Abstract
Background
Quantitative reaction norm theory proposes that genotype-by-environment interaction (GxE) results from inter-individual differences of expression in adaptive suites of genes in distinct environments. However, environmental norms for actual gene suites are poorly documented. In this study, we investigated the effects of GxE interactions on levels of gene transcription and growth by documenting the impact of rearing environment (freshwater vs. saltwater), sex and genotypic (low vs. high estimated breeding value EBV) effects on the transcription level of insulin-like growth factor (IGF-1) and growth hormone receptor (GHR) in brook charr (Salvelinus fontinalis).
Results
Males grew faster than females (μ♀ = 1.20 ± 0.07 g·d-1, μ♂ = 1.46 ± 0.06 g·d-1) and high-EBV fish faster than low-EBV fish (μLOW = 0.97 ± 0.05 g·d-1, μHIGH = 1.58 ± 0.07 g·d-1; p < 0.05). However, growth was markedly lower in saltwater-reared fish than freshwater sibs (μFW = 1.52 ± 0.07 g·d-1, μSW = 1.15 ± 0.06 g·d-1), yet GHR mRNA transcription level was significantly higher in saltwater than in freshwater (μSW = 0.85 ± 0.05, μFW = 0.61 ± 0.05). The ratio of actual growth to units in assayed mRNA ('individual transcript efficiency', iTE; g·d-1·u-1) also differed among EBV groups (μLOW = 2.0 ± 0.24 g·d-1·u-1; μHIGH = 3.7 ± 0.24 g·d-1·u-1) and environments (μSW = 2.0 ± 0.25 g·d-1·u-1; μFW = 3.7 ± 0.25 g·d-1·u-1) for GHR. Males had a lower iTE for GHR than females (μ♂ = 2.4 ± 0.29 g·d-1·u-1; μ♀ = 3.1 ± 0.23 g·d-1·u-1). There was no difference in IGF-1 transcription level between environments (p > 0.7) or EBV groups (p > 0.15) but the level of IGF-1 was four times higher in males than females (μ♂ = 2.4 ± 0.11, μ♀ = 0.58 ± 0.09; p < 0.0001). We detected significant sexual differences in iTE (μ♂ = 1.3 ± 0.59 g·d-1·u-1; μ♀ = 3.9 ± 0.47 g·d-1·u-1), salinities (μSW = 2.3 ± 0.52 g·d-1·u-1; μFW = 3.7 ± 0.53 g·d-1·u-1) and EBV-groups (μLOW = 2.4 ± 0.49 g·d-1·u-1; μHIGH = 3.8 ± 0.49 g·d-1·u-1). Interaction between EBV-group and environment was detected for both GHR (p = 0.027) and IGF-1 (p = 0.019), and for iTE in the two genes (p < 0.0001; p < 0.05, respectively), where increased divergence in levels of GHR and IGF-1 transcription occurred among EBV-groups in the saltwater environment.
Conclusion
Our results show that both environment and sex have major impacts on the expression of mRNA for two key genes involved in the physiological pathway for growth. We also demonstrate for the first time, at least in fish, genotype-by-environment interaction at the level of individual gene transcription. This work contributes significantly to ongoing efforts towards documenting environmentally and sexually induced variance of gene activity and understanding the resulting phenotypes.
Background
An increasing number of studies has documented differential expression of genes induced by environmental change (gene expression plasticity) [1-4]. However, only a handful of them have applied a quantitative genetic framework to document how this phenomenon differs between distinct genotypes, that is genotype-by-environment interaction (GxE) [5,6]. As a fundamental quantitative or qualitative change in gene activity, the physiological genetic basis of GxE interaction to environmental change should be discernable at the molecular level among animals with similar genetic background exposed to different environments [7]. Precise quantification of intracellular processes at the level of individual genes could provide a new insight into events occurring between the gene and the appearance of the trait. Such associations should be particularly effective for genes in physiological pathways known to affect particular phenotypes.
Brook charr (Salvelinus fontinalis; Osteichthyes: Salmonidae) life history ranges in habitat from permanent residency in streams or lakes to anadromous incursions [8], where individuals make summer migrations into intermediate-salinity estuaries and coastal marine waters [9,10]. Despite potential tradeoffs with survival [11], annual saltwater migration is considered advantageous because it provides access to superior food resources compared to the freshwater environment, translating into higher final weight-at-age (>4 kg) at the end of the saltwater period and greater longevity (8+ years) compared to freshwater residents (<1 kg and 3–4 years) [9,10]. There is evidence of genetic differentiation between the resident and anadromous life history types, both at the population level [12,13], and as distinct quantitative genetic units within the same population [14]. Exposure to the saline environment should, therefore, involve differentiation in the activity of genes in the growth pathway.
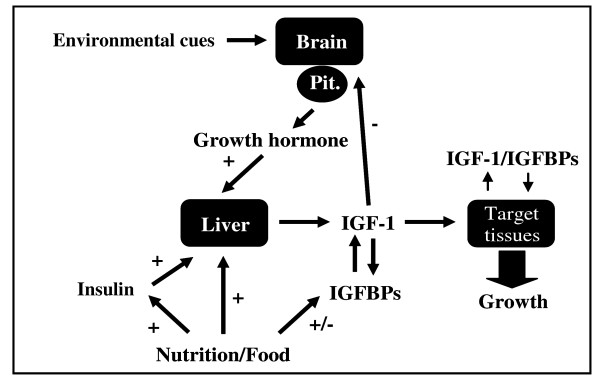
As in mammals, growth hormone (GH) and insulin-like growth factor (IGF) are two major molecular targets in the potential endocrine regulation of growth in teleost fishes [15-18]. GH is a pluripotent hormone produced by the pituitary gland in teleosts, and acts by binding to a single-transmembrane receptor, the GH receptor (GHR). Ligand binding induces receptor dimerization producing an active trimeric complex [19]. This active complex stimulates the transcription and production of insulin-like growth factor 1 (IGF-1), a hormone which plays a central role by mediating the growth-promoting actions of pituitary growth hormone [20] (Fig. 1). The direct versus indirect nature of GH action remains to be clarified, but GH appears to act both locally at the target tissue level to stimulate the autocrine/paracrine action of IGF-1, as well as on the liver to increase plasma IGF-1 levels [21]. Expression of both IGF-1 and growth hormone receptor (GHR) mRNA are detected in multiple fish tissues, but expression appears to be greatest in the liver [22]. The impact of environment on expression level of GH/IGF-1 pathway genes is well documented in fish. For instance, work in rainbow trout (Oncorhynchus mykiss) [23,24] showed that higher water temperature increases the level IGF-1 and GHR mRNA in liver tissues. Other work on rainbow trout [25] and coho salmon (O. kisutch) [26] also demonstrated that the feeding period is associated with increased IGF-1 mRNA expression in muscle and liver, respectively. In several fish species, tissue levels of IGF-1 mRNA positively correlate with body growth rate [19,20,27,28]. Similarly, work with coho salmon [20] and gilthead sea bream (Sparus aurata) [29] indicated that the level of hepatic GHR was related to growth performance. However, investment in somatic growth carries inherent metabolic costs and may be limited by energy investment towards maintenance, reproduction and activity, including differential osmoregulation costs between freshwater and saltwater environments [30]. Besides environment and genotypic effects, gene expression may also be influenced by sex. Indeed, differences between sexes in gene activity in the GH/IGF-1 axis have been documented [31-33] and associated with physiological systems (i.e. cortisol receptor, tilapia, [50]). Moreover, it has also been demonstrated that males and females may vary in their transcriptional response to different environmental conditions [34-36].
Figure 1.
GH/IGF-1 axis. Illustration of the endocrine axis controlling growth in teleost fish. Multiple hormonal and nutritional factors may stimulate (+) the production and/or modify (+/-) the activity of IGF-1. Negative feedback (-) by IGF-1 inhibits growth hormone secretion by the pituitary.
In this study, we used a quantitative genetic approach to investigate the effects of GxE interactions on levels of gene expression and growth by documenting the impact of rearing environment (freshwater vs. saltwater), sex and genotypic effects on the transcription level of insulin-like growth factor (IGF-1) and growth hormone receptor (GHR) in brook charr. We also predicted that i) differences in transcription level for IGF-1 and GHR should positively correlate with differences in growth, and ii) the production of IGF-1 and GHR should differ between sexes.
