatively spliced exons
All alternatively spliced exons have been aligned manually. Some kind of relationship is already obvious from these sequence alignments. To get a more quantitative description, sequence identity matrices have been calculated for each set of aligned exons. Subsequently, sets of homologous exons from all Mhc1 genes have been clustered by sequence similarity. We have visualized the results in graphs that have to be read in columns. The highest identity between an exon listed on top and any variant of a certain Mhc1 protein listed on the left side has been set to 1 (red colour) while the differences between the values of the lower identity exons and the value of the highest identity have been plotted for the other combinations of exons. Thus, in every column the highest identity of the named exon to one of the variants of the other Mhc1 proteins is visualized.
Building trees
The phylogenetic tree was generated using neighbour joining and the Bootstrap (1,000 replicates) method as implemented in ClustalW (standard settings) [51] and drawn by using TreeView [52]. The sequence of DapMhc1 has been used as outgroup.
List of abbreviations
Mhc, myosin heavy chain; for abbreviations of species names see Table 1.
Authors' contributions
F.O. performed data analysis. M.K. assembled all sequences, performed data analysis and wrote the manuscript. Both authors read and approved the manuscript.
Supplementary Material
Mhc1 sequence alignment. The file contains the aligned arthropod Mhc1 protein sequences. Also included are all variants of the alternatively spliced exons.
Sequence alignment and analysis of the alternatively spliced exons. The file contains the aligned alternative exons of the arthropod Mhc1 protein sequences. Also included are the graphical representations of the sequence identities.
Acknowledgments
Acknowledgements
M.K. was supported by a Liebig Stipendium of the Fonds der Chemischen Industrie, which was in part financed by the BMBF. This work has been funded by grant I80798 of the VolkswagenStiftung and grant KO 2251/3-1 of the Deutsche Forschungsgemeinschaft. We thank the DOE Joint Genome Institute [48] and the Daphnia Genomics Consortium [49] for providing access to the assembly of the Daphnia pulex genome, and the Human Genome Sequencing Center at Baylor College of Medicine for providing access to the assembly of the Nasonia vitripennis genome preliminary to publication. Also, we would like to thank all the reviewers for their very helpful and thoughtful comments that improved the manuscript considerably.
Contributor Information
Florian Odronitz, Email: flod@nmr.mpibpc.mpg.de.
Martin Kollmar, Email: mako@nmr.mpibpc.mpg.de.
References
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/S0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell. 2000;103:367–370. doi: 10.1016/S0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- Thanaraj TA, Stamm S, Clark F, Riethoven JJ, Le Texier V, Muilu J. ASD: the Alternative Splicing Database. Nucleic Acids Res. 2004;32:D64–9. doi: 10.1093/nar/gkh030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Koonin EV. Origin of alternative splicing by tandem exon duplication. Hum Mol Genet. 2001;10:2661–2669. doi: 10.1093/hmg/10.23.2661. [DOI] [PubMed] [Google Scholar]
