Abstract
Purpose
To evaluate the feasibility of genetic testing of uveal melanoma using fine-needle aspiration biopsy (FNAB).
Methods
Noncomparative case series of 140 patients in which FNAB was performed immediately prior to plaque radiotherapy. The specimen was sent for genetic analysis using DNA amplification and microsatellite assay for evaluation for monosomy 3.
Results
Monosomy 3 was found in 44 cases (32%) and disomy 3 in 76 cases (54%); genomic DNA yield was insufficient for genetic analysis in 20 cases (14%). Monosomy 3 was found in 26% of small melanomas (16/61), 36% of medium melanomas (24/67), and 33% of large melanomas (4/12). Adequate DNA was achieved in 97% of cases using a 27-gauge needle via transvitreal tumor apex approach and in 75% of cases using a 30-gauge needle via transscleral tumor base approach. Factors predictive of monosomy 3 included greater tumor basal dimension (P = .016) and greater distance from the optic disc (P = .019). Transient localized vitreous hemorrhage was found in 46% of eyes. There was no case of diffuse vitreous hemorrhage, retinal detachment, or tumor recurrence along the biopsy tract.
Conclusions
FNAB provides adequate DNA in most cases for genetic analysis of uveal melanoma using microsatellite assay.
INTRODUCTION
The search for information on host, environmental, and genetic factors in uveal melanoma is well under way.1–2 Most of the published work on genetic testing of uveal melanoma has been performed on enucleated eyes in which a macroscopic sample of tumor was retrieved and studied using one of several techniques.3–10 However, enucleation is typically reserved for eyes with large tumors or those with circumpapillary tumor location, extrascleral tumor extension, secondary glaucoma, extensive retinal detachment, or poor visual prognosis. Currently, most eyes with uveal melanoma are managed with nonenucleation measures, such as plaque radiotherapy or charged-particle radiotherapy. Tissue sampling for genetic testing in this cohort of patients has been preliminarily investigated in a series of 8 patients using transscleral fine-needle aspiration biopsy (FNAB) and fluorescence in situ hybridization (FISH).13 In this report, the feasibility of genetic testing of uveal melanoma in a large cohort of 140 patients is evaluated using microscopic sampling with FNAB immediately prior to plaque radiotherapy and testing using DNA amplification and microsatellite assay.
METHODS
The clinical records of all patients on the Ocular Oncology Service at Wills Eye Hospital, Philadelphia, Pennsylvania, with the diagnosis of uveal melanoma who underwent FNAB for genetic testing for chromosome 3 status between November 1, 2005, and March 1, 2006, were reviewed. Wills Eye Hospital Institutional Review Board issued approval for this retrospective study.
Data were gathered regarding clinical and genetic features of the tumor. The clinical data at initial examination included age, race (African American, Hispanic, Asian, Caucasian), gender (female, male), affected eye (right, left), visual acuity, and symptoms. The tumor data included location (iris, ciliary body, choroid), quadrant location (inferior, temporal, superior, nasal, macula), anteroposterior location (macula, macula-equator, equator-ora, ciliary body, iris), distance to the optic nerve (millimeters), distance to the foveola (millimeters), tumor basal dimension (millimeters), tumor thickness (millimeters by ultrasonography), subretinal fluid, orange pigment on the tumor surface, and previous documented tumor growth. The FNAB parameters included needle gauge, route (transscleral tumor base approach, transvitreal tumor apex approach), and fundus findings immediately after biopsy. Follow-up data included needle biopsy complications and recurrence at the site of biopsy.
TECHNIQUE OF FNAB
The technique of FNAB for genetic analysis of uveal melanoma is shown in Figure 1. After informed patient consent for FNAB and genetic testing was obtained, tissue sampling was performed. A 3-mL blood sample in a purple-topped EDTA tube at room temperature was obtained for isolation of constitutional DNA to be used as a control for comparison to microsatellite alleles detected in tumor DNA. The intraocular tumor sample was obtained at the time of plaque radiotherapy with the patient under retrobulbar anesthesia. After localization of the intraocular tumor and placement of intrascleral nylon sutures before securing the plaque, FNAB was performed.
FIGURE 1.