Results
Freshwater and saltwater growth
We observed marked differences in growth between progeny from the same families in the two salinity conditions (0‰ and 20‰) (Table 1, Figure 2a, see also Table 2 for details on each family). Freshwater-reared individuals grew almost 30% faster than their saltwater-reared full-sibs at the same feeding regime (μFW = 1.52 ± 0.0689 g·d-1; μSW = 1.15 ± 0.0572 g·d-1). Males had a significantly higher growth rate than females in both environments (μmale = 1.46 ± 0.0597 g·d-1, μfemale = 1.20 ± 0.0666 g·d-1). HIGH-EBV families also exhibited higher growth (μ = 1.58 ± 0.0663 g·d-1) relative to those of the LOW-EBV group (μ = 0.970 ± 0.0537 g·d-1). There was also evidence of GxE interactions in the form of variable growth responses between families belonging to different EBV groups in different environments, as detailed below (Table 1).
Table 1.
Results of the mixed-model maximum likelihood analysis of variance for factors explaining absolute growth (g·d-1).
| Absolute growth | |||
| Source of variation | d.f | F | P |
| Intercept | 143 | 718.239 | <.0001* |
| Environment | 1 | 22.552 | <.0001* |
| EBV group | 1 | 51.569 | 0.002* |
| Sex | 1 | 9.757 | 0.002* |
| EBV group × environment | 2 | 19.945 | <.0001* |
Asterisks indicate significant effects (α = 0.05).
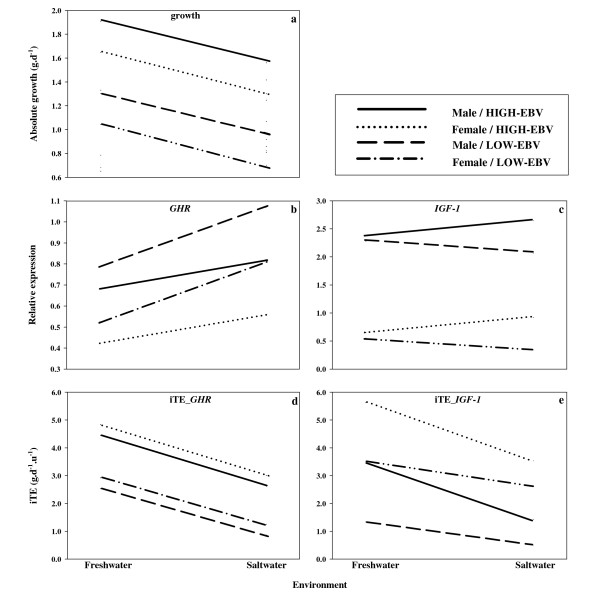
Figure 2.
Reaction norm for growth, gene expression and individual transcript efficiencies. Predicted linear means for a) absolute growth, b) relative expression of GHR, c) relative expression of IGF-1 and d, e) individual transcript efficiencies (iTE) for GHR and IGF-1 of males and females of HIGH- and LOW-EBV group reared in freshwater and saltwater (20 ppt) environments. Gene-by-environment interaction between EBVgroup-by-environment for growth and iTEGHR is not clearly illustrated by non-parallel reaction norms because the difference in EBV group within each environment was small, albeit statistically significant.
Table 2.
Absolute growth for male and female of HIGH- and LOW-EBV families reared in freshwater and saltwater environments.
| Absolute growth (g·d-1) | ||||
| Freshwater | Saltwater | |||
| Family | Female | Male | Female | Male |
| LOW-EBV-1 | 0.832 ± 0.127 | 1.518 ± 0.323 | 0.870 ± 0.187 | 0.843 ± 0.162 |
| LOW-EBV-13 | 1.130 ± 0.152 | 1.517 ± 0.187 | 0.806 ± 0.138 | 0.855 ± 0.264 |
| LOW-EBV-20 | 1.491 ± 0.457 | 1.291 ± 0.227 | 0.729 ± 0.162 | 0.961 ± 0.187 |
| HIGH-EBV-7 | 1.361 ± 0.205 | 1.784 ± 0.145 | 1.244 ± 0.173 | 1.291 ± 0.187 |
| HIGH-EBV-15 | 1.712 ± 0.132 | 2.325 ± 0.264 | 1.395 ± 0.145 | 1.901 ± 0.229 |
| HIGH-EBV-24 | 1.856 ± 0.152 | 1.096 ± 0.229 | 1.520 ± 0.264 | 1.750 ± 0.264 |
Values are LS means ± SE. LS means were estimated as linear predictors based on the most parsimonious model. Standard errors for each estimate were determined from a bootstrap distribution (1,000 iterations) of model predictions.
Sex, family and environmental effects on level of gene transcription
GHR level of gene transcription of individuals in the HIGH-EBV group was significantly less (μ = 0.620 ± 0.0819) than that observed in the LOW-EBV group (μ = 0.839 ± 0.0855) (Table 3, Figure 2b; see also Table 4). However, the level of GHR transcription was also significantly higher for fish reared in saltwater (μ = 0.848 ± 0.0460) than their full-sibs reared in freshwater (μ = 0.611 ± 0.0510; p < 0.05), despite the fact that growth in saltwater was slower. There were also marked effects of sex on gene transcription for GHR. GHR expression was almost twice as high in males (μ♂ = 0.907 ± 0.0537) than females (μ♀ = 0.551 ± 0.0485; p < 0.05). Similarly, IGF-1 transcription level was almost four times higher in males (μ♂ = 2.38 ± 0.114) than in females (μ♀ = 0.583 ± 0.0910). There was no evidence of environmental effects, or impact of genotypic value, on the expression of IGF-1 (Table 3, Figure 2c).
Table 3.
Results of the maximum likelihood analysis of variance in mixed models for factors explaining gene expression.
| GHR | IGF-1 | |||||
| Source of variation | d.f. | F | P | d.f. | F | P |
| Intercept | 145 | 388.001 | <.0001* | 145 | 299.664 | <.0001* |
| Environment | 1 | 12.005 | 0.001* | 1 | 0.093 | 0.761 |
| EBV group | 1 | 7.170 | 0.055* | 1 | 2.444 | 0.193 |
| Sex | 1 | 13.603 | 0.001* | 1 | 134.533 | <.0001* |
| EBV group × environment | 2 | 3.689 | 0.027* | 2 | 4.066 | 0.019* |
Asterisks indicate significant effects (α = 0.05).
Table 4.
Gene expression of GHR and IGF-1 genes for male and female of HIGH- and LOW-EBV families reared in freshwater and saltwater environments.
| GHR | IGF-1 | |||||||
| Freshwater | Saltwater | Freshwater | Saltwater | |||||
| Family | Female | Male | Female | Male | Female | Male | Female | Male |
| LOW-EBV-1 | 0.687 ± 0.102 | 1.310 ± 0.260 | 0.893 ± 0.150 | 1.09 ± 0.130 | 0.651 ± 0.217 | 1.821 ± 0.554 | 0.557 ± 0.320 | 1.440 ± 0.277 |
| LOW-EBV-13 | 0.400 ± 0.123 | 0.626 ± 0.150 | 0.697 ± 0.106 | 1.83 ± 0.213 | 0.419 ± 0.260 | 2.611 ± 0.320 | 0.365 ± 0.226 | 2.291 ± 0.452 |
| LOW-EBV-20 | 0.205 ± 0.260 | 0.679 ± 0.184 | 0.636 ± 0.130 | 1.02 ± 0.150 | 0.196 ± 0.554 | 2.140 ± 0.391 | 0.338 ± 0.277 | 2.530 ± 0.320 |
| HIGH-EBV-7 | 0.500 ± 0.165 | 0.579 ± 0.116 | 0.679 ± 0.139 | 0.506 ± 0.150 | 0.489 ± 0.350 | 2.503 ± 0.248 | 1.990 ± 0.296 | 1.784 ± 0.320 |
| HIGH-EBV-15 | 0.322 ± 0.106 | 0.549 ± 0.213 | 0.664 ± 0.116 | 1.18 ± 0.184 | 0.495 ± 0.226 | 2.472 ± 0.452 | 0.649 ± 0.248 | 3.362 ± 0.391 |
| HIGH-EBV-24 | 0.612 ± 0.122 | 0.861 ± 0.213 | 0.320 ± 0.184 | 0.659 ± 0.213 | 0.577 ± 0.261 | 2.980 ± 0.452 | 0.273 ± 0.391 | 2.670 ± 0.452 |
Values are LS means ± SE. LS means were estimated as linear predictors based on the most parsimonious model. Standard errors for each estimate were determined from a bootstrap distribution (1,000 iterations) of model predictions.
