Synopsis
Post-translational modification plays crucial roles in signal transduction in eukaryotic cells. To elucidate the biological function of a protein with a specific post-translational modification, it is necessary to isolate the modified protein. However, it is difficult to incorporate a modified amino acid into a specific position of a protein, in particular, in a large-scale preparation. In order to prepare post-translationally modified proteins in Escherichia coli (E. coli), we have constructed co-expression vectors that contain protein and corresponding enzyme genes. The protein and enzyme are co-expressed in the same E. coli cells and the protein is post-translationally modified in vivo. By using this system, the transcriptional activator cyclic-AMP-response-element-binding protein (CREB) was phosphorylated at Ser-133 and the hypoxia-inducible factor-1α (HIF-1α) was hydroxylated at Asn-803 in E. coli. Although the constructs of the proteins we used are very flexible and susceptible to degradation by proteases in E. coli when they are expressed alone, the B1 domain of streptococcal protein G (GB1) fused to the N-terminus of the proteins increased the yields dramatically. Site-specific phosphorylation of CREB and hydroxylation of HIF-1α were confirmed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) and NMR. Our GB1-fusion co-expression system can be used in the same way as conventional protein expression in E. coli, making it a flexible and economical method to produce a large amount of a post-translationally modified protein.
Keywords: phosphorylation, hydroxylation, co-expression, Escherichia coli, B1 domain of protein G (GB1), CREB, HIF-1α
INTRODUCTION
Post-translational modification plays a crucial role in signal transduction in eukaryotic cells by introducing diversity and versatility into proteins translated from genomic DNA sequences [1]. Post-translational modification extends the range of function of the proteins by attaching other biochemical functional groups such as phosphate, hydroxyl group, acetate, lipids, or carbohydrates, causing changes in intrinsic activities of the proteins such as activation, inactivation or change in other properties such as conformational change. Despite the importance of post-translational modifications in biology, the production of large quantities of modified proteins for biophysical characterization is difficult. Here we report a general method for overexpression of specifically modified proteins in E. coli and illustrate its application to two key post-translational modifications involved in eukaryotic signal transduction: Ser-133-phosphorylation of the kinase inducible domain (KID) of the transcriptional activator cyclic-AMP-response-element-binding protein (CREB), and Asn-803-hydroxylation of the C-terminal activation domain (CAD) of the hypoxia-inducible factor-1α (HIF-1α).
The transcriptional activator CREB regulates transcription of target genes in response to hormonal stimuli that activate protein kinase A (PKA) [2]. PKA phosphorylates CREB at Ser-133 in the KID domain by using ATP as the substrate, enabling CREB to recruit the transcriptional co-activator CREB-binding protein (CBP) and its paralogue p300 through direct interaction between the phosphorylated KID domain and the KIX domain of CBP/p300. Among its many biological functions, CREB enhances long-term memory; consequently the CREB pathway has been targeted with the aim of developing new drugs for Alzheimer’s disease [3].
HIF is a major transcriptional mediator of the hypoxic response in eukaryotic cells, regulating the expression of myriad genes involved in oxygen transport, glucose uptake, glycolysis and angiogenesis [4]. Hypoxic regions often observed in many tumor tissues require the formation of new blood vessels, enabling tumors to develop their own blood supply. The HIF-1α CAD interacts with the transcriptional adapter zinc finger (TAZ1) domain of CBP/p300 in hypoxia, resulting in transcription of oxygen stress genes. Under normoxic conditions, the factor inhibiting HIF-1 (FIH), an asparaginyl hydroxylase, catalyzes hydroxylation of HIF-1α at the β-carbon of Asn-803 [5]. Hydroxylation of this specific asparagine residue in HIF-1α impairs the interaction with CBP/p300. The HIF pathway has been extensively studied for the development of new cancer therapies.
In order to characterize the function and interactions of phosphorylated KID (pKID) and hydroxylated HIF-1α CAD (HIF-OH) by biophysical methods, it is necessary to isolate site-specifically modified pKID and HIF-OH. In particular, structural studies by NMR and X-ray crystallography require large quantities (tens of milligrams) of purified proteins. However, it is difficult to incorporate an unusual amino acid into a specific position of a protein in a large-scale preparation using the conventional protein-expression system with Escherichia coli (E. coli), favored with respect to ease of handling and low costs. Signal transduction pathways in E. coli cells are significantly different from those in eukaryotic cells, namely E. coli cells do not possess enzymes such as kinases or hydroxylases involved in eukaryotic post-translational modification. While it is possible to obtain post-translationally modified proteins using eukaryotic expression systems, yields of isotopically labeled proteins are typically lower than with prokaryotic expression systems and there is the potential to obtain additional undesired modifications. Short peptides containing an unusual amino acid at a specific position can be prepared by solid-phase peptide synthesis, but it is expensive to synthesize isotope-labeled forms (13C and 15N) for the purpose of NMR studies and synthesis of large proteins remains technically challenging.