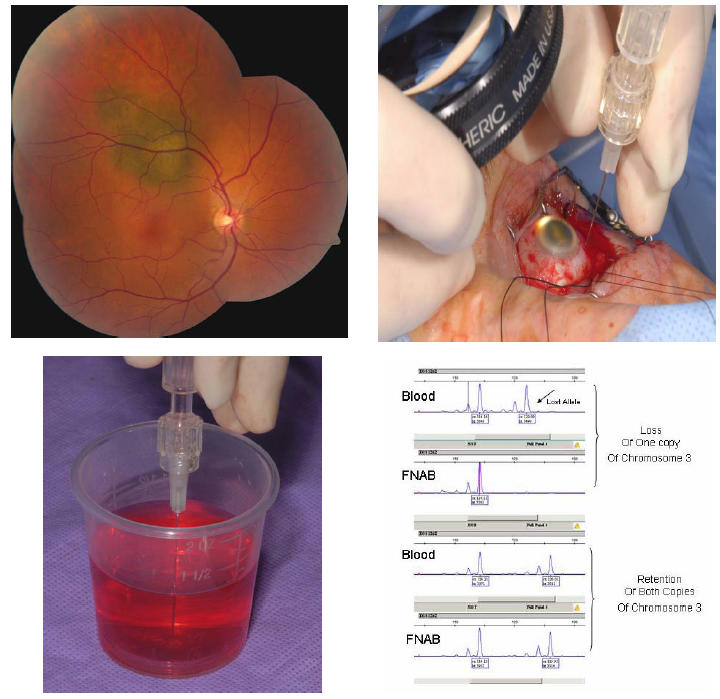
Technique of fine-needle aspiration biopsy (FNAB) for genetic analysis of uveal melanoma. Top left, Small choroidal melanoma with overlying orange pigment and subretinal fluid. Genetic testing confirmed monosomy 3. Top right, At the time of surgery, the tumor is localized and nylon sutures for securing the radioactive plaque are placed in the sclera. Immediately before placement of the radioactive plaque, FNAB is performed through the par plana using a 27-gauge long needle. The tumor sample is taken from an extramacular portion of the tumor, taking care to avoid the major retinal vessels. A small amount of localized vitreous hemorrhage at the site of tumor penetration is expected. Bottom left, The microscopic sample is aspirated into the syringe with pink Hanks solution and then flushed into a test tube for analysis by the genetics laboratory. Bottom right, Microsatellite assay displaying two different samples. The top example shows blood results (normal peripheral blood lymphocyte DNA) with both copies of chromosome 3, but the FNAB of the choroidal melanoma showed loss of 1 copy (monosomy 3). The bottom sample shows blood results with both copies of chromosome 3 and the FNAB of the choroidal melanoma showed retention of both copies (disomy 3).
If the tumor was postequatorial, sampling was done via trans–pars plana, transvitreal approach using a 27-gauge long needle on a connector tubing and a 10-mL syringe.14,15 If the tumor was preequatorial, sampling was performed via transscleral approach using a 30-gauge short needle on a connector tubing and a 10-mL syringe.
For the pars plana approach, the needle was entered along the meridian of the tumor 4 mm posterior to the limbus and directed into the extrafoveal apical portion of the tumor using indirect ophthalmoscopic guidance. For the transscleral approach, the tumor transillumination shadow was outlined on the sclera and the needle was entered nearly perpendicular to the sclera at the tumor base, but with a slight bevel to make the wound self-sealing. The depth of penetration through the sclera depended on the measured tumor thickness, and the needle was aimed to sample the tumor base or midportion.
With both approaches, the needle was held securely for the 10-mL syringe aspiration, and after needle withdrawal, pressure was applied to the globe puncture site with a cotton-tipped applicator. The microscopic cells were aspirated up the needle tip into the syringe using Hanks solution and then flushed into a test tube and stored in refrigeration until analysis by the genetic laboratory. The radioactive plaque was then applied to the eye using standard technique, and the remainder of the procedure was completed.
TECHNIQUE OF GENETIC TESTING
DNA extractions from blood and the FNAB samples were performed using commercially available isolation kits (Qiagen; Valencia, California) following manufacturer-suggested protocols. Polymerase chain reaction (PCR)–based diagnosis for monosomy of chromosome 3 was performed by evaluating 10 polymorphic microsatellite markers on chromosome 3. These markers were purchased from Applied Biosystem Inc (ABI; http://www.appliedbiosystems.com/) human genome mapping kit, v2.5, and used according to the manufacturer’s instructions. The amplification products were analyzed on ABI 3100 fragment analyzer. The data analysis was performed using ABI GeneMapper software v3.0.
STATISTICAL METHODS
The clinical data were then analyzed with regard to the single outcome of presence of monosomy 3. The effect of each individual clinical variable on this outcome was analyzed by Fisher’s exact test and logistic regression analysis. All variables were analyzed as discrete variables except for patient age, tumor base, tumor thickness, proximity to optic disc, and proximity to foveola, which were analyzed as continuous variables. The average age, tumor base, and tumor thickness in eyes with monosomy 3 vs disomy 3 were compared using independent sample t test. The distributions of proximity to optic disc and proximity to foveola were compared using the Wilcoxon rank sum test. Statistical significance was assigned at P < .05.
RESULTS
There were 140 eyes of 140 patients with uveal melanoma sampled for chromosome 3 abnormalities using FNAB. The patient and tumor findings are listed in Table 1. The median patient age at FNAB was 59 years (mean, 59 years; range, 24–3 years). There were 139 Caucasians (99%), 1 Hispanic (<1%), 75 males (54%), and 65 females (46%).