Individual transcript efficiency (iTE)
We detected highly significant differences in growth relative to GHR expression among environments with substantially lower gains per unit GHR investment (iTEGHR) in fish reared in saltwater (μiTE = 2.00 ± 0.246 g·d-1·u-1) compared to their freshwater-reared sibs (μiTE = 3.71 ± 0.253 g·d-1·u-1) (Table 5, Figure 2d; see also Table 6). Females had significantly higher iTEGHR (μiTE = 3.12 ± 0.229 g·d-1·u-1) than males (μiTE = 2.362 ± 0.286 g·d-1·u-1). Differences among EBV groups for iTEGHR were also highly significant where the LOW-EBV group (μiTE = 1.97 ± 0.241 g·d-1·u-1) showed a much lower effective growth per unit GHR production than the HIGH-EBV group (μiTE = 3.73 ± 0.239 g·d-1·u-1). Also, there was evidence of interaction between EBV group and environment for iTEGHR (see below; Table 5).
Table 5.
Results of the maximum likelihood analysis of variance in mixed models for factors explaining individual transcript efficiency (iTE).
| iTE_GHR | iTE_IGF-1 | |||||
| Source of variation | d.f. | F | P | d.f. | F | P |
| Intercept | 146 | 246.948 | <.0001* | 146 | 29.991 | <.0001* |
| Environment | 1 | 60.215 | <.0001* | 1 | 6.096 | 0.014* |
| EBV group | 1 | 27.786 | 0.006* | 1 | 5.599 | 0.077 |
| Sex | 1 | 3.991 | 0.048* | 1 | 8.699 | 0.004* |
| EBV group × environment | 2 | 17.484 | <.0001* | 2 | 3.060 | 0.050* |
Asterisks indicate significant effects (α = 0.05).
Table 6.
Individual transcript efficiency (iTE) of GHR and IGF-1 for male and female of HIGH- and LOW-EBV families reared in freshwater and saltwater environments.
| iTE_GHR | iTE_IGF-1 | |||||||
| Freshwater | Saltwater | Freshwater | Saltwater | |||||
| Family | Female | Male | Female | Male | Female | Male | Female | Male |
| LOW-EBV-1 | 2.509 ± 0.545 | 1.162 ± 1.39 | 1.210 ± 0.802 | 0.804 ± 0.695 | 3.364 ± 1.112 | 0.831 ± 2.854 | 2.620 ± 1.648 | 0.665 ± 1.427 |
| LOW-EBV-13 | 3.451 ± 0.655 | 2.963 ± 0.802 | 1.380 ± 0.593 | 0.740 ± 1.135 | 4.285 ± 1.345 | 0.588 ± 1.648 | 2.924 ± 1.217 | 0.510 ± 2.330 |
| LOW-EBV-20 | 4.522 ± 1.390 | 1.891 ± 0.983 | 1.441 ± 0.695 | 0.988 ± 0.802 | 4.194 ± 2.854 | 0.676 ± 2.018 | 3.190 ± 1.427 | 0.379 ± 1.648 |
| HIGH-EBV-7 | 3.983 ± 0.879 | 4.272 ± 0.622 | 2.212 ± 0.743 | 4.000 ± 0.802 | 4.880 ± 1.805 | 4.955 ± 1.276 | 1.631 ± 1.525 | 3.920 ± 1.648 |
| HIGH-EBV-15 | 6.122 ± 0.567 | 4.823 ± 1.140 | 2.802 ± 0.622 | 1.860 ± 0.983 | 6.310 ± 1.165 | 0.960 ± 2.330 | 3.661 ± 1.276 | 0.574 ± 2.018 |
| HIGH-EBV-24 | 4.471 ± 0.655 | 2.032 ± 1.140 | 3.391 ± 0.983 | 2.831 ± 1.135 | 5.365 ± 1.345 | 0.557 ± 2.330 | 4.120 ± 2.018 | 0.701 ± 2.323 |
Values are LS means ± SE. LS means were estimated as linear predictors based on the most parsimonious model. Standard errors for each estimate were determined from a bootstrap distribution (1,000 iterations) of model predictions.
Similar to findings for GHR, fish in saltwater had significantly lower growth per unit IGF-1 mRNA (μiTE = 2.34 ± 0.516 g·d-1·u-1) than full-sibs reared in freshwater (μiTE = 3.67 ± 0.530 g·d-1·u-1 Table 5 and Figure 2e). Like GHR also, females had significantly higher iTE (μ♀ = 3.88 ± 0.471 g·d-1·u-1) than males (μ♂ = 1.28 ± 0.588 g·d-1·u-1) (Table 5). Likewise, growth per unit IGF-1 was higher in HIGH-EBV (μiTE = 3.78 ± 0.488 g·d-1·u-1) than in LOW-EBV (μiTE = 2.35 ± 0.491 g·d-1·u-1) group. We also detected evidence of GxE for iTEIGF-1 (see below; Figure 2e).
Genotype-by-environment interaction
We detected significant gene-by-environment interaction, supported by non-parallel reaction norms (Figure 2), for absolute growth (Table 1), level of gene transcription, as well as individual transcript efficiency for both GHR and IGF-1 genes (Table 3, Table 5). Also, genotype-by-environment interaction within each sex was observed for both GHR and IGF-1, whereby LOW-EBV males produced almost 50% more GHR in saltwater compared to a 20% increase for the HIGH-EBV males. Similarly, LOW-EBV females produced 70% more GHR in saltwater compared to a 20% increase for the HIGH-EBV females (Figure 2b). All EBV groups, however, experienced a net increase in GHR on exposure to saltwater. A similar but reversed association was seen for IGF-1: divergence in transcription level between EBV groups was highest in saltwater and HIGH-EBV individuals produced higher levels of IGF-1 (Figure 2c). In this case, however, both males and females of the LOW-EBV group experienced a net decrease in IGF-1 production.
Net changes in individual weight per unit GHR production (iTE) were approximately parallel among the four sex-EBV groups, although changes in iTEGHR among environments were marked (Figure 2d). The iTEIGF-1 reaction norms into saltwater were more convergent, so that males and females in the LOW- and HIGH-EBV groups were more similar than in freshwater. Differences in iTE between freshwater and saltwater were more pronounced for HIGH-EBV than LOW-EBV fish, and the increased difference between EBV groups in saltwater was about 30% for females and > 50% for males (Figure 2e).
Discussion
The aim of this study was to determine the impact of interactions between environments, sexes and genetic values (EBV) on the transcription level of two key genes in the GH/IGF-1 axis and their association with growth in brook charr. Our results provided evidence of GxE interactions for individual gene transcription, as well as pronounced sex effects. We observed that freshwater-reared individuals grew almost 30% faster than saltwater-reared full-sibs under the same feeding regime. A first explanation for such differences could be intolerance of moderate salinity by the Rupert R. charr population, either as a failure of saltwater acclimation, or because of inappropriate genetic architecture for saltwater tolerance. Studies on Atlantic salmon (Salmo salar) [37] and kokanee (O. nerka) [38] showed that physiological shifts from non-anadromy to anadromy are possible for populations confined to freshwater for thousands of generations. There is also a well documented case of the adoption of anadromy in a population of introduced freshwater rainbow trout (O. mykiss) [39]. Therefore, we expected a priori that similar plasticity would apply to brook charr from the Rupert R. In contrast, our results suggest that the capacity for osmoregulatory efficiency in saltwater may not be ubiquitous among resident populations of salmonid species, and that this capacity may have been partially lost in the Rupert R. brook charr population. Secondly, we cannot completely refute the possibility that Rupert R. brook charr grew more slowly in the saltwater environment because of the ration used in this study (0.8% body weight/d). A more extensive range of feeding regimes was not feasible due to space constraints. Consequently, a single standardized commercial ration level was chosen for ensuring equal feeding opportunity to all members of the same tank/treatment group, and to avoid confounding effects of growth with excess feed availability. Thus, while this study revealed important sexual, genetic and environmental effects on the activity and efficiency of the genomic transcripts for the same diet, poor mechanical physiological ability might be compensated for through increased consumption rate. This, however, remains to be experimentally tested. We also observed that females had lower overall growth than males, but no evidence of sex-by-environment interaction for growth specifically. The higher overall growth for males compared to females could be the result of differences in bioenergetic costs associated with sexual maturation. Given that our sampling was conducted during late summer and early fall, the period immediately preceding spawning, it is plausible that females were allocating proportionally more energy to gonadal tissues, and less to skeletal growth, than males [40-42].