A protein can be modified site-specifically by in vitro enzymatic reaction. However, enzymes are often expensive for a large-scale preparation or commercially unavailable, thus such enzymes also need to be overexpressed and isolated. In vitro modification is time intensive because it usually requires two rounds of chromatographic purification before and after the in vitro enzymatic reaction to remove the enzyme, substrates, by-products and unmodified protein. Moreover, it is often difficult to separate the unmodified protein since it has a similar chromatographic retention time as the modified protein. A major difficulty in expressing proteins, including KID and HIF-1α CAD, subject to post-translational modification is that those proteins are often intrinsically flexible in solution, in part because such proteins interact with both the enzymes modifying them and other target biomolecules with different conformations [6;7], thus they are insufficiently resistant against proteolytic degradation in the cells, causing low yields.
In this study, we have designed co-expression vectors that contain both protein (KID or HIF-1α CAD) and corresponding enzyme (PKA or FIH) genes to obtain a large amount of a post-translationally modified protein (pKID or HIF-OH) in E. coli. Both the protein and enzyme are expressed in the same host cells with an appropriate co-factor so that the protein is modified post-translationally in E. coli using standard culture conditions. To overcome the proteolytic degradation issue causing low yields of intrinsically disordered proteins, we have utilized the B1 domain of streptococcal protein G (GB1) as a fusion tag linked to the N-terminus of the protein. Fusion with GB1 has been shown to greatly increase protein expression levels without the need for extensive optimization of the expression conditions [8;9]. The GB1 can be easily excised from the fusion protein with a protease such as thrombin by inserting a cleavable linker between GB1 and the protein of interest. The GB1-fusion co-expression vectors, GB1KID-PKA and GB1HIF-FIH, enabled us to prepare large quantities of isotopically labeled pKID and HIF-OH, >30 mg/L. Since prepared pKID and HIF-OH are quantitatively phosphorylated or hydroxylated at the correct position, Ser-133 and Asn-803, respectively, as confirmed by NMR and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF), it is not necessary to separate unmodified KID or HIF. It should be noted, however, that the expression conditions, particularly temperature and length of induction, should be optimized to avoid non-specific modifications by the enzyme and to increase the yield, as demonstrated for Ser-133-phosphorylation of KID. Using the method described here, the modified KID or HIF-1α CAD can be used directly after purification from the E. coli lysate or after cleavage with a protease to remove the GB1 fragment. A major advantage of this method is that the modifying enzyme, PKA or FIH, does not have to be isolated and purified separately.
Our protein-enzyme co-expression system utilizing the GB1 tag facilitates studies requiring a large amount of an intrinsically disordered protein with a site-specific post-translational modification. It is also suitable for easy and inexpensive labeling with 13C and/or 15N for NMR studies.
EXPERIMENTAL
Co-expression vector with a GB1 tag
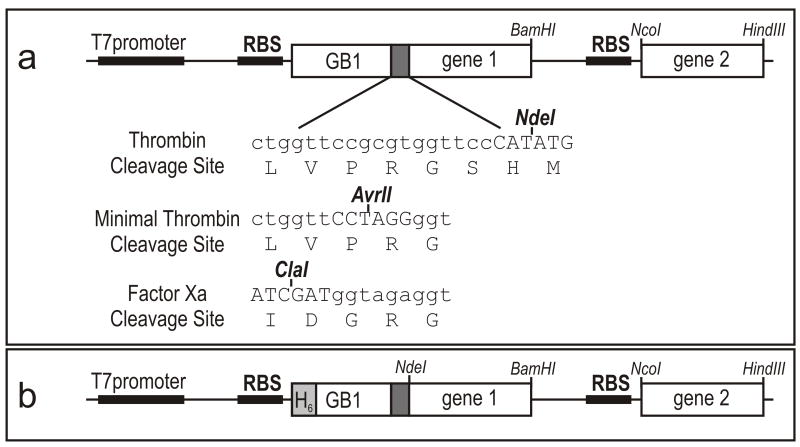
Figure 1 outlines the GB1-fusion co-expression system, a derivative of pET22b (Novagen) with two cloning and ribosomal binding sites. The co-expression vector was originally constructed to avoid degradation of intrinsically disordered proteins in E. coli by co-expressing the protein and its target protein that can form a complex resistant to proteolytic degradation [10;11]. Expression of both substrate protein as a C-terminal GB1 fusion and modifying enzyme is driven from a single T7 promoter. The enzyme gene is inserted into the second cloning site since somewhat lower expression levels are usually obtained at this position, having the effect of enhancing the specificity of the post-translational modification. The substrate protein is connected with GB1 through a thrombin-cleavable linker, Leu-Val-Pro-Arg-Gly-Ser. In the case of the co-expression of GB1KID and PKA, the serine residue in the thrombin cleavage site can be phosphorylated since it is contained within a PKA recognition site, Arg (or X)-Arg-X-Ser/Thr-X [12]. The serine residue is not necessary for the thrombin cleavage reaction as long as an acidic residue is not placed at the position of the serine residue in the thrombin cleavage site [13]. As the N-terminal residue of our KID construct is (non-acidic) histidine that originates from the NdeI restriction enzyme site, the serine residue was deleted by QuickChange site-directed mutagenesis method (Stratagene).
Figure 1.
(a) Design of the GB1-fusion co-expression vector with two cloning and ribosomal binding sites (RBS). The coding sequences of the substrate protein and enzyme are respectively inserted into the gene1 and 2 cloning sites. The DNA sequence of linkers containing protease cleavage and cloning sites are listed. (b) Design of His6-tag (H6) GB1 fusion co-expression vector.