TABLE 1.
MONOSOMY 3 ANALYSIS OF UVEAL MELANOMA IN 140 CASES USING FINE-NEEDLE ASPIRATION BIOPSY (FNAB): CLINICAL FEATURES, BIOPSY APPROACH, AND RESULTS
| FEATURE | ALL PATIENTS NO. (%)n = 140 (100) | MONOSOMY 3 NO. (%)n = 44 (32) | DISOMY 3 NO. (%)n = 76 (54) | QUANTITY NOT SUFFICIENT FOR GENETIC TESTING NO. (%)n = 20 (14) |
|---|---|---|---|---|
| Patient age (yr) median (mean, range) | 59 (59, 24–93) | 60 (60, 24–89) | 57 (58, 26–85) | 61 (61, 42–93) |
| Tumor quadrant | ||||
| Macula | 13 (9%) | 7 (16%) | 6 (8%) | 0 (0%) |
| Inferior | 37 (26%) | 13 (29%) | 15 (20%) | 9 (45%) |
| Temporal | 39 (28%) | 10 (23%) | 22 (29%) | 7 (35%) |
| Superior | 41 (29%) | 11 (25%) | 27 (35%) | 3 (15%) |
| Nasal | 9 (6%) | 3 (7%) | 5 (6%) | 1 (5%) |
| Diffuse | 1 (1%) | 0 (0%) | 1 (2%) | 0 (%) |
| Tumor epicenter | ||||
| Macula | 13 (9%) | 7 (16%) | 6 (8%) | 0 (0%) |
| Macula – equator | 75 (54%) | 18 (41%) | 49 (64%) | 8 (40%) |
| Anterior to equator | 52 (37%) | 19 (43%) | 21 (28%) | 12 (60%) |
| Tumor proximity (mm), median (mean, range) | ||||
| To foveola | 3.5 (5.43, 0–25) | 4.0 (5.6, 0–25) | 3.0 (4.1, 0–25) | 7.0 (9.7, 0–25) |
| To optic disc | 4.0 (5.6, 0–28) | 4.8 (6.0, 0–25) | 2.0 (4.2, 0–28) | 7.2 (10, 2–25) |
| Tumor size (mm) median (mean, range) | ||||
| Basal dimension | 11 (10.6, 3–20) | 12 (11.4, 5–18) | 9.5 (10, 5–18) | 11.5 (11.3, 3–20) |
| Thickness | 3.9 (4.6, 1.6–11.6) | 4.1 (4.9, 2.1–10) | 3.8 (4.6 2–11.6) | 3.1 (4.0, 1.7–8.7) |
| Tumor thickness (mm) | ||||
| 0–3 | 61 (44%) | 16 (36%) | 33 (44%) | 12 (60%) |
| 3–8 | 67 (48%) | 24 (55%) | 38 (50%) | 5 (25%) |
| > 8 | 12 (8%) | 4 (9%) | 5 (6%) | 3 (15%) |
| Tumor features | ||||
| Subretinal fluid | 109 (78%) | 35 (79%) | 64 (84%) | 10 (50%) |
| Orange pigment | 69 (49%) | 23 (52%) | 40 (53%) | 6 (30%) |
| FNAB approach | ||||
| Transvitreal into tumor apex | 67 (48%) | 16 (36%) | 49 (64%) | 2 (10%) |
| Transscleral into tumor base | 73 (52%) | 28 (64%) | 27 (36%) | 18 (90%) |
| FNAB needle size | ||||
| 27 gauge | 67 (48%) | 16 (36%) | 49 (64%) | 2 (10%) |
| 30 gauge | 73 (52%) | 28 (64%) | 27 (36%) | 18 (90%) |
| FNAB complications | ||||
| Vitreous blood localized* | 64 (46%) | 15 (34%) | 48 (63%) | 1 (5%) |
| Vitreous blood diffuse | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Retinal detachment | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Tumor recurrence along tract | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) |
CI, confidence interval.
The vitreous blood spontaneously resolved in all cases.
Preoperative visual acuity was 20/20 to 20/50 in 106 eyes (76%), 20/60 to 20/100 in 15 (11%), and 20/200 or worse in 19 eyes(14%). Patient symptoms included blurred vision in 54 (39%), flashes/floaters in 17 (12%), metamorphopsia in 4 (3%), visual field loss in 10 (7%), color vision loss in 1 (<1%), and pain in 1 (<1%); 53 patients (38%) were asymptomatic. The tumor was located predominantly in the choroid in 129 eyes (92%), ciliary body in 9 (6%), and iris in 2 eyes (1%). The median largest tumor basal dimension measured with ophthalmoscopy, transillumination, and ultrasonography was 11 mm (mean, 10.6 mm; range 3–20 mm), and median tumor thickness by ultrasonography was 3.9 mm (mean, 4.6 mm; range, 1.6–11.6 mm).