Many studies have documented the effects of the environment on levels of gene expression [23,24,43,44]. Our results add to these studies by revealing differential gene expression under distinct environments in the brook charr. However, they refuted our working hypothesis since we observed that fish reared in saltwater expressed more GHR mRNA, despite the fact that they had a slower growth rate than fish reared in freshwater. As a consequence, effective growth relative to transcript production – individual transcript efficiency (iTE) – was also considerably higher in freshwater-reared individuals than their saltwater-reared siblings for both genes. At any given point during their development, organisms must invest energy into basal and active metabolism, reproduction, catabolic processes and stress resistance. Our findings suggest that the relative investment to produce the same amount of tissue in saltwater-reared brook charr is considerably higher than in the freshwater environment, particularly so for GHR. This suggests either an environmental inhibition of molecular elements of growth below GHR and/or a failure of control in the production of GHR and/or precursors to it in saltwater. The direct ratio of growth with gene production on the g-per-unit mRNA scale implicit in the iTE index assumes proportionality and uniformity in pathways of the GH/IGF-1 system candidate genes themselves and in the surrounding metabolic systems along the translational and transcriptional axes. As such, the ratio is analogous to a single-gene assay just above the level of candidate gene analysis, or of a single-QTL effect on phenotypic variance observed in contrasting environments which partitions phenotypic variance into locus-specific effects without knowledge of all physiological processes occurring between genetic and phenotypic expression. While our family-replicated design should have sufficiently accounted for most sources of background variance, we cannot exclude the possibility that full-sib means were partially mediated by dominant genetic effects which may partially bias estimates of additive variance [45,46]. Future studies should emphasize a more complete analysis with a full range of physiological products of the growth pathway and associated members. However, the genes analyzed here are ubiquitous in the process of growth, and represent relevant candidates for the description of this pathway.
Partial discrepancies between the transcription level of genes associated with GH/IGF-1 and growth rate may have several physiological explanations. The GH/IGF-1 system has long been recognized as an important participant in the osmoregulatory physiology of fishes, at least for euryhaline species [21,22]. The absence of differences for the transcription level of liver mRNA IGF-1 between salt and freshwater has previously been observed in rainbow trout, for which an abrupt transfer to 80% seawater resulted in increased IGF-1 mRNA levels in gills and kidneys, but not in the liver [47]. In contrast, and similar to our observations for GHR, Sakamoto and Hirano [48] showed that rainbow trout acclimated to seawater experienced an initial decrease in liver GHR followed by a significant increase after four days. Thus, our results, and those of Sakamoto and Hirano [48], indicate the likelihood of at least partial mediation by the liver in seawater adaptation. Unlike Sakamoto and Hirano [48], however, we did not evaluate the level of active GHR in the membrane, but rather estimated the number of copies of the gene product. This suggests that the density of the receptor at the cell surface may depend not only on the rate of gene expression and the stability of the GHR mRNA, but also on the removal of the protein [21].
We also detected a sex effect on gene activity in which the expression of IGF-1 and GHR was significantly higher for brook charr males. Differences between sex in IGF and GH production have also been reported in other species, such as chickens [49,50], and pigs [31], where males typically have higher production rates than females, as in this study. Similarly, these authors observed that male growth per GHR production was actually lower than that of females, which also corroborates our results. Sex steroid hormones may influence the GH/IGF-1 axis. Riley et al. [33] found that in sexually dimorphic tilapia, injection of 17β-estradiol into males resulted in a GH/IGF-1 profile more closely resembling that of female fish (namely a lower plasma level of IGF-1 and higher plasma levels of GH), whereas administering di-hydrotestosterone to females elicited a serum GH/IGF-1 profile resembling that of males. From these observations [33,51], Wood et al. [22] suggested cross-involvement between sex steroid hormones and the GH/IGF-1 axis, possibly at the level of hepatic GHR. GH resistance induced by sex steroids may, thus, contribute to the developmental switch between somatic and reproductive development associated with sexual maturation in fishes.
Genotypic effect on gene expression, measured as the difference between EBV groups, was observed for both GHR and IGF-1. This corroborates previous observations (e.g. Drosophila melanogaster, [52]; Fundulus heteroclitus, [53]) that the genotype has a significant impact on variation in gene expression between individuals within population. To our knowledge, however, our study represents one of very few reports of a GxE interaction for levels of gene expression [5,6], whereby the effect of genotypic value on gene expression changed in relation to environmental condition. As such, non-parallel reaction norms for expression of GHR and IGF-1 genes indicate genetic variance in reaction norm for growth at the level of individual genes in the GH/IGF-1 axis as a function of the environment. Given its link with fitness, genetic variation for growth could be maintained by a form of balancing selection: the presence of GxE interaction for individual genes of the GH/IGF-1 axis can change the genetic target for selection because different genotypes may produce optimal phenotypes under different environmental conditions. Both mutations and environmental shock during developmental processes [54,55], as well as quantitative genetic variance for environmental reaction, have been proposed for explaining phenotypically plastic responses to different environments [56,57].
Conclusion
In summary, we documented that both environment and sex had major impacts on the expression of mRNA for two key genes, GHR and IGF, involved in the physiological pathway for growth. We also demonstrated for the first time, at least in fish, genotype-by-environment interaction at the level of individual gene transcription. As such, this work contributes significantly to ongoing efforts towards documenting environmentally and sexually induced variance of gene activity, and to understand the resulting phenotypes. However, to achieve a more complete understanding of the molecular architecture responsible for the variation of a quantitative trait such as growth, future research should quantify both gene expression and the resultant proteins implicated in the physiological pathway underlying the trait. The continued increase in the use of methods such as RT-qPCR and microarrays in different fields of biology and physiology should greatly improve our understanding of the functional and evolutionary significance of variation in gene expression.
Methods
Husbandry and strain history
In 2001, 20 mixed half-sib families were generated at the Laboratoire Régional des Sciences Aquatiques (LARSA, Université Laval, QC, Canada) from 15 sires and 10 dams (Table 7) originating from a strain of brook charr derived from the Rupert River which drains into the James-Hudson Bay in northwestern Québec. Individuals, marked by external T-tags (Floy Inc., Seattle, Washington, USA), were randomly divided and assigned to one of two re-circulation units, each composed of three 3,000 L tanks. All fish were maintained at 10°C and 90% oxygen saturation, and on external photoperiod (50°C25'N, 73°53'W, Québec, CA). Fish were fed 0.8% of their body weight (commercial feed pellets, Corey Feed Mills, Inc., NB) throughout the experiment. At the end of June 2004 (2+ age class), salinity was increased in one of the units (20‰) (SW; n = 415) while being maintained at freshwater (≈0‰) (FW; n = 433) in the other for a period of five months. This salinity is typical of conditions commonly encountered by brook charr in estuarine and coastal marine waters.
Table 7.
Sample information.
| Dam | EBV | Sire | EBV | Family | n | EBV Group |
| 126 | -0.253 | 196 | -0.205 | 1 | 102 | LOW |
| 126 | -0.253 | 100 | -0.042 | 2 | 52 | |
| 126 | -0.253 | 162 | -0.204 | 3 | 78 | |
| 193 | 0.029 | 162 | -0.204 | 4 | 88 | |
| 193 | 0.029 | 100 | -0.042 | 5 | 90 | |
| 193 | 0.029 | 138 | -0.178 | 6 | 82 | |
| 252 | 0.184 | 88 | 0.276 | 7 | 90 | HIGH |
| 252 | 0.184 | 249 | 0.313 | 8 | 84 | |
| 252 | 0.184 | 196 | -0.205 | 9 | 79 | |
| 252 | 0.184 | 138 | -0.178 | 10 | 13 | |
| 217 | -0.262 | 250 | -0.262 | 13 | 102 | LOW |
| 242 | 0.218 | 84 | 0.185 | 15 | 93 | HIGH |
| 251 | 0.159 | 177 | 0.081 | 16 | 44 | |
| 231 | -0.049 | 114 | -0.049 | 17 | 93 | |
| 38 | -0.188 | 247 | -0.197 | 20 | 27 | LOW |
| 38 | -0.188 | 148 | 0.185 | 21 | 134 | |
| 38 | -0.188 | 177 | 0.081 | 22 | 127 | |
| 94 | 0.212 | 148 | 0.185 | 24 | 88 | HIGH |
| 135 | -0.048 | 177 | 0.081 | 26 | 55 | |
| 135 | -0.048 | 235 | 0.030 | 27 | 84 |
Rupert strain pedigree, estimated breeding value (EBV) for each dam and sire, number of individuals and family selected in reference to HIGH- and LOW EBV group (in bold characters).