Preparation of phosphorylated KID
The gene sequences of the KID domain corresponding to residues 110–147 of rat CREB and the catalytic subunit of mouse PKA corresponding to residues 1–350 were PCR amplified and cloned into the co-expression vector using NdeI and BamHI cloning sites and NcoI and HindIII cloning sites, respectively. E. coli BL21 (DE3) [DNAY] cells harboring the GB1KID-PKA co-expression vector were grown at 37 °C in LB medium or minimal M9 medium containing 0.5g/L [15N]-ammonium sulfate, 0.5g/L [15N]-ammonium chloride and 4g/L D-glucose as nitrogen and carbon sources for preparation of NMR samples. ATP used by PKA as the co-factor for the phosphorylation of KID is biosynthesized in E. coli cells in sufficient quantity to allow quantitative phosphorylation of overexpressed GB1KID. Isopropyl-1-thio-β-D-galactopyranoside (IPTG) was added to a final concentration of 1 mM for induction of GB1KID and PKA expression when OD600nm reached ~0.8. After addition of IPTG, the cells were incubated 3 hours during which Ser-133 in GB1KID was phosphorylated by PKA in vivo. The cells were harvested, resuspended in 25 mM Tris buffer (pH 8.0) and lysed by sonication. The cell lysate was diluted three fold using 0.1% (v/v) acetonitrile/trifluoroacetic acid (TFA) and precipitated debris were removed by centrifugation. GB1pKID in the supernatant was purified to homogeneity via reversed-phase HPLC on a 25mm C4 cartridge (Waters) using standard acetonitrile/0.1% TFA mobile phase and lyophilized. The purified GB1pKID was dissolved into thrombin-cleavage buffer (20 mM Tris (pH 8), 150 mM NaCl and 2.5 mM CaCl2) to a final concentration of 2 mg/ml and was cleaved by 1 unit/mg thrombin at 25 °C for 24 hours. Cleaved pKID was purified to homogeneity via reversed-phase HPLC. The identity and integrity of the proteins were confirmed by MALDI-TOF. A KID-PKA co-expression vector lacking the GB1 gene was also constructed to compare the yields of modified GB1pKID and pKID.
A GB1KID-KIX co-expression vector was constructed to prepare unphosphorylated KID to test in vitro phosphorylation of KID. Co-expression of KIX can increase the proteolytic resistance of GB1KID in E. coli since KID can bind to KIX weakly even in the unphosphorylated form [14]. The gene sequence of the KIX domain of mouse CBP, corresponding to residues 586–672, was PCR amplified and inserted into the second gene cloning site instead of PKA. GB1KID was expressed and purified using the same method as for GB1pKID. PKA was prepared using the GB1KID-PKA co-expression vector in the same way as GB1pKID except that the cells were induced overnight at 15 °C in order to enhance the yield of soluble PKA. PKA was purified using FPLC with a Hi-Trap SP column (GE Healthcare) in 20 mM Tris buffer (pH 7.0) containing 5 mM dithiothreitol (DTT). KID was phosphorylated by PKA in vitro as described previously [15], and the phosphorylated KID was purified to homogeneity via reversed-phase HPLC. The identity and integrity of KID and pKID were confirmed by MALDI-TOF.
For NMR studies, [15N]-pKID phosphorylated in vivo and in vitro and unphosphorylated [15N]-KID were dissolved in 90% H2O/10% D2O containing 20 mM Tris-d11-acetate-d4 (pH 5.5), 50 mM NaCl and 2 mM NaN3. NMR spectra were measured at 25 °C on Bruker Avance500 and DRX800 spectrometers. 1H-15N heteronuclear single quantum coherence (HSQC) spectra [16] were measured for all samples. The cross peaks of [15N]-KID were identified based on our previous assignments [17].
Preparation of hydroxylated HIF-1α
The gene sequence of the human HIF-1α CAD corresponding to residues 776–826 and human asparaginyl hydroxylase FIH were PCR amplified from a human liver cDNA library and cloned into the co-expression vector using NdeI and BamHI cloning sites and NcoI and HindIII cloning sites, respectively. E. coli BL21 (DE3) [DNAY] cells harboring the GB1HIF-FIH co-expression vector were grown at 37 °C in LB medium or minimal M9 medium containing [15N]-ammonium sulfate, [15N]-ammonium chloride and D-glucose (or [13C6]-D-glucose) as nitrogen and carbon sources. FeSO4 was added, to 1 mM final concentration, to the medium at OD600nm of ~0.4 as Fe2+ is the essential co-factor of FIH. IPTG was added to a final concentration of 0.8 mM for induction of GB1HIF and FIH expression when OD600nm reached ~1. After addition of IPTG, the cells were incubated at 15°C for 15 hours during which Asn-803 in HIF was hydroxylated in E. coli by FIH using O2 abundant in the air. The cells were harvested and resuspended in 20 mM Tris buffer (pH 8.0) containing 2 mM DTT and 1 mM EDTA. Soluble GB1HIF-OH was purified by anion exchange chromatography on a HiTrap Q column (GE Healthcare). GB1HIF-OH was eluted using 20 mM Tris buffer (pH 8.0) containing 0.6 M NaCl and 1 mM EDTA and subsequently purified to homogeneity via reversed-phase HPLC. GB1HIF-OH was cleaved by thrombin and cleaved HIF-OH was purified to homogeneity via reversed-phase HPLC. The identity and integrity of HIF-OH were confirmed by MALDI-TOF.