- Anastassiou D, Liu H, Varadan V. Variable window binding for mutually exclusive alternative splicing. Genome Biol. 2006;7:R2. doi: 10.1186/gb-2006-7-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123:65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Bork P. Common exon duplication in animals and its role in alternative splicing. Hum Mol Genet. 2002;11:1561–1567. doi: 10.1093/hmg/11.13.1561. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Kaur A, Gunning D, Zipursky SL, Rowen L, Clemens JC. The organization and evolution of the dipteran and hymenopteran Down syndrome cell adhesion molecule (Dscam) genes. Rna. 2004;10:1499–1506. doi: 10.1261/rna.7105504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George EL, Ober MB, Emerson CP., Jr. Functional domains of the Drosophila melanogaster muscle myosin heavy-chain gene are encoded by alternatively spliced exons. Mol Cell Biol. 1989;9:2957–2974. doi: 10.1128/mcb.9.7.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/S0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Schliwa M, Woehlke G. Molecular motors. Nature. 2003;422:759–765. doi: 10.1038/nature01601. [DOI] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol Biol Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TN, Berg JS, Cheney RE. Tails of unconventional myosins. Cell Mol Life Sci. 1999;56:243–257. doi: 10.1007/s000180050426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC. Introduction. Philos Trans R Soc Lond B Biol Sci. 2004;359:1813–1818. doi: 10.1098/rstb.2004.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita RA, Sellers JR, Anderson JB. Identification and analysis of the myosin superfamily in Drosophila: a database approach. J Muscle Res Cell Motil. 2000;21:491–505. doi: 10.1023/A:1026589626422. [DOI] [PubMed] [Google Scholar]
- Chiba S, Awazu S, Itoh M, Chin-Bow ST, Satoh N, Satou Y, Hastings KE. A genomewide survey of developmentally relevant genes in Ciona intestinalis. IX. Genes for muscle structural proteins. Dev Genes Evol. 2003;213:291–302. doi: 10.1007/s00427-003-0324-x. [DOI] [PubMed] [Google Scholar]
- Zhang S, Bernstein SI. Spatially and temporally regulated expression of myosin heavy chain alternative exons during Drosophila embryogenesis. Mech Dev. 2001;101:35–45. doi: 10.1016/S0925-4773(00)00549-9. [DOI] [PubMed] [Google Scholar]
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Siden-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, WoodageT. Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai Z, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chaturverdi K, Christophides GK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu Z, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke Z, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin JJ, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O'Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, Shao H, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun J, Thomasova D, Ton LQ, Topalis P, Tu Z, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang X, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang H, Zhao Q, Zhao S, Zhu SC, Zhimulev I, Coluzzi M, della Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork P, Brey PT, Venter JC, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O'Leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita K, Kasahara M, Sasaki S, Nagayasu Y, Yamada T, Kanamori H, Namiki N, Kitagawa M, Yamashita H, Yasukochi Y, Kadono-Okuda K, Yamamoto K, Ajimura M, Ravikumar G, Shimomura M, Nagamura Y, Shin IT, Abe H, Shimada T, Morishita S, Sasaki T. The genome sequence of silkworm, Bombyx mori. DNA Res. 2004;11:27–35. doi: 10.1093/dnares/11.1.27. [DOI] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, Pollard