Growth measurements and tissue sampling
Absolute growth (GA = ((W2-W1)/d), where W1 and W2 represent weight for the two successive samplings and d represents the interval in days [10] were calculated for the entire population for the period of July to October. In November 2004, 6 – 15 fish were selected at random within each half-sib family (see below) in the freshwater and saltwater treatments. Fish were killed by rapid decapitation and ≈200 mg of liver tissue was removed and immediately frozen in liquid nitrogen for subsequent analysis of gene expression by reversed transcribed quantitative PCR (RT-qPCR). Individuals were sexed by examination of the gonads at euthanization.
Quantitative genetic modeling
We used a reduced animal model [58-61] for the estimation of additive quantitative genetic variance (σ2a) and best linear unbiased predictions (BLUP; [58]) of breeding value (estimated breeding value; EBV) for absolute growth for all individuals (Perry et al., unpublished). Parent-offspring relationship and growth rate (as a single-vector phenotype) were coded using PEST [62] for REML in VCE5.1 [61] with the iteration of analytical gradients [60] in the animal model
| y = Xβ + Za + e |
where y is the phenotypic vector for growth rate, X is the design matrix (n × p) of fixed effects, β the fixed effects coefficient vector (p × 1), Z is the incidence/relationship matrix (n × q) of genetic effects, a is the vector (u × 1) for additive genetic effects and e is random error. The above REML model was fit separately for each treatment. Rearing tank and group intercept within treatment tank were fit as fixed effects, and animal was the sole random effect. Two groups of three full-sib families having the most extreme mean EBV for growth were selected to represent the 'HIGH' and 'LOW' growth categories, respectively (see Appendix 1)
Total RNA extraction and reverse transcription
Total RNA was extracted from approximately 25 mg of liver tissue using a Qiazol isolation reagent (RNeasy 96 Universal Tissue, Qiagen), following the manufacturer's instructions. RNA concentrations were determined by spectrophotometry (GeneQuant, Pharmacia). Following DNAse treatment to remove residual genomic DNA, 10 μg total RNA was used in a 100 μl reaction to obtain first-strand cDNA by reverse transcriptase reaction (cDNA Archive Kit, Applied Biosystems).
Oligonucleotide design and RT-qPCR analysis of gene expression
The mRNA sequences for IGF-1 and GHR were not available for brook charr in EMBL nor NCBI GeneBank [63]. Consequently, we used the mRNA IGF-1 sequence from chum salmon (O. keta) (Acc. No. AF063216) and the mRNA GHR sequence from coho salmon (O. kisutch) (Acc. No. AF403539) to design non-specific primers. We used these primer pairs to amplify and sequence a region (≈200 bp) of brook charr IGF-1 and GHR mRNA. Gene specific primers and probes were designed from this sequence using Primer Express® software (Table 8). Primers were tested using conventional PCR and tested by amplifying a single band of approximately 90 bp. Primer concentrations were optimized following the manufacturer's instructions (ABI PRISM ®7000 Sequence Detection System (SDS), Applied Biosystems). Relative quantification of gene expression was achieved by concurrent amplification of the eukaryotic 18S rRNA endogenous control (Applied Biosystems).
Table 8.
Primers and probes used in RT-qPCR assays of gene expression.
| Target gene | Primer Set (5'→3') | Probe (5'→3') |
| IGF-1 | Forward: CAGGCATCCAGATTGTGCAA | CAGCCATTACTCTCTG |
| Reverse: ACCATGTTCTGAGAATTCCTGTGTT | ||
| GHR | Forward: CCCACTGCCCCCTGTATCT | ACCATGGTGGAAGGAG |
| Reverse: CTTCAGAAGGAGGCTGTTTTGC |
Each reaction (25 μl) was run in triplicate and contained 5 μl of cDNA (diluted 1: 10 for target genes and 1: 100 for 18S rRNA gene), 12.5 μl Taqman Universal PCR master mix (Applied Biosystem), and 0.9 μM F/R primers. The thermocycling profile used was the default from the sds 2.0 software (50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min). For each primer and probe set, two negative controls were also amplified: non-reverse transcribed total RNA treated with DNase (as a control for contamination by genomic DNA) and a template negative sample, to control for any contamination of the reagents. Amplification efficiencies for all primer/probe sets were calculated following the manufacturer's instructions, and all values proved to be sufficient to allow direct comparison of amplification plots according to the ΔΔCt method (see Sequence Detection Systems Quantitative Assay Design and Optimization, Applied Biosystems).
Statistical modeling
All data were modeled under a linear mixed-effects framework (S-Plus 6.1; Insightful Corporation), in which each family was nested as a random factor within their respective EBV group (HIGH/LOW). Thus, the underlying covariance structure inherent within the data set due to non-replicable nesting of one grouping factor within another was accounted for as random variation between EBV group and among families within each EBV group. Effects of rearing environment (saltwater (SW) vs. freshwater (FW)) and sex (male vs. female), and their interactions with genotypic value (HIGH vs. LOW EBV) were modeled as fixed-effects, with parameters estimated by maximum likelihood. Although this analysis captures interaction effects with EBV group, the significance of the main effect of genotypic value cannot directly be estimated from the model due to insufficient (0) degrees of freedom associated with the nesting factor (EBV group). Consequently, the EBV group effect was estimated separately using a linear mixed effects model incorporating random variation among families and fixed-effect differences between EBV groups. Underlying assumptions of normality and homogeneity of variance were evaluated with diagnostic plots. In one case (growth relative to GHR expression; see subsequent section), heteroscedasticity of within group errors was incorporated into the model via separate variance estimators for each grouping stratum.
Model selection followed a backwards step-wise procedure. A maximal model incorporating all simple and interaction terms was initially defined, of the form
| yijk = (β0 + bi + bij) + β1E + β2S + β3GxE + β4GxS + β5ExS + β6GxExS + εijk |
where yijk is mRNA expression (GHR or IGF-1) or individual absolute growth for the kth individual from the ith family of the kth EBV group, betas (β) corresponding to fixed-effects coefficients for the terms environment (E), sex (S) and EBV group (G), herein also representative of genotypic effects, bi and bij represent random effects vectors describing variation around the model intercepts (i.e. whole-experimental mean) attributable to the ith family nested within the kth EBV group, and εijk is random error. The significance of each term (i.e. model coefficient) was evaluated by conditional F-tests (Table 9, 10, 11). Progressively simpler models were subsequently defined by removing non-significant terms from precedent models beginning with interaction effects. Nested models were then evaluated using likelihood ratio tests to select the most parsimonious model. The reported p-values correspond to results of the likelihood ratio tests wherein a value greater than the nominal level of significance (α = 0.05) indicates that removal of the model term does not increase model deviance significantly [64]. In the event that term removal resulted in a likelihood ratio test approaching a marginal level of significance (0.05 ≤ p ≤ 0.10), final model selection was made through use of the Akaike Information Criterion. Finally, group means were estimated as linear predictors based on the most parsimonious model. Standard errors for each estimate were determined from a bootstrap distribution (1,000 iterations) of model predictions.
Table 9.