In order to confirm that FIH specifically hydroxylates Asn-803 in HIF, a series of co-expression vectors, HIF-FIH, HIF-TAZ1, [N803A]-HIF-FIH and [N803A]-HIF-TAZ1, were constructed. It should be noted that those expression vectors do not have the GB1 gene, but the resulting HIF-1α CAD expression levels are sufficient to quantitate their molecular weights by MALDI-TOF. TAZ1 does not modify HIF-1α or [N803A]-HIF-1α CAD although it can increase the yields of HIF-1α CAD by forming a HIF:TAZ1 complex that is more resistant to degradation by proteases in E. coli.
For NMR studies, [15N]-HIF-OH and [13C,15N]-HIF-OH were dissolved in 90% H2O/10% D2O containing 20mM MES (pH 6.12), 2 mM DTT and 2 mM NaN3.
RESULTS
In vivo phosphorylation of KID
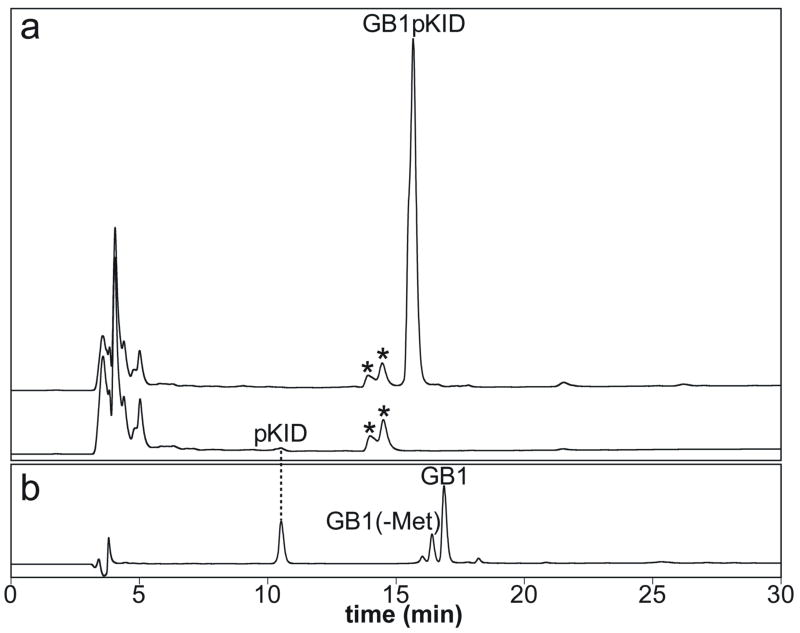
pKID and GB1pKID were expressed in 20 ml LB medium at 37°C using the KID-PKA and GB1KID-PKA co-expression vectors, respectively, in order to test how much the GB1 fusion can increase the yield of pKID. The cells were lysed in 1 ml lysis buffer and 50 μl supernatants of the lysate were diluted three fold using 0.1% (v/v) TFA and loaded onto a C4 column for HPLC analysis (Figure 2). Remarkably, the yield of pKID increased by approximately 150-fold upon fusion with GB1. The non-fused pKID peak intensity is near the detection limit (Figure 2a), consistent with the fact that intrinsically disordered proteins are susceptible to degradation by proteases in E. coli. Phosphorylation of KID in the non-fusion construct was confirmed by MALDI-TOF. The non-fused KID was doubly phosphorylated to some extent (data not shown). Although we have not yet identified which Ser or Thr is phosphorylated in addition to Ser-133, this double phosphorylation probably occurs because the PKA concentration is much higher than the optimum concentration ratio, given the extremely low expression level of unfused KID in E. coli. GB1pKID was expressed in [15N]-M9 medium as well for the NMR studies. [15N]-GB1pKID was cleaved by thrombin completely after 24-hour incubation at 25 °C despite the lack of the serine residue in the thrombin cleavages site (Figure 2b). The cleaved [15N]-pKID was singly phosphorylated, as confirmed by MALDI-TOF. The measured molecular weight of [15N]-pKID was the same as the theoretical value, 4776 Da. The molecular weight increased by that of one phosphate group (79 Da) from unphosphorylated [15N]-KID. One might expect that, even as a GB1 fusion protein, KID would be flexible and susceptible to proteolytic degradation; however, MALDI-TOF and reversed-phase HPLC did not detect shorter GB1pKID fragments proteolyzed in the KID region. It appears that, by some unknown mechanism, the GB1 fusion protects KID from proteolytic degradation. Koenig et. al. have also reported an increase in the stability and yield of a flexible protein fused with GB1 in E. coli [9]. It should be noted that even as a GB1 fusion, KID was multiply phosphorylated if the cells were incubated at 15 °C overnight after addition of IPTG since the expression level of soluble PKA is much higher at lower temperature. In the case of the GB1KID-PKA co-expression, 3-hour incubation at 37 °C after addition of IPTG gives the best yield of Ser-133 site-specifically phosphorylated GB1pKID.