DA, Sackton TB, Larracuente AM, Singh ND, Abad JP, Abt DN, Adryan B, Aguade M, Akashi H, Anderson WW, Aquadro CF, Ardell DH, Arguello R, Artieri CG, Barbash DA, Barker D, Barsanti P, Batterham P, Batzoglou S, Begun D, Bhutkar A, Blanco E, Bosak SA, Bradley RK, Brand AD, Brent MR, Brooks AN, Brown RH, Butlin RK, Caggese C, Calvi BR, Bernardo de Carvalho A, Caspi A, Castrezana S, Celniker SE, Chang JL, Chapple C, Chatterji S, Chinwalla A, Civetta A, Clifton SW, Comeron JM, Costello JC, Coyne JA, Daub J, David RG, Delcher AL, Delehaunty K, Do CB, Ebling H, Edwards K, Eickbush T, Evans JD, Filipski A, Findeiss S, Freyhult E, Fulton L, Fulton R, Garcia AC, Gardiner A, Garfield DA, Garvin BE, Gibson G, Gilbert D, Gnerre S, Godfrey J, Good R, Gotea V, Gravely B, Greenberg AJ, Griffiths-Jones S, Gross S, Guigo R, Gustafson EA, Haerty W, Hahn MW, Halligan DL, Halpern AL, Halter GM, Han MV, Heger A, Hillier L, Hinrichs AS, Holmes I, Hoskins RA, Hubisz MJ, Hultmark D, Huntley MA, Jaffe DB, Jagadeeshan S, Jeck WR, Johnson J, Jones CD, Jordan WC, Karpen GH, Kataoka E, Keightley PD, Kheradpour P, Kirkness EF, Koerich LB, Kristiansen K, Kudrna D, Kulathinal RJ, Kumar S, Kwok R, Lander E, Langley CH, Lapoint R, Lazzaro BP, Lee SJ, Levesque L, Li R, Lin CF, Lin MF, Lindblad-Toh K, Llopart A, Long M, Low L, Lozovsky E, Lu J, Luo M, Machado CA, Makalowski W, Marzo M, Matsuda M, Matzkin L, McAllister B, McBride CS, McKernan B, McKernan K, Mendez-Lago M, Minx P, Mollenhauer MU, Montooth K, Mount SM, Mu X, Myers E, Negre B, Newfeld S, Nielsen R, Noor MA, O'Grady P, Pachter L, Papaceit M, Parisi MJ, Parisi M, Parts L, Pedersen JS, Pesole G, Phillippy AM, Ponting CP, Pop M, Porcelli D, Powell JR, Prohaska S, Pruitt K, Puig M, Quesneville H, Ram KR, Rand D, Rasmussen MD, Reed LK, Reenan R, Reily A, Remington KA, Rieger TT, Ritchie MG, Robin C, Rogers YH, Rohde C, Rozas J, Rubenfield MJ, Ruiz A, Russo S, Salzberg SL, Sanchez-Gracia A, Saranga DJ, Sato H, Schaeffer SW, Schatz MC, Schlenke T, Schwartz R, Segarra C, Singh RS, Sirot L, Sirota M, Sisneros NB, Smith CD, Smith TF, Spieth J, Stage DE, Stark A, Stephan W, Strausberg RL, Strempel S, Sturgill D, Sutton G, Sutton GG, Tao W, Teichmann S, Tobari YN, Tomimura Y, Tsolas JM, Valente VL, Venter E, Venter JC, Vicario S, Vieira FG, Vilella AJ, Villasante A, Walenz B, Wang J, Wasserman M, Watts T, Wilson D, Wilson RK, Wing RA, Wolfner MF, Wong A, Wong GK, Wu CI, Wu G, Yamamoto D, Yang HP, Yang SP, Yorke JA, Yoshida K, Zdobnov E, Zhang P, Zhang Y, Zimin AV, Baldwin J, Abdouelleil A, Abdulkadir J, Abebe A, Abera B, Abreu J, Acer SC, Aftuck L, Alexander A, An P, Anderson E, Anderson S, Arachi H, Azer M, Bachantsang P, Barry A, Bayul T, Berlin A, Bessette D, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Bourzgui I, Brown A, Cahill P, Channer S, Cheshatsang Y, Chuda L, Citroen M, Collymore A, Cooke P, Costello M, D'Aco K, Daza R, De Haan G, DeGray S, DeMaso C, Dhargay N, Dooley K, Dooley E, Doricent M, Dorje P, Dorjee K, Dupes A, Elong R, Falk J, Farina A, Faro S, Ferguson D, Fisher S, Foley CD, Franke A, Friedrich D, Gadbois L, Gearin G, Gearin CR, Giannoukos G, Goode T, Graham J, Grandbois E, Grewal S, Gyaltsen K, Hafez N, Hagos B, Hall J, Henson C, Hollinger A, Honan T, Huard MD, Hughes L, Hurhula B, Husby ME, Kamat A, Kanga B, Kashin S, Khazanovich D, Kisner P, Lance K, Lara M, Lee W, Lennon N, Letendre F, LeVine R, Lipovsky A, Liu X, Liu J, Liu S, Lokyitsang T, Lokyitsang Y, Lubonja R, Lui A, MacDonald P, Magnisalis V, Maru K, Matthews