Model selection for absolute growth using a backwards step-wise procedure.
| Absolute growth | |||
| Model | Log likelihood | Test | P |
| Growth ~Env + Sex + EBV·Env + EBV·Sex +Env·Sex +EBV·Env·Sex (1) | -107.460 | ||
| ~Env + Sex + EBV·Env + EBV·Sex +Env·Sex (2) | -108.443 | 1 vs. 2 | 0.169 |
| ~Env + Sex + EBV·Env + EBV·Sex (3) | -108.499 | 2 vs. 3 | 0.737 |
| ~Env + Sex + EBV·Env (4) | -108.607 | 3 vs. 4 | 0.643 |
| ~Env + Sex (5) | -112.465 | 4 vs. 5 | 0.021 |
Progressively simpler models are subsequently defined by removing terms from previous models beginning with interaction effects (e.g. 1 vs. 2). Nested models were tested evaluated using likelihood ratio tests to select the most parsimonious model. The reported p-values correspond to results of the likelihood ratio tests wherein a value greater than the nominal level of significance (α = 0.05) indicates that removal of the model term does not increase model deviance significantly. The final, parsimonious model that was selected is indicated in bold characters (Env = environment, EBV = EBV group).
Table 10.
Model selection for gene expression using a backwards step-wise procedure.
| GHR | IGF-1 | |||||
| Model | Log likelihood | Test | P | Log likelihood | Test | P |
| Gene ~Env + Sex + EBV·Env + EBV·Sex +Env·Sex +EBV·Env·Sex (1) | -82.617 | -198.918 | ||||
| ~Env + Sex + EBV·Env + EBV·Sex +Env·Sex (2) | -83.111 | 1 vs. 2 | 0.320 | -199.142 | 1 vs. 2 | 0.502 |
| ~Env + Sex + EBV·Env + EBV·Sex (3) | -83.460 | 2 vs. 3 | 0.403 | -199.170 | 2 vs. 3 | 0.815 |
| ~Env + Sex + EBV·Env (4) | -84.513 | 3 vs. 4 | 0.147 | -200.233 | 3 vs. 4 | 0.145 |
| ~Env + Sex (5) | -87.055 | 4 vs. 5 | 0.079 | -203.634 | 4 vs. 5 | 0.033 |
Progressively simpler models are subsequently defined by removing terms from previous models beginning with interaction effects (e.g. 1 vs. 2). Nested models were tested evaluated using likelihood ratio tests to select the most parsimonious model. The reported p-values correspond to results of the likelihood ratio tests wherein a value greater than the nominal level of significance indicates that removal of the model term does not increase model deviance significantly. The final, parsimonious model that was selected is indicated in bold characters (Env = environment, EBV = EBV group). In the case of GHR, removal of the EBVxEnv term (model 5) resulted in a marginally non-significant likelihood ratio test. However, the interaction term was retained in the final model (model 4 AIC = 185.02), given that its exclusion resulted in a greater penalised likelihood score (model 5 AIC = 186.11).
Table 11.
Model selection for individual transcript efficiency using a backwards step-wise procedure.
| iTE_GHR | iTE_IGF-1 | |||||
| Model | Log likelihood | Test | P | Log likelihood | Test | P |
| iTE ~Env + Sex + EBV·Env + EBV·Sex +Env·Sex +EBV·Env·Sex (1) | -297.317 | -436.651 | ||||
| ~Env + Sex + EBV·Env + EBV·Sex +Env·Sex (2) | -297.689 | 1 vs. 2 | 0.388 | -436.657 | 1 vs. 2 | 0.906 |
| ~Env + Sex + EBV·Env + EBV·Sex (3) | -297.945 | 2 vs. 3 | 0.474 | -436.897 | 2 vs. 3 | 0.489 |
| ~Env + Sex + EBV·Env (4) | -298.417 | 3 vs. 4 | 0.331 | -437.236 | 3 vs. 4 | 0.410 |
| ~Env + Sex (5) | -302.856 | 4 vs. 5 | 0.012 | -439.672 | 4 vs. 5 | 0.088 |
Progressively simpler models are subsequently defined by removing terms from previous models beginning with interaction effects (e.g. 1 vs. 2). Nested models were tested evaluated using likelihood ratio tests to select the most parsimonious model. The reported p-values correspond to results of the likelihood ratio tests wherein a value greater than the nominal level of significance (α = 0.05) indicates that removal of the model term does not increase model deviance significantly. The final, parsimonious model that was selected is indicated in bold characters (Env = environment, EBV = EBV group). In the case of iTEIGF-1, removal of the EBVxEnv term (model 5) resulted in a marginally non-significant likelihood ratio test. However, the interaction term was retained in the final model (model 4 AIC = 890.47), given that its exclusion resulted in a greater penalised likelihood score (model 5 AIC = 891.34).
Individual transcript efficiency (iTE)
Given the direct role of both IGF-1 and GHR in the physiological process of vertebrate growth [16,18-20,27,29,51], the effect of environmental modification on their ultimate phenotypic expression might be most effectively surveyed from the direct ratio of gram-to-gain for inferred mRNA production to phenotype. Such an analysis would reflect that of a simple single-QTL design or candidate gene analysis without control for physiological/genomic background. Here, mRNA production was inherently evaluated relative to that of an endogenous control molecule (eukaryotic 18S rRNA; see above), which at the least controls for gross effects of general physiological background. Thus, in order to evaluate relative association between mRNA production and phenotype, we propose to measure a ratio termed 'individual transcript efficiency' (iTE). This iTE ratio represents the effective growth per unit of mRNA production (GA/X in g·d-1·u-1, where GA is the absolute growth and X is the production in units of IGF-1 or GHR (units is arbitrary, herein the value representing the fold difference between a given gene and individual control). We tested iTE for GHR and IGF-1 using the same general model (see above under Statistical modeling) to evaluate effects associated with integral genetic value (families from either HIGH or LOW growth category), saline environment (FW/SW), sex and GxE interaction on the relative genomic efficiency for each gene, i.e. the dependence of their specific capacity to produce phenotype depending on environmental and genetic group effects.
Authors' contributions
GC participated in the design of the study, was responsible for collecting samples, carried out the molecular work, drafted and wrote the manuscript. GP participated in the design of the study and the collection of the samples, participated in the analysis and interpretation of data, and contributed to writing the manuscript. PB commented on an earlier draft of the paper. LB initiated the idea of testing GxE interactions at the gene transcription level, participated in the design of the study and the interpretation of data, and contributed to writing the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This research was funded through the auspices of AquaNet (National Centres of Excellence; National Sciences and Engineering Research Council, Canada) and funds provided by the Société de recherche et de développement en aquaculture continentale (SORDAC, Inc.), the Société de développement de l'industrie maricole (SODIM, Inc.) and Valorisation Recherche Québec (VRQ) through the Réseau Aquaculture Québec (RAQ). We are grateful to C. Audet for her helpful comments on an earlier version of this manuscript, and to S. McCairns for his advice regarding statistical analyses, and critique of the manuscript. S. Higgins and the staff of LARSA (Université Laval) were instrumental in the design of the experimental facilities, the transfer of fish to and from the salinity experiment and general husbandry. We thank R. Gélinas and P.-P. Dupont for the spawning of the experimental families in autumn 2001 and early period husbandry, and V. Albert, L. Papillon and V. Bourret for their assistance during sampling.
Contributor Information
Guillaume Côté, Email: guillaume.cote@giroq.ulaval.ca.
Guy Perry, Email: gperry@usc.edu.
Pierre Blier, Email: pierre_blier@uqar.qc.ca.
Louis Bernatchez, Email: louis.bernatchez@bio.ulaval.ca.