Figure 2.
Expression of pKID and GB1pKID and thrombin-cleavage reaction of GB1pKID monitored by reversed-phase HPLC. (a) Comparison of expression levels between pKID and GB1pKID in E. coli cell lysates. Other E. coli proteins are indicated by asterisks (*). (b) Thrombin cleavage of GB1pKID using 1 unit/mg thrombin after 24-hour incubation at 25 °C. N-terminal Met of approximately 28% GB1 was truncated in E. coli cells.
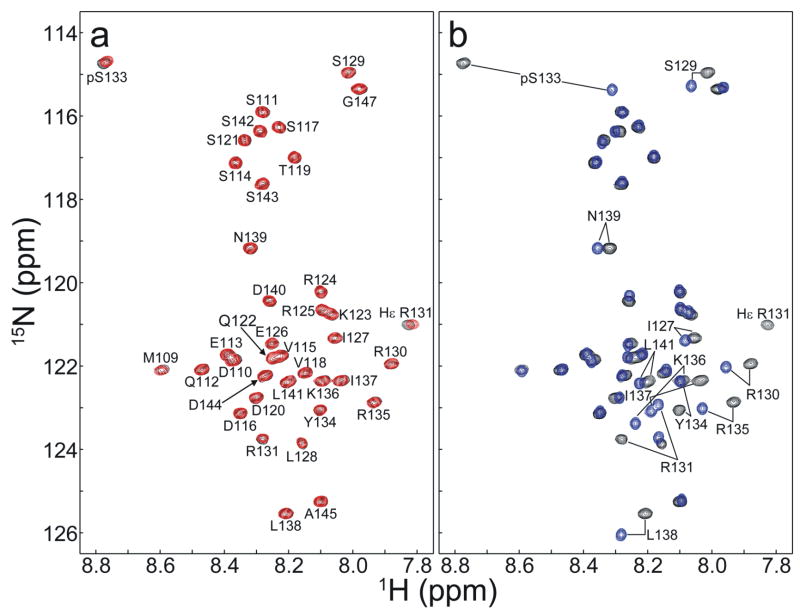
Figure 3 shows the 1H-15N HSQC spectra of [15N]-KID (blue) and [15N]-pKID phosphorylated in vivo (black) and in vitro (red). Both KID and pKID are unstructured since all amide peaks including pSer133 exhibit random coil chemical shifts [18–20]. The HSQC spectrum of pKID phosphorylated in vivo and in vitro are virtually identical (Figure 3a). NMR signals of residues around Ser-133 are shifted by the Ser-133-phosphorylation (Figure 3b). As in vitro phosphorylation has been previously shown to phosphorylate KID specifically at Ser-133 [15;17], we conclude that Ser-133 of KID is correctly phosphorylated by PKA in E. coli. pKID produced using the in vivo phosphorylation method binds KIX with identical affinity as pKID produced via in vitro phosphorylation (data not shown). A study of KIX:pKID binding interactions using pKID prepared in this study is discussed elsewhere [21].
Figure 3.
1H-15N HSQC spectra of Ser-133-phosphorylated KID and unphosphorylated KID. Residue-specific assignments are indicated. (a) Overlay of the 1H-15N HSQC spectra of pKID phosphorylated in vivo (black) and in vitro (red). (b) Overlay of the 1H-15N HSQC spectra of pKID phosphorylated in vivo (black) and unphosphorylated KID (blue). NMR peaks shifted by Ser-133 phosphorylation are labeled.
In order to further optimize the GB1-fusion co-expression vector for expression of short peptide fragments, we constructed another GB1KID-PKA co-expression vector that utilizes a Factor Xa cleavage site (Ile-Asp-Gly-Arg) instead of the thrombin cleavage site and deleted the NdeI cloning site. As Factor Xa cleaves after the C-terminal arginine residue in the Factor Xa cleavage site, no additional residues (Gly-His-Met from the thrombin and NdeI restriction sites) remain at the N-terminus of the expressed protein after cleavage of the GB1 fusion tag. We obtained correctly phosphorylated KID in high yield using this new co-expression vector as well.
In vivo hydroxylation of HIF
HIF-1α CAD and [N803A]-HIF-1α CAD were expressed using a series of co-expression vectors, HIF-FIH, HIF-TAZ1, [N803A]-HIF-FIH and [N803A]-HIF-TAZ1, to test whether FIH specifically hydroxylates Asn-803 of HIF. The molecular weights of HIF and [N803A]-HIF measured by MALDI-TOF are listed in Table 1. The molecular weight of HIF increased by that of one hydroxyl group when co-expressed with FIH while that of [N803A]-HIF did not change with FIH co-expression. Co-expressed TAZ1 did not change the molecular weight of either HIF or [N803A]-HIF, as expected. Therefore, we conclude that Asn-803 in HIF is specifically hydroxylated by FIH in E. coli when HIF-1α CAD and FIH are co-expressed. Subsequently, the co-expression vector of GB1HIF-FIH was used to maximize the yield for NMR studies. Similar to GB1pKID, GB1HIF-OH expressed much more than HIF-OH itself and GB1 was cleaved efficiently by thrombin (data not shown). It should be noted that, in contrast to phosphorylation of KID by PKA, FIH specifically hydroxylates Asn-803 even when the cells are incubated at 15 °C overnight. This suggests that the duration of incubation and the temperature should be optimized carefully for each protein-enzyme pair of interest.