C, McCusker W, McDonough S, Mehta T, Meldrim J, Meneus L, Mihai O, Mihalev A, Mihova T, Mittelman R, Mlenga V, Montmayeur A, Mulrain L, Navidi A, Naylor J, Negash T, Nguyen T, Nguyen N, Nicol R, Norbu C, Norbu N, Novod N, O'Neill B, Osman S, Markiewicz E, Oyono OL, Patti C, Phunkhang P, Pierre F, Priest M, Raghuraman S, Rege F, Reyes R, Rise C, Rogov P, Ross K, Ryan E, Settipalli S, Shea T, Sherpa N, Shi L, Shih D, Sparrow T, Spaulding J, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Strader C, Tesfaye S, Thomson T, Thoulutsang Y, Thoulutsang D, Topham K, Topping I, Tsamla T, Vassiliev H, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Young G, Yu Q, Zembek L, Zhong D, Zimmer A, Zwirko Z, Jaffe DB, Alvarez P, Brockman W, Butler J, Chin C, Gnerre S, Grabherr M, Kleber M, Mauceli E, MacCallum I. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, Thornton K, Hubisz MJ, Chen R, Meisel RP, Couronne O, Hua S, Smith MA, Zhang P, Liu J, Bussemaker HJ, van Batenburg MF, Howells SL, Scherer SE, Sodergren E, Matthews BB, Crosby MA, Schroeder AJ, Ortiz-Barrientos D, Rives CM, Metzker ML, Muzny DM, Scott G, Steffen D, Wheeler DA, Worley KC, Havlak P, Durbin KJ, Egan A, Gill R, Hume J, Morgan MB, Miner G, Hamilton C, Huang Y, Waldron L, Verduzco D, Clerc-Blankenburg KP, Dubchak I, Noor MA, Anderson W, White KP, Clark AG, Schaeffer SW, Gelbart W, Weinstock GM, Gibbs RA. Comparative genome sequencing of Drosophila pseudoobscura: chromosomal, gene, and cis-element evolution. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Errico I, Gadaleta G, Saccone C. Pseudogenes in metazoa: origin and features. Brief Funct Genomic Proteomic. 2004;3:157–167. doi: 10.1093/bfgp/3.2.157. [DOI] [PubMed] [Google Scholar]
- Balakirev ES, Ayala FJ. Pseudogenes: are they "junk" or functional DNA? Annu Rev Genet. 2003;37:123–151. doi: 10.1146/annurev.genet.37.040103.103949. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. Pseudogenes. Nature. 1980;286:840–841. doi: 10.1038/286840a0. [DOI] [PubMed] [Google Scholar]
- Dhawan P, Yang E, Kumar A, Mehta KD. Genetic complexity of the human geranylgeranyltransferase I beta-subunit gene: a multigene family of pseudogenes derived from mis-spliced transcripts. Gene. 1998;210:9–15. doi: 10.1016/S0378-1119(98)00042-0. [DOI] [PubMed] [Google Scholar]
- Suzuki E, Lowry J, Sonoda G, Testa JR, Walsh K. Structures and chromosome locations of the human MEF2A gene and a pseudogene MEF2AP. Cytogenet Cell Genet. 1996;73:244–249. doi: 10.1159/000134348. [DOI] [PubMed] [Google Scholar]
- Breathnach R, Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Rozek CE, Davidson N. Differential processing of RNA transcribed from the single-copy Drosophila myosin heavy chain gene produces four mRNAs that encode two polypeptides. Proc Natl Acad Sci U S A. 1986;83:2128–2132. doi: 10.1073/pnas.83.7.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein SI, Hansen CJ, Becker KD, Wassenberg DR, 2nd, Roche ES, Donady JJ, Emerson CP., Jr. Alternative RNA splicing generates transcripts encoding a thorax-specific isoform of Drosophila melanogaster myosin heavy chain. Mol Cell Biol. 1986;6:2511–2519. doi: 10.1128/mcb.6.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible F, Tessmar-Raible K, Osoegawa K, Wincker P, Jubin C, Balavoine G, Ferrier D, Benes V, de Jong P, Weissenbach J, Bork P, Arendt D. Vertebrate-type intron-rich genes in the marine annelid Platynereis dumerilii. Science. 2005;310:1325–1326. doi: 10.1126/science.1119089. [DOI] [PubMed] [Google Scholar]