References
- Fisher MA, Oleksiak MF. Convergence and divergence in gene expression among natural populations exposed to pollution. BMC Genomics. 2007;8 doi: 10.1186/1471-2164-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucassen M, Koschnick N, Eckerle LG, Portner HO. Mitochondrial mechanisms of cold adaptation in cod (Gadus morhua) populations from different climatic zones. Journal of Experimental Biology. 2006;209:2462–2471. doi: 10.1242/jeb.02268. [DOI] [PubMed] [Google Scholar]
- Picard DJS, PM Variation in gene expression in response to stress in two populations of Fundulus heteroclitus. Comparative Biochemistry and Physiology a-Molecular & Integrative Physiology. 2004;137:205–216. doi: 10.1016/S1095-6433(03)00292-7. [DOI] [PubMed] [Google Scholar]
- Larsen P, Nielsen EE, Williams TD, Hemmer-Hansen J, Chipman JK, Kruhoffer M, Gronkjaer P, George SG, Dyrskjot L, Loeschcke V. Adaptive differences in gene expression in European flounder(Platichthys flesus) Molecular Ecology. 2007;16:4674–4683. doi: 10.1111/j.1365-294X.2007.03530.x. [DOI] [PubMed] [Google Scholar]
- Li Y, Lvarez OAA, Gutteling EW, Tijsterman M, Fu JJ, Riksen JAG, Hazendonk E, Prins P, Plasterk RHA, Jansen RC, Breitling R, Kammenga JE. Mapping determinants of gene expression plasticity by genetical genomics in C-elegans. Plos Genetics. 2006;2:2155–2161. doi: 10.1371/journal.pgen.0020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Oh J, Hartl DL, Cavalieri D. Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene. 2006;366:343–351. doi: 10.1016/j.gene.2005.10.042. [DOI] [PubMed] [Google Scholar]
- Schulte PM. Environmental adaptations as windows on molecular evolution. Comparative Biochemistry and Physiology B-Biochemistry Molecular Biology. 2001;128:597–611. doi: 10.1016/S1096-4959(00)00357-2. [DOI] [PubMed] [Google Scholar]
- Groot C. Salmonid life histories. In: Pennell WaB BA, editor. Developments in Aquaculture and Fisheries Science no29: Principles of salmonids culture. New York: Elsevier Press; 1996. pp. 97–128. [Google Scholar]
- Power G. The brook charr, Salvelinus fontinalis. In: EK B, editor. Charrs: Salmonid Fishes of the Genus Salvelinus. The Hague, Netherlands: Dr W. Junk Publischers; 1980. pp. 141–203. [Google Scholar]
- Barton BA. Growth of salmonids. In: Pennell WaB BA, editor. Developments in Aquaculture and Fisheries Science no29: Principles of salmonids culture. New York: Elsevier Press; 1996. pp. 29–86. [Google Scholar]
- Lenormand S, Dodson JJ, Menard A. Seasonal and ontogenetic patterns in the migration of anadromous brook charr (Salvelinus fontinalis) Canadian Journal of Fisheries and Aquatic Sciences. 2004;61:54–67. doi: 10.1139/f03-137. [DOI] [Google Scholar]
- Perry GML, Audet C, Bernatchez L. Maternal genetic effects on adaptive divergence between anadromous and resident brook charr during early life history. Journal of Evolutionary Biology. 2005;18:1348–1361. doi: 10.1111/j.1420-9101.2005.00954.x. [DOI] [PubMed] [Google Scholar]
- Boula D, Castric V, Bernatchez L, Audet C. Physiological, endocrine, and genetic bases of anadromy in the brook charr, Salvelinus fontinalis, of the Laval River (Quebec, Canada) Environmental Biology of Fishes. 2002;64:229–242. doi: 10.1023/A:1016054119783. [DOI] [Google Scholar]
- Thériault V, Bernatchez L, Dodson JJ. Mating patterns and individual reproductive success of sympatric anadromous and resident brook charr, Salvelinus fontinalis, under natural conditions. Behav Ecol Sociobiol. 2007;62:51–65. doi: 10.1007/s00265-007-0437-8. [DOI] [Google Scholar]
- Dechiara TM, Efstratiadis A, Robertson EJ. A Growth-Deficiency Phenotype in Heterozygous Mice Carrying an Insulin-Like Growth Factor-Ii Gene Disrupted by Targeting. Nature. 1990;345:78–80. doi: 10.1038/345078a0. [DOI] [PubMed] [Google Scholar]
- Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of Insulin-Like Growth-Factors in Embryonic and Postnatal-Growth. Cell. 1993;75:73–82. [PubMed] [Google Scholar]
- Moriyama S, Swanson P, Nishii M, Takahashi A, Kawauchi H, Dickhoff WW, Plisetskaya EM. Development of a Homologous Radioimmunoassay for Coho Salmon Insulin-Like Growth-Factor-I. General and Comparative Endocrinology. 1994;96:149–161. doi: 10.1006/gcen.1994.1167. [DOI] [PubMed] [Google Scholar]
- Le Bail PY, Gentil V, Noel O, Gomez JM, Carre F, Le Goff P, Weil C. Structure, function, and regulation of insulin-like growth factors in fish. Ann N Y Acad Sci. 1998;839:157–161. doi: 10.1111/j.1749-6632.1998.tb10750.x. [DOI] [PubMed] [Google Scholar]
- Reinecke M, Bjornsson BT, Dickhoff WW, McCormick SD, Navarro I, Power DM, Gutierrez J. Growth hormone and insulin-like growth factors in fish: Where we are and where to go. General and Comparative Endocrinology. 2005;142:20–24. doi: 10.1016/j.ygcen.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Duan CM. The insulin-like growth factor system and its biological actions in fish. American Zoologist. 1997;37:491–503. [Google Scholar]
- Bjornsson BT, Johansson V, Benedet S, Einarsdottir IE, Hildahl J, Agustsson T, Jonsson E. Growth hormone endocrinology of salmonids: regulatory mechanisms and mode of action. Fish Physiology and Biochemistry. 2002;27:227–242. doi: 10.1023/B:FISH.0000032728.91152.10. [DOI] [Google Scholar]
- Wood AW, Duan C, Bern HA. Insulin-like growth factor signaling in fish. Int Rev Cytol. 2005;243:215–285. doi: 10.1016/S0074-7696(05)43004-1. [DOI] [PubMed] [Google Scholar]
- Gabillard JC, Weil C, Rescan PY, Navarro I, Gutierrez J, Le Bail PY. Effects of environmental temperature on IGF1, IGF2, and IGF type I receptor expression in rainbow trout (Oncorhynchus mykiss) General and Comparative Endocrinology. 2003;133:233–242. doi: 10.1016/S0016-6480(03)00167-9. [DOI] [PubMed] [Google Scholar]
- Gabillard JC, Yao K, Vandeputte M, Gutierrez J, Le Bail PY. Differential expression of two GH receptor mRNAs following temperature change in rainbow trout (Oncorhynchus mykiss) Journal of Endocrinology. 2006;190:29–37. doi: 10.1677/joe.1.06695. [DOI] [PubMed] [Google Scholar]
- Chauvigne F, Gabillard JC, Weil C, Rescan PY. Effect of refeeding on IGFI, IGFII, IGF receptors, FGF2, FGF6, and myostatin mRNA expression in rainbow trout myotomal muscle. General and Comparative Endocrinology. 2003;132:209–215. doi: 10.1016/S0016-6480(03)00081-9. [DOI] [PubMed] [Google Scholar]
- Duan C, Plisetskaya EM. Nutritional Regulation of Insulin-Like Growth Factor-I Messenger-Rna Expression in Salmon Tissues. Journal of Endocrinology. 1993;139:243–252. doi: 10.1677/joe.0.1390243. [DOI] [PubMed] [Google Scholar]
- Beckman BR, Shimizu M, Gadberry BA, Cooper KA. Response of the somatotropic axis of juvenile coho salmon to alterations in plane of nutrition with an analysis of the relationships among growth rate and circulating IGF-I and 41 kDa IGFBP. General and Comparative Endocrinology. 2004;135:334–344. doi: 10.1016/j.ygcen.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Uchida K, Kajimura S, Riley LG, Hirano T, Aida K, Grau EG. Effects of fasting on growth hormone/insulin-like growth factor I axis in the tilapia, Oreochromis mossambicus. Comparative Biochemistry and Physiology a-Molecular & Integrative Physiology. 2003;134:429–439. doi: 10.1016/S1095-6433(02)00318-5. [DOI] [PubMed] [Google Scholar]
- Calduch-Giner JA, Mingarro M, de Celis SVR, Boujard D, Perez-Sanchez J. Molecular cloning and characterization of gilthead sea bream, (Sparus aurata) growth hormone receptor (GHR). Assessment of alternative splicing. Comparative Biochemistry and Physiology B-Biochemistry & Molecular Biology. 2003;136:1–13. doi: 10.1016/S1096-4959(03)00150-7. [DOI] [PubMed] [Google Scholar]
- Iwama GK. Growth of salmonids. In: Pennell WaB BA, editor. Developments in Aquaculture and Fisheries Science no29: Principles of salmonids culture. New York: Elsevier Press; 1996. pp. 467–505. [Google Scholar]