Table 1.
Molecular weights of HIF-1α and [N803A]-HIF-1α CADs co-expressed with FIH or TAZ1
| co-expression partner | ||
|---|---|---|
| protein | FIH | TAZ1 |
| measured mass (MALDI-TOF) | ||
| HIF | 5578 | 5562 |
| [N803A]-HIF | 5521 | 5520 |
| theoretical mass | ||
| HIF | 5563 | |
| HIF-OH | 5579 | |
| [N803A]-HIF | 5520 | |
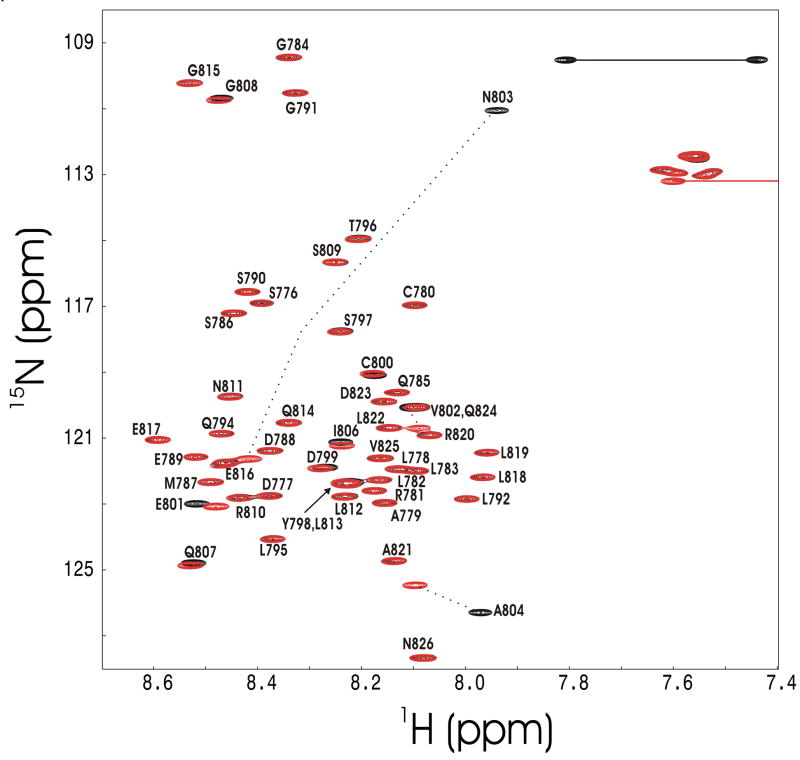
Figure 4 shows the 1H-15N HSQC spectrum of [15N]-HIF-OH prepared using the GB1HIF-FIH co-expression vector. HIF-OH is clearly unstructured since all amide peaks appear within a narrow range of 1H chemical shifts, < 1 ppm. The NMR peak of the backbone amide of Asn-803, however, appeared in an unusual position, providing direct evidence for Asn-803 hydroxylation. The observed amide 1H and 15N chemical shifts of Asn-803 are 7.94 and 110.96 ppm, respectively, but random coil chemical shifts of Asn corrected for the HIF sequence are 8.56 and 122.73 ppm [18;19]. The NMR peaks of the Asn-803 side chain NH2 group are also different from those of other side chain NH2groups due to hydroxylation at the β-carbon.
Figure 4.
1H-15N HSQC spectra of Asn-803-hydroxylated HIF-1α CAD (black) and unmodified HIF-1α CAD (red). Residue-specific assignments are indicated (unpublished data). Amide peaks that undergo significant chemical shift change upon hydroxylation are designated by a dotted line. Side chain NH2 groups of Asn-803 are denoted by a solid line. Only one of two NMR peaks of each side chain NH2 group except for hydroxylated Asn-803 is shown for clarity.
The preferred method of purification for small peptides is reversed-phase HPLC (RPHPLC) but it is sometimes the case that the GB1-fusion and the cleaved peptide have very similar RPHPLC retention times, complicating the final purification. In order to further enhance the ease, efficiency and cost-effectiveness of large scale purification of modified proteins using this system, we have also constructed a His6GB1-fusion co-expression vector (Figure 1). Yields of modified His6GB1 fusion proteins are similar or higher than those obtained with the corresponding non-His6-tag versions and the induction conditions necessary to obtain high yields of specifically modified protein are identical. Using this system, the soluble His6GB1-fusion protein is isolated from the cell lysate by metal affinity chromatography on Ni-NTA (Qiagen) under native or denaturing conditions. Contaminants including the co-expressed modifying enzyme are removed by washing the resin with buffer containing a low concentration (10–30 mM) of imidazole and the His6GB1 fusion tag is cleaved off directly on the resin with 1 U/mg thrombin at 25°C for 15 hours. Cleaved, modified peptide present in the supernatant or column flow through is then purified to homogeneity by reversed-phase HPLC chromatography. This method eliminates the initial purification of the GB1-fusion protein by RPHPLC, dramatically shortening the purification time requirement and reducing solvent and resin usage.