- Bernstein SI, Milligan RA. Fine tuning a molecular motor: the location of alternative domains in the Drosophila myosin head. J Mol Biol. 1997;271:1–6. doi: 10.1006/jmbi.1997.1160. [DOI] [PubMed] [Google Scholar]
- Sweeney HL, Rosenfeld SS, Brown F, Faust L, Smith J, Xing J, Stein LA, Sellers JR. Kinetic tuning of myosin via a flexible loop adjacent to the nucleotide binding pocket. J Biol Chem. 1998;273:6262–6270. doi: 10.1074/jbc.273.11.6262. [DOI] [PubMed] [Google Scholar]
- Kollmar M, Durrwang U, Kliche W, Manstein DJ, Kull FJ. Crystal structure of the motor domain of a class-I myosin. Embo J. 2002;21:2517–2525. doi: 10.1093/emboj/21.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda TQ, Ruppel KM, Spudich JA. Enzymatic activities correlate with chimaeric substitutions at the actin-binding face of myosin. Nature. 1994;368:567–569. doi: 10.1038/368567a0. [DOI] [PubMed] [Google Scholar]
- Joel PB, Trybus KM, Sweeney HL. Two conserved lysines at the 50/20-kDa junction of myosin are necessary for triggering actin activation. J Biol Chem. 2001;276:2998–3003. doi: 10.1074/jbc.M006930200. [DOI] [PubMed] [Google Scholar]
- Furch M, Geeves MA, Manstein DJ. Modulation of actin affinity and actomyosin adenosine triphosphatase by charge changes in the myosin motor domain. Biochemistry. 1998;37:6317–6326. doi: 10.1021/bi972851y. [DOI] [PubMed] [Google Scholar]
- Langkjaer RB, Cliften PF, Johnston M, Piskur J. Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature. 2003;421:848–852. doi: 10.1038/nature01419. [DOI] [PubMed] [Google Scholar]
- Littlefield KP, Swank DM, Sanchez BM, Knowles AF, Warshaw DM, Bernstein SI. The converter domain modulates kinetic properties of Drosophila myosin. Am J Physiol Cell Physiol. 2003;284:C1031–8. doi: 10.1152/ajpcell.00474.2002. [DOI] [PubMed] [Google Scholar]
- Swank DM, Knowles AF, Suggs JA, Sarsoza F, Lee A, Maughan DW, Bernstein SI. The myosin converter domain modulates muscle performance. Nat Cell Biol. 2002;4:312–316. doi: 10.1038/ncb776. [DOI] [PubMed] [Google Scholar]
- Coulombe-Huntington J, Majewski J. Intron Loss and Gain in Drosophila. Mol Biol Evol. 2007 doi: 10.1093/molbev/msm235. [DOI] [PubMed] [Google Scholar]
- NCBI BLAST with arthropoda genomes http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=insects
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. 10.1101/gr.229202. Article published online before March 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, Hillman-Jackson J, Kuhn RM, Pedersen JS, Pohl A, Raney BJ, Rosenbloom KR, Siepel A, Smith KE, Sugnet CW, Sultan-Qurraie A, Thomas DJ, Trumbower H, Weber RJ, Weirauch M, Zweig AS, Haussler D, Kent WJ. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–8. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UCSC Genome Bioinformatics http://genome.cse.ucsc.edu/
- DOE Joint Genome Institute http://www.jgi.doe.gov/
- Daphnia Genomics Consortium http://daphnia.cgb.indiana.edu/wfleabase/
- Human Genome Sequencing Center at Baylor College of Medicine http://www.hgsc.bcm.tmc.edu/projects/nasonia/ [DOI] [PubMed]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mhc1 sequence alignment. The file contains the aligned arthropod Mhc1 protein sequences. Also included are all variants of the alternatively spliced exons.
Sequence alignment and analysis of the alternatively spliced exons. The file contains the aligned alternative exons of the arthropod Mhc1 protein sequences. Also included are the graphical representations of the sequence identities.