The FNAB approach was transscleral at the site of the tumor into the tumor base in 73 cases (52%) and trans–pars plana through the vitreous into the tumor apex in 67 cases (48%). Localized vitreous/subretinal blood at the biopsy site occurred immediately at the time of FNAB in 64 cases (46%) and resolved in all cases. In no case was there extensive intraocular hemorrhage or retinal detachment. There have been no cases of tumor recurrence at the needle biopsy site over a median 8 months of follow-up.
Based on tumor size, adequate yield was found in 49 (80%) of 61 small melanomas 3 mm or less in thickness, in 62 (93%) of 67 medium melanomas between 3 and 8 mm in thickness, and in 9 (75%) of 12 large melanomas 8 mm or greater in thickness. Based on biopsy approach, adequate yield was found in 55 (75%) of 73 cases using the transscleral tumor base approach and 65 (97%) of 67 cases using the transvitreal tumor apex approach. In all cases the yield was microscopic, and no cells were visible after aspiration with the exception of a large necrotic melanoma that yielded visible cellular debris; however, because of extensive tumor necrosis, genetic studies were not possible.
Based on tumor size, monosomy 3 was found in 16 (26%) of 61 small melanomas, 24 (36%) of 67 medium melanomas, and 4 (33%) of 12 large melanomas (Figures 2 and 3). Based on melanoma quadrant location, monosomy 3 was found in 7 (54%) of 13 tumors in the macular region, 13 (35%) of 37 inferiorly, 10 (26%) of 39 temporally, 11 (27%) of 41 superiorly, and 3 (33%) of 9 nasally (Table 1). Based on melanoma anteroposterior location, monosomy 3 was found in 7 of 13 (54%) in the macular region, 18 of 75 (24%) between the macular area and equator, and 19 of 52 (37%) anterior to the equator. Melanomas with monosomy 3 were a median of 4.0 mm from the foveola and 4.8 mm from the optic disc compared to those with disomy 3, which were a median of 3.0 mm from the foveola and 2.0 mm from the optic disc. Melanomas with monosomy 3 had a median basal diameter of 12 mm and a median thickness of 4.1 mm relative to those with disomy 3, which had a median basal diameter of 9.5 mm and a median thickness of 3.8 mm.
FIGURE 2.
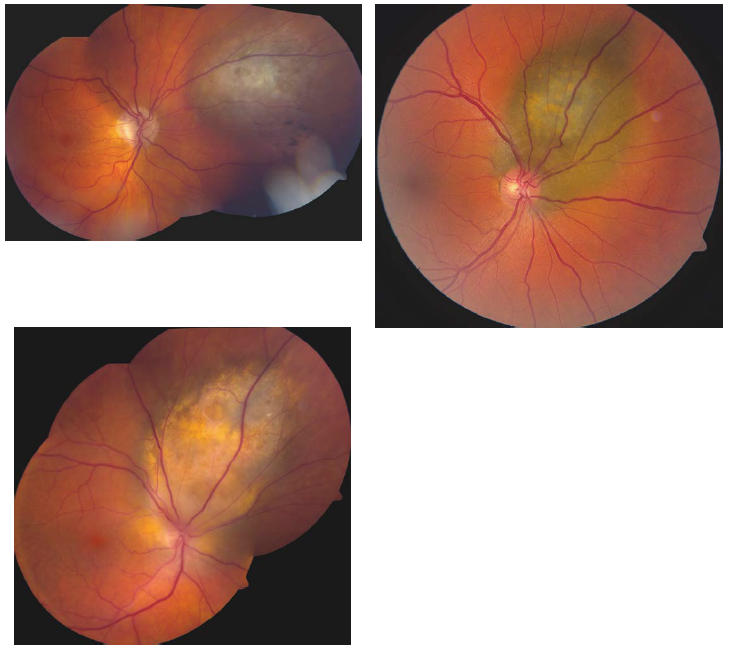
Examples of choroidal melanomas sampled by FNAB that revealed disomy 3.
FIGURE 3.