DISCUSSION
We have demonstrated Ser-133-phosphorylation of KID and Asn-803-hydroxylation of HIF in E. coli by using GB1KID-PKA and GB1HIF-FIH co-expression vectors, respectively. The GB1 tag is very useful for increasing the yield of the protein fused with GB1, especially for proteins like KID and HIF-1α CAD that are intrinsically disordered and are highly susceptible to degradation in E. coli. The GB1 is easily removed by thrombin cleavage, and the post-translationally modified protein can be purified by reversed-phase HPLC. Specific Ser-133-phosphorylation of KID and Asn-803-hydroxylation of HIF-1α CAD were confirmed by NMR and MALDI-TOF.
Brown et. al. [22] produced a short fragment of phosphorylated cyclin-dependent kinase 2 (CDK2) in E. coli by co-expression of human GST-CDK2 and S. cerevisiae GST-Cak1, which phosphorylates CDK2 at Thr-160. Their method is presumably similar to ours but it has not yet been described in detail. Several groups have reported expression of post-translationally modified proteins in E. coli but their methods were not optimized for over-expression of peptides and required either fusion of the substrate protein directly to the modifying enzyme or co-transfection with two different plasmids, decreasing the ease of purification and requiring use of multiple antibiotics [23, 24, 25]. Liu and Schultz [26] reported a sophisticated method to incorporate sulfotyrosine into a specific position (residue 63) of hirudin in E. coli. They generated a unique tRNA/aminoacyl-tRNA synthetase pair that leads to the in vivo incorporation of the synthetic sulfotyrosine into the protein in response to an amber nonsense codon. The same group previously succeeded in the in vivo incorporation of synthetic O-methyl-L-tyrosine into dihydrofolate reductase in a similar manner [27]. This technique is very powerful as it does not require the enzyme that modifies the protein, but there is a limitation in that it requires a unique transfer RNA (tRNA)/aminoacyl-tRNA synthetase pair that does not cross-react with endogenous host tRNA, aminoacyl-tRNA synthetase or amino acids. Recently, Uchimura et. al. [28] prepared small ubiquitin-related modifier (SUMO-1 and SUMO-2) conjugated (sumoylated) proteins in E. coli. They constructed a co-expression vector of a linear fusion of E1 and E2 ligases and SUMO-1 (or SUMO-2), and the co-expression vector was co-transformed with a substrate protein vector into E. coli BL21 (DE3). A wide variety of substrate proteins, including RanGAP1, RanBP2-IR, thymine DNA glycosylase, p53 and TONAS-ring were sumoylated. Our results together with this in vivo sumoylation study suggest that a wide variety of post-translational modifications can be achieved in E. coli by co-expression of a protein-enzyme pair. Co-expression vectors are commercially available, e.g. pETDuet™ (Novagen), so that a new protein-enzyme co-expression vector can be easily constructed. It should be noted, however, that the expression conditions should be optimized carefully because various factors, such as the incubation time, the temperature, and the molar ratio between the protein and modifying enzyme, can affect the yield and specificity of the modification, as described in the case of the GB1KID and PKA.
Our GB1-fusion co-expression system is a flexible and economical method to obtain a large amount of a post-translationally modified protein. In addition to structural studies using NMR and X-ray, our method is suitable for generating large quantities of modified proteins for applications in screening for new drug candidates.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (CA96865) and by the Skaggs Institute for Chemical Biology. We thank Dr. Susan S. Taylor (University of California, San Diego) for providing the PKA gene, Jonathan Lansing for NMR analysis of HIF-OH, H. Jane Dyson for a critical reading of the manuscript, Roberto De Guzman for the GB1 DNA template and Leonard Kaljevic, Linda Tennant and Euvel Manlapaz for technical assistance. Plasmids available upon request.