- Parvizi N, Ellendorff F, Elsaesser F. Neuroendocrine regulation of growth hormone and luteinizing hormone in fetal and neonatal pig. Reproduction in Domestic Animals. 2001;36:261–266. doi: 10.1046/j.1439-0531.2001.00292.x. [DOI] [PubMed] [Google Scholar]
- Chowen JA, Gonzalez-Parra S, Garcia-Segura LM, Argente J. Sexually dimorphic interaction of insulin-like growth factor (IGF)-I and sex steroids in lactotrophs. Journal of Neuroendocrinology. 1998;10:493–502. doi: 10.1046/j.1365-2826.1998.00228.x. [DOI] [PubMed] [Google Scholar]
- Riley LG, Richman NH, Hirano T, Grau EG. Activation of the growth hormone/insulin-like growth factor axis by treatment with 17 alpha-methyltestosterone and seawater rearing in the tilapia, Oreochromis mossambicus. General and Comparative Endocrinology. 2002;127:285–292. doi: 10.1016/S0016-6480(02)00051-5. [DOI] [PubMed] [Google Scholar]
- Andrade PM, Silva I, Borra RC, Lima GR, Baracat EC. Estrogen and selective estrogen receptor modulator regulation of insulin-like growth factor binding protein 5 in the rat uterus. Gynecological Endocrinology. 2002;16:265–270. doi: 10.1080/713603094. [DOI] [PubMed] [Google Scholar]
- Goya L, Garcia-Segura LM, Ramos S, Pascual-Leone AM, Argente J, Martin MA, Chowen JA. Interaction between malnutrition and ovarian hormones on the systemic IGF-I axis. European Journal of Endocrinology. 2002;147:417–424. doi: 10.1530/eje.0.1470417. [DOI] [PubMed] [Google Scholar]
- Han VKM. The ontogeny of growth hormone, insulin-like growth factors and sex steroids: Molecular aspects. Hormone Research. 1996;45:61–66. doi: 10.1159/000184761. [DOI] [PubMed] [Google Scholar]
- Staurnes M, Lysfjord G, Berg OK. Parr-Smolt Transformation of a Nonanadromous Population of Atlantic Salmon (Salmo salar) in Norway. Canadian Journal of Zoology-Revue Canadienne De Zoologie. 1992;70:197–199. [Google Scholar]
- Foote CJ, Mayer I, Wood CC, Clarke WC, Blackburn J. On the Developmental Pathway to Nonanadromy in Sockeye-Salmon, Oncorhynchus-Nerka. Canadian Journal of Zoology-Revue Canadienne De Zoologie. 1994;72:397–405. [Google Scholar]
- Pascual M, Bentzen P, Rossi CR, Mackey G, Kinnison MT, Walker R. First documented case of anadromy in a population of introduced rainbow trout in Patagonia, Argentina. Transactions of the American Fisheries Society. 2001;130:53–67. doi: 10.1577/1548-8659(2001)130<0053:FDCOAI>2.0.CO;2. [DOI] [Google Scholar]
- Jonsson N, Hansen LP, Jonsson B. Variation in Age, Size and Repeat Spawning of Adult Atlantic Salmon in Relation to River Discharge. Journal of Animal Ecology. 1991;60:937–947. doi: 10.2307/5423. [DOI] [Google Scholar]
- Jonsson N, Jonsson B. Energy allocation in polymorphic brown trout. Functional Ecology. 1997;11:310–317. doi: 10.1046/j.1365-2435.1997.00083.x. [DOI] [Google Scholar]
- Hutchings JA. Survival consequences of sex-biased growth and the absence of a growth-mortality trade-off. Functional Ecology. 2006;20:347–353. doi: 10.1111/j.1365-2435.2006.01092.x. [DOI] [Google Scholar]
- Scott GR, Richards JG, Forbush B, Isenring P, Schulte PM. Changes in gene expression in gills of the euryhaline killifish Fundulus heteroclitus after abrupt salinity transfer. American Journal of Physiology-Cell Physiology. 2004;287:C300–C309. doi: 10.1152/ajpcell.00054.2004. [DOI] [PubMed] [Google Scholar]
- Weber TE, Bosworth BG. Effects of 28 day exposure to cold temperature or feed restriction on growth, body composition, and expression of genes related to muscle growth and metabolism in channel catfish. Aquaculture. 2005;246:483–492. doi: 10.1016/j.aquaculture.2005.02.032. [DOI] [Google Scholar]
- Roff DA. Evolutionary quantitative genetics. Toronto: Chapman & Hall, International Thomson Pub; 1997. [Google Scholar]
- Pante MJR, Gjerde B, McMillan I, Misztal I. Estimation of additive and dominance genetic variances for body weight at harvest in rainbow trout, Oncorhynchus mykiss. Aquaculture. 2002;204:383–392. doi: 10.1016/S0044-8486(01)00825-0. [DOI] [Google Scholar]
- Sakamoto T, Hirano T. Expression of Insulin-Like Growth Factor-I Gene in Osmoregulatory Organs During Seawater Adaptation of the Salmonid Fish – Possible Mode of Osmoregulatory Action of Growth-Hormone. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:1912–1916. doi: 10.1073/pnas.90.5.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Hirano T. Growth-Hormone Receptors in the Liver and Osmoregulatory Organs of Rainbow-Trout – Characterization and Dynamics During Adaptation to Seawater. Journal of Endocrinology. 1991;130:425–433. doi: 10.1677/joe.0.1300425. [DOI] [PubMed] [Google Scholar]
- Goddard C, Johnson R, Gilhooley HJ, Gardner JO, Gray A, Wilkie RS, Butterwith SC. Decreased muscle cell proliferation in chicks with a deletion in the GH receptor gene. Journal of Molecular Endocrinology. 1996;17:67–78. doi: 10.1677/jme.0.0170067. [DOI] [PubMed] [Google Scholar]
- Romero GS, Stephan DA, Sperling MA, Menon RK. Distinct sexual dimorphism in the effect of hypothyroidism on the expression of the growth hormone receptor and growth hormone-binding protein gene in rat liver. Hormone Research. 1996;45:273–278. doi: 10.1159/000184805. [DOI] [PubMed] [Google Scholar]
- Moriyama S, Shimma H, Tagawa M, Kagawa H. Changes in plasma insulin-like growth factor-I levels in the precociously maturing amago salmon, Oncorhynchus masou ishikawai. Fish Physiology and Biochemistry. 1997;17:253–259. doi: 10.1023/A:1007725010022. [DOI] [Google Scholar]
- Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, Gibson G. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nature Genetics. 2001;29:389–395. doi: 10.1038/ng766. [DOI] [PubMed] [Google Scholar]
- Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nature Genetics. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- West-Eberhard MJ. Developmental plasticity and evolution. Oxford, New York: Oxford University Press; 2003. [Google Scholar]
- Pigliucci M, Murren CJ. Perspective: Genetic assimilation and a possible evolutionary paradox: Can macroevolution sometimes be so fast as to pass us by? Evolution. 2003;57:1455–1464. doi: 10.1111/j.0014-3820.2003.tb00354.x. [DOI] [PubMed] [Google Scholar]
- DeWitt TjaS SM. Phenotypic Plasticity Functional and Conceptual Approaches. New York: Oxford University Press; 2004. [Google Scholar]
- de Jong G. Evolution of phenotypic plasticity: patterns of plasticity and the emergence of ecotypes. New Phytologist. 2005;166:101–117. doi: 10.1111/j.1469-8137.2005.01322.x. [DOI] [PubMed] [Google Scholar]
- Lynch MaW B. Genetics and analysis of quantitative traits. Sunderland, Massachusetts: Sinauer Associates; 1998. [Google Scholar]
- Kruuk LE. Estimating genetic parameters in natural populations using the "animal model". Philos Trans R Soc Lond B Biol Sci. 2004;359:873–890. doi: 10.1098/rstb.2003.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrode RA, Swanson GJT, Winters MS. Method and effects of incorporating foreign information into United Kingdom production evaluations. Animal Science. 1996;63:373–379. [Google Scholar]
- Kovac M, Groeneveld E, Gracia-Cortés LA. 7th World Congress on Genetics Applied to Livestock Production. Montpellier, France; 2002. VCE-5: A package for the optimization of dispersion parameters. [Google Scholar]
- Groeneveld E, Kovac M, Wang T. Proceeding of the 4th World Congress on Genetics Applied to Livestock Production. Edinburgh, United Kingdom; 1990. PEST, a general purpose BLUP package for multivariate prediction and estimation; pp. 488–491. [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Research. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley MJ. Statical computing An introduction to data analysis using S-Plus. England: Jonh Wiley and Sons; 2002. [Google Scholar]