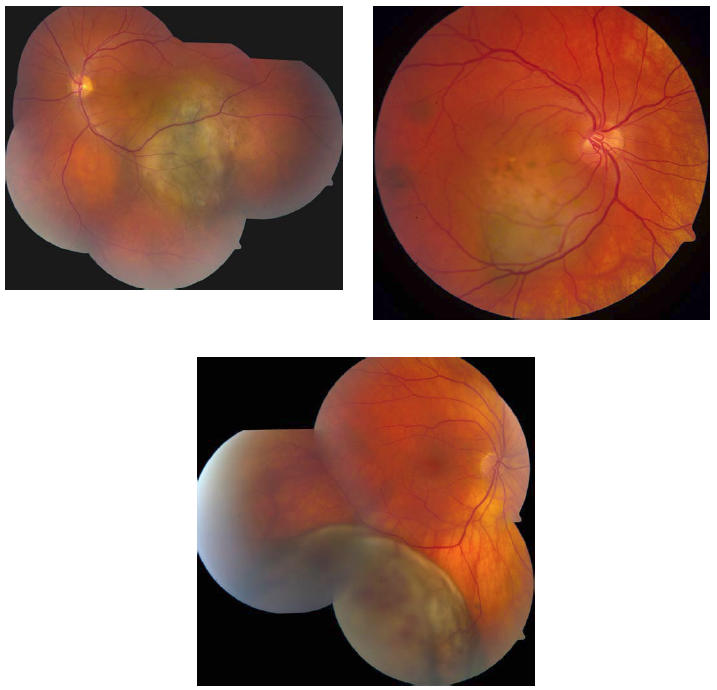
Examples of choroidal melanomas sampled by FNAB that revealed monosomy 3.
Statistical analysis of the impact of each clinical variable on the single outcome of presence of monosomy 3 revealed significant factors of greater basal dimension (OR = 1.17 per every 1-mm increase, P = .016) (Table 2). A trend toward presence of monosomy 3 was found with melanoma location anterior to the equator (OR = 4.54, P = .053) and within the macular region (OR = 3.18, P = .063) (compared to location at macula to equator). The median distance to optic disc was also found to be greater among those with monosomy 3 (P = .019; Wilcoxon rank sum test).
TABLE 2.
MONOSOMY 3 ANALYSIS OF UVEAL MELANOMA IN 140 CASES USING FINE-NEEDLE ASPIRATION BIOPSY (FNAB): FACTORS PREDICTIVE OF MONOSOMY 3*
| VARIABLE | MONOSOM Y 3 (n = 44) | DISOMY 3 (n = 76) | P VALUE† | P VALUE‡ | OR | 95% CI |
|---|---|---|---|---|---|---|
| Univariable Analysis | ||||||
| Age (yr), mean (SD) | 60.2 (12.8) | 58.6 (13.9) | 0.531§ | 0.288 | 1.01¶ | 0.98, 1.04 |
| Race, white vs Hispanic | 44 (100%) | 75 (98.7%) | 1.000 | 1.000 | ||
| Sex, male vs female | 28 (63.6%) | 40 (52.6%) | 0.258 | 0.242 | 1.58 | 0.74, 3.37 |
| Eye, left vs right | 22 (50.0%) | 32 (42.1%) | 0.179 | 0.403 | 1.38 | 0.65, 2.90 |
| Visual acuity, 20/20 vs <20/20 | 12 (27.3%) | 20 (26.3%) | 1.000 | 0.909 | 1.05 | 0.46, 2.43 |
| Symptoms | ||||||
| Floaters vs loss visual field | 1 (2.3%) | 4 (5.3%) | 1.000 | 1.000 | 1.00 | 0.07, 14.64 |
| Asymptomatic vs loss visual field | 15 (34.1%) | 28 (36.8%) | 0.471 | 0.372 | 2.14 | 0.40, 11.40 |
| Blurred vision vs loss visual field | 19 (43.2%) | 29 (39.2%) | 0.301 | 0.254 | 2.62 | 0.50, 13.70 |
| Photopsia vs loss visual field | 4 (9.1%) | 5 (6.6%) | 0.350 | 0.262 | 3.20 | 0.42, 24.42 |
| Metamorphopsia vs loss visual field | 2 (4.5%) | 2 (2.6%) | 0.521 | 0.277 | 4.00 | 0.33, 48.66 |
| Location | ||||||
| Ciliary body vs choroid | 2 (4.8%) | 2 (2.7%) | 0.601 | 0.488 | 2.03 | 0.27, 15.03 |
| Ciliochoroidal vs choroid | 6 (14.3%) | 4 (5.3%) | 0.162 | 0.101 | 3.04 | 0.81, 11.51 |
| Tumor quadrant | ||||||
| Macula vs superior | 7 (15.9%) | 6 (7.9%) | 0.177 | 0.112 | 2.86 | 0.78, 10.47 |
| Inferior vs superior | 13 (29.5%) | 15 (19.7%) | 0.197 | 0.147 | 2.13 | 0.77, 5.91 |
| Temporal vs superior | 10 (22.7%) | 22 (28.9%) | 1.000 | 0.834 | 1.12 | 0.40, 3.11 |
| Nasal vs superior | 3 (6.8%) | 5 (6.6%) | 0.684 | 0.634 | 1.47 | 0.30, 7.25 |
| Anteroposterior location | ||||||
| Macula vs macula to equator | 7 (15.9%) | 6 (7.9%) | 0.098 | 0.063 | 3.18 | 0.94, 10.72 |
| Equator to ora vs macula to equator | 9 (20.5%) | 14 (18.4%) | 0.299 | 0.271 | 1.75 | 0.65, 4.74 |
| Ciliary body + equator to ora vs macula to equator | 5 (11.4%) | 3 (3.9%) | 0.053 | 0.053 | 4.54 | 0.98, 20.95 |
| Ciliary body vs macula to equator | 2 (4.5%) | 2 (2.6%) | 0.314 | 0.334 | 2.72 | 0.36, 20.79 |
| Ciliary body + ora vs macula to equator | 1 (2.3%) | 1 (1.3%) | 0.478 | 0.487 | 2.72 | 0.16, 45.85 |
| Distance optic disc (mm), mean(SD), median | 5.97 (5.8), 4.8 | 4.18 (5.8), 2.0 | 0.019# | 0.122 | 1.05** | 0.99, 1.12 |
| Distance foveola (mm), mean(SD), median | 5.61 (6.3), 4.0 | 4.12 (5.4), 3.0 | 0.214# | 0.184 | 1.05** | 0.98, 1.12 |
| Largest base (mm), mean (SD) | 11.40 (3.4) | 9.93 (2.9) | 0.013§ | 0.016 | 1.17** | 1.03, 1.32 |
| Thickness (mm), mean (SD) | 4.92 (2.4) | 4.64 (2.3) | 0.517§ | 0.514 | 1.06** | 0.90, 1.24 |
| Thickness > 2 mm, positive vs negative | 40 (90.9%) | 67 (88.2%) | 0.088 | 1.000 | ||
| Fluid (negative vs positive) | 5 (11.4%) | 9 (11.8%) | 1.000 | 0.979 | 1.02 | 0.32, 3.27 |
| Symptoms (positive vs negative) | 26 (59.1%) | 47 (61.8%) | 1.000 | 0.948 | 1.03 | 0.46, 2.30 |
| Orange pigment (positive vs negative) | 23 (52.3%) | 40 (52.6%) | 0.844 | 0.782 | 1.13 | 0.51, 2.43 |
| Documented growth (positive vs negative) | 14 (31.8%) | 20 (26.3%) | 0.535 | 0.520 | 1.31 | 0.58, 2.95 |
CI, confidence interval.
Of the 140 samples, 120 had sufficient DNA for analysis and 20 samples were insufficient for analysis.
Fisher’s exact test.
Logistic regression analysis/l.
Independent sample t test.
Per 10-year increase.
Wilcoxon rank sum test.
Per 1-mm increase.
The only complication of needle biopsy was localized transient vitreous hemorrhage at the tumor site in 64 eyes (46%). There was no case of diffuse vitreous hemorrhage, retinal detachment, or tumor recurrence along the biopsy tract.
DISCUSSION
In 1990, Sisley and coworkers5 in England published cytogenetic findings in 6 eyes with posterior uveal melanoma that showed monosomy 3 and 8q abnormalities (n = 3, 50%), chromosome 1 abnormality (n = 2, 33%), and chromosome 6 abnormality (n = 4, 67%). Two years later, Sisley and coworkers6 analyzed 10 cases of uveal melanoma, and abnormalities of chromosomes 3, 6, and 8 were found in 5 cases (50%), chromosome 11 in 3 cases (30%), and chromosome 13 in 2 cases (20%). One tumor showed normal chromosome complement. In 1992, Horsthemke and coworkers3 from Germany found loss of chromosome 3 alleles and multiplication of chromosome 8 alleles in uveal melanoma.
Later, Prescher and coworkers,4 from the same laboratory in Germany, published the prognostic implications of monosomy 3. They evaluated 54 patients enucleated for uveal melanoma and found monosomy 3 in 30 tumors (56%) and disomy 3 in 24 tumors (44%). By 3 years, 50% of the patients with monosomy 3 showed metastasis, whereas those with disomy 3 showed no metastatic disease. They concluded that monosomy 3 was a significant predictor of poor life prognosis. Similar findings were published in 1997 by Sisley and associates,7 who discovered that monosomy 3 and additional copies of 8q statistically correlated with reduced patient survival. Three years later, they recognized that the amount of chromosomal abnormalities increased with increasing tumor size.8
More refined global gene expression patterns of uveal melanoma have been recently studied by Onken and associates9 from the United States using fresh tumor samples taken at the time of enucleation. They performed gene expression microarray analysis of 3075 significant genes in 25 enucleated eyes and found a distinctive separation of uveal melanoma into 2 groups, class 1 (low-grade tumor) in 14 cases (56%) and class 2 (high-grade tumor) in 11 cases (44%). They found that class 2 tumors displayed down-regulated gene clusters on chromosome 3 and up-regulated clusters on chromosome 8q. These findings paralleled those of Sisley and associates.7 Onken and associates9 further evaluated prognostic implications of the 2 classes and found patient survival in 95% of class 1 patients and 31% of class 2 patients at 8 years.
The above studies have been performed on fresh or paraffin-embedded tissue from eyes with melanoma following enucleation. Most of these studies have concluded that loss (monosomy) or down-regulation of chromosome 3 is the most important genetic factor related to prognosis of patients with uveal melanoma. Our analysis specifically focused on our ability to detect abnormalities of chromosome 3 using only an FNAB sample of the tumor without a solid tissue sample. We were able to harvest adequate cells for analysis in 86% of cases, despite the fact that 61 (44%) of 140 melanaomas were 3 mm or less in thickness. Of the 20 cases in which genetic testing was not feasible, most (12) were 3 mm or less in thickness. Of the 61 tumors that were 3 mm or less in thickness, genetic testing was feasible in 49 (80%). We suspect that failure to obtain adequate sample in 20 of our cases was due to one or more reasons, including small tumor size, small needle bore, tightly cohesive spindle cells not yielding to aspiration, necrotic cells without intact DNA, and loss of cells during transfer to test tube. Of the 79 melanomas measuring more than 3 mm in thickness, adequate aspirate was obtained in 71 (90%).
Naus and coworkers16 validated that FISH analysis for uveal melanoma obtained by needle aspirate was reliable. They evaluated 40 eyes with uveal melanoma managed with enucleation. The mean tumor thickness was not evaluated, but the mean tumor diameter was 12.9 mm. After eye removal, a 25-gauge needle attached to a 10-mL syringe was inserted through the sclera into the tumor (transscleral approach), and cells were aspirated. In 39 of 40 eyes (98%), the FISH of both chromosomes 3 and 8 could be analyzed. In 11 of 249 hybridizations, discrepancies between the results of FNAB and solid tumor were detected, but the overall weighted kappa statistic was 0.95, indicating good agreement between the FNAB and solid tumor results. Such discrepancies included mostly differences in the extent of chromosome or subclone abnormality on FNAB vs solid tumor analysis. These investigators concluded that FNAB of uveal melanoma provided sufficient sample and was reliable for FISH analysis for chromosome 3 or 8 abnormalities. Sisley and coworkers17 found similar accurate correlation of cytogenetic analysis in FNAB compared with solid tumor samples following enucleation in 10 cases.
In our series of 140 consecutive cases, FNAB was used at the time of plaque radiotherapy rather than enucleation. In 67 cases (48%) the route was transvitreal using indirect ophthalmoscopy guidance of a 27-gauge needle, whereas in 73 cases (52%) the route was transscleral, directly through the sclera into the tumor base after transillumination, using a 30-gauge needle. We preferred the small needle bore for the transscleral route to minimize possible tumor seeding through the site of scleral perforation. Results were obtained in 65 of 67 melanomas (97%) using the transvitreal route and 55 of 73 melanomas (75%) using the transscleral route. Since most uveal melanomas currently are treated with radiotherapy rather than enucleation, genetic analysis of FNAB specimens provides a method of obtaining important genetic information on all patients with uveal melanoma. For eyes that come to enucleation, fresh tissue can be harvested for the same genetic studies.
In this analysis, greater basal dimension and greater distance from the optic disc were factors associated with monosomy 3. Monosomy 3 was noted in 26% of small melanomas, 36% of medium melanomas, and 33% of large melanomas. These findings could indicate that monosomy 3 mutation develops in the course of tumor enlargement, but further analysis is warranted because this could reflect the difficulty in obtaining sufficient DNA in the smaller tumors.
A microsatellite assay, rather than FISH analysis, was used for our FNAB specimens. The microsatellite assay is more refined and provides more information on chromosomal segments than does FISH analysis.18,19 The microsatellite-based assay is more robust from the inherent amplification of signals due to PCR. The main advantage of this assay is ready adaptability of this test in any molecular biology laboratory. In contrast, FISH requires access to specialized microscopy and operator skills to correctly identify and quantify the signals that infer monosomy vs disomy. For microsatellite-based assays, the analysis software aids in making automated calls for loss of heterozygosity (LOH) and infers monosomy vs disomy. A drawback of the latter assay might be the inability to distinguish between LOH and copy neutral amplification (LOH followed by reduplication of the lost allele). There has been only 1 report of an alternate form of chromosome 3 abnormality for uveal melanoma, and thus this concern is minimal.20
Percutaneous biopsy for prognostication via genetic testing has been found feasible and is employed in other fields of medicine. Teixeira and coworkers21 found that genomic analysis of prostate carcinoma obtained by ultrasound-guided needle biopsy was possible in 34 of 35 cases using chromosome banding analysis and comparative genomic hybridization. They found aberrations in 69% of samples and noted that specific imbalances, such as 16q and 8q abnormalities, imparted worse prognosis. Hoffer and associates22 found that percutaneous needle biopsy in 21 children with neuroblastoma provided genetic prognostic information in 95%, DNA index (ploidy) in 90%, and N-myc gene expression in 70%. They concluded that percutaneous biopsy of advanced neuroblastoma was a feasible alternative to open biopsy. Our study has shown that FNAB of uveal melanoma for genetic information is possible, and this, combined with previous knowledge that needle biopsy specimens correlate with open biopsy specimens, suggests that this technique may be useful in assessing ultimate patient prognosis.
PEER DISCUSSION
DR DENNIS M. ROBERTSON
In this paper from the Wills Ocular Oncology service, the authors investigate the utility of polymerase chain reaction (PCR) technology in an effort to determine something about the chromosomal characteristics of presumed ocular melanoma. They studied 10 polymorphic microsatellite markers on chromosome 3 in cell samples obtained from fine-needle aspirates of melanocytic uveal tumors. In all cases the tumors were managed with radioactive plaque therapy immediately after aspirating the cells. The reference control for the chromosomal assay was the patient’s own blood.
This paper generates several questions and some concerns:
Is an investigation of the chromosomal status of a melanoma using fine-needle aspiration of the tumor in advance of treatment with radiation a good idea? There is evidence from some studies that patients who have in their tumors certain chromosomal abnormalities (including monosomy 3) appear to have higher tumor-related mortality rates than those patients who do not have this abnormality. An investigation attempting to learn about the chromosomal status of a tumor, therefore, seems to be a good idea, particularly if it is safe, if it is not costly, and if it provides useful information.
-
Is it safe?
Is it safe regarding operative complications? Since no significant operative complications were observed in these cases, we can conclude that this investigative approach carries only a small surgical risk.
Is it safe regarding local and systemic long-term outcomes? One concern relates to the possibility of seeding tumor cells along the tract of the needle. Since for anteriorly located tumors the needle tract site is immediately treated with radiation from the episcleral plaque, any cells left along the tract site should be annihilated; therefore, this is probably not a clinically significant problem. Another concern relates to the possibility of long-term complications caused by disruption of tumor tissue when the tumor is invaded with a needle; this concern is not unique to ophthalmology, for as we know, it is common practice to perform needle biopsies in the evaluation of many cancers (breast, for example). Since there are so many other factors that relate to the establishment of metastatic disease, it is likely that needle biopsies do not significantly affect outcomes. However, it is almost certain that we will never have a final answer regarding any effect needle biopsies have on long-term outcomes.
Presumably, the cost of this investigation was borne by research funds.
Is the technique of DNA amplification with the commercial kit valid? Here the authors must rely on the manufacturer’s specification. The authors indicate they evaluated 10 polymorphic microsatellite markers on chromosome 3. There is some question how their data was interpreted. The authors do not provide any information regarding how many of the 10 microsatellite markers demonstrated a monosomy pattern. For example, if one or two of the 10 microsatellite markers were abnormal, how was this considered in the statistics? If all 10 markers were abnormal, does this imply a different level of confidence?
Does the small sample of cells obtained by needle aspirate reflect the general characteristics of the tumor? We all are aware that cellular characteristics within the same tumor may differ greatly from one site to another; benign-appearing spindle cells may be present in one part, whereas highly malignant appearing epithelioid cells may be present in another part.
Do the results reflect the true proportion of tumors associated with monosomy 3? In this study, the authors report that approximately 32% of the eyes demonstrated monosomy 3 in the cellular aspirates. Other studies of choroidal melanoma in enucleated eyes show higher percentages of monosomy 3 (50% to 56%). In this study, it is reasonable to accept that those samples that showed monosomy 3 were obtained from cells aspirated from tumor tissue; however, there is no way of knowing whether the absence of monosomy 3 is because monosomy 3 was verily not present within the tumor or that the absence of monosomy 3 was because cells other than melanoma cells were sampled; for example, a sample of cells from normal uveal tissue or sclera would likely demonstrate disomy 3 (a normal result), so a cell sample showing a normal chromosome 3 could come from tumor tissue or nontumor tissue. Despite extensive experience with fine-needle biopsy, other than obtaining larger tissue samples and performing both histology and DNA amplification, the authors cannot know for certain whether they were studying tumor tissue or nontumor tissue. This problem, coupled with the 20 eyes in which an adequate sample was not retrieved for study, adds further uncertainty to the results. This is especially important if there is any consideration to use the data to correlate with future systemic outcomes for these patients.
How do the authors share the information with their patients?
Whereas in this study the authors used indirect ophthalmoscopy to guide placement of the aspirating needle in tumors located posterior to the equator, I am wondering if they have not considered using the biomicroscope with a wide-angle viewing system. With biomicroscopy and wide-angle viewing, placement of the needle can be made with greater ease and greater precision.
Finally, I believe some of the uncertainties in the results of the present study could be rather easily addressed if the authors