Abbreviations used
- CAD
C-terminal activation domain
- CBP
CREB-binding protein
- CDK2
cyclin-dependent kinase 2
- CREB
cyclic-AMP-response-element-binding protein
- DTT
dithiothreitol
- E. coli
Escherichia coli
- FIH
factor inhibiting HIF-1
- GB1
B1 domain of streptococcal protein G
- HIF-1α
hypoxia-inducible factor-1α
- HIF-OH
hydroxylated HIF-1α
- HSQC
heteronuclear single quantum coherence
- IPTG
isopropyl-1-thio-β-D-galactopyranoside
- KID
kinase inducible domain
- MALDI-TOF
matrix-assisted laser desorption/ionization time-of-flight
- PKA
protein kinase A
- pKID
phosphorylated KID
- RBS
ribosomal binding sites
- SUMO
small ubiquitin-related modifier
- TAZ
transcriptional adapter zinc-binding
- TFA
trifluoroacetic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Graves DJ, Martin BL, Wang JH. Co- and Post-Translational Modification of Proteins: Chemical Principles and Biological Effects. Oxford University Press; New York: 1994. [Google Scholar]
- 2.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 3.Tully T, Bourtchouladze R, Scott R, Tallman J. Targeting the CREB pathway for memory enhancers. Nat Rev Drug Discov. 2003;2:267–277. doi: 10.1038/nrd1061. [DOI] [PubMed] [Google Scholar]
- 4.Bracken CP, Whitelaw ML, Peet DJ. The hypoxia-inducible factors: key transcriptional regulators of hypoxic responses. Cell Mol Life Sci. 2003;60:1376–1393. doi: 10.1007/s00018-003-2370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNeill LA, Hewitson KS, Claridge TD, Seibel JF, Horsfall LE, Schofield CJ. Hypoxia-inducible factor asparaginyl hydroxylase (FIH-1) catalyses hydroxylation at the beta-carbon of asparagine-803. Biochem J. 2002;367:571–575. doi: 10.1042/BJ20021162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Current Opinion in Structural Biology. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 7.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 8.Huth JR, Bewley CA, Jackson BM, Hinnebusch AG, Clore GM, Gronenborn AM. Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Science. 1997;6:2359–2364. doi: 10.1002/pro.5560061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koenig BW, Rogowski M, Louis JM. A rapid method to attain isotope labeled small soluble peptides for NMR studies. Journal of Biomolecular NMR. 2003;26:193–202. doi: 10.1023/a:1023887412387. [DOI] [PubMed] [Google Scholar]
- 10.Demarest SJ, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson HJ, Evans RM, Wright PE. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature. 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- 11.Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci USA. 2002;99:5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennelly PJ, Krebs EG. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. Journal of Biological Chemistry. 1991;266:15555–15558. [PubMed] [Google Scholar]
- 13.Le Bonniec BF, Myles T, Johnson T, Knight CG, Tapparelli C, Stone SR. Characterization of the P2' and P3' specificities of thrombin using fluorescence-quenched substrates and mapping of the subsites by mutagenesis. Biochemistry. 1996;35:7114–7122. doi: 10.1021/bi952701s. [DOI] [PubMed] [Google Scholar]
- 14.Zor T, Mayr BM, Dyson HJ, Montminy MR, Wright PE. Roles of Phosphorylation and Helix Propensity in the Binding of the KIX Domain of CREB-binding Protein by Constitutive (c-Myb) and Inducible (CREB) Activators. Journal of Biological Chemistry. 2002;277:42241–42248. doi: 10.1074/jbc.M207361200. [DOI] [PubMed] [Google Scholar]
- 15.Radhakrishnan I, Pérez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:Coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 16.Grzesiek S, Bax A. The importance of not saturating H2O in protein NMR. Application to sensitivity enhancement and NOE measurements. Journal of the American Chemical Society. 1993;115:12593–12594. [Google Scholar]
- 17.Radhakrishnan I, Pérez-Alvarado GC, Dyson HJ, Wright PE. Conformational preferences in the Ser133-phosphorylated and non-phosphorylated forms of the kinase inducible transactivation domain of CREB. FEBS Letters. 1998;430:317–322. doi: 10.1016/s0014-5793(98)00680-2. [DOI] [PubMed] [Google Scholar]
- 18.Schwarzinger S, Kroon GJA, Foss TR, Wright PE, Dyson HJ. Random coil chemical shifts in acidic 8 M urea: implementation of random coil chemical shift data in NMRView. Journal of Biomolecular NMR. 2000;18:43–48. doi: 10.1023/a:1008386816521. [DOI] [PubMed] [Google Scholar]
- 19.Schwarzinger S, Kroon GJA, Foss TR, Chung J, Wright PE, Dyson HJ. Sequence dependent correction of random coil NMR chemical shifts. Journal of the American Chemical Society. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 20.Bienkiewicz EA, Lumb KJ. Random-coil chemical shifts of phosphorylated amino acids. Journal of Biomolecular NMR. 1999;15:203–206. doi: 10.1023/a:1008375029746. [DOI] [PubMed] [Google Scholar]
- 21.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 22.Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol. 1999;1:438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- 23.Ray S, Zozulya S, Niemi GA, Flaherty KM, Brolley D, Dizhoor A, McKay D, Hurley J, Stryer L. Cloning, expression, and crystallization of recoverin, a calcium sensor in vision. Proc Natl Acad Sci USA. 1992;89:5705–5709. doi: 10.1073/pnas.89.13.5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thurmond JM, Hards RG, Seipelt CT, Leonard AE, Hansson L, Stromqvist M, Bystrom M, Enquist K, Xu BC, Kopchick JJ, Mukerji P. Expression and characterization of phosphorylated recombinant human β-casein in Escherichia coli. Protein Expression Purif. 1997;10:202–208. doi: 10.1006/prep.1997.0737. [DOI] [PubMed] [Google Scholar]
- 25.Acharya A, Xu XJ, Husain-Ponnampalam RD, Hoffmann-Benning S, Kuo MH. Production of constitutively acetylated recombinant p53 from yeast and Escherichia coli by tethered catalysis. Protein Expression Purif. 2005;41:417–425. doi: 10.1016/j.pep.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Liu CC, Schultz PG. Recombinant expression of selectively sulfated proteins in Escherichia coli. Nat Biotechnol. 2006;24:1436–1440. doi: 10.1038/nbt1254. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 28.Uchimura Y, Nakamura M, Sugasawa K, Nakao M, Saitoh H. Overproduction of eukaryotic SUMO-1- and SUMO-2-conjugated proteins in Escherichia coli. Analytical Biochemistry. 2004;331:204–206. doi: 10.1016/j.ab.2004.04.034. [DOI] [PubMed] [Google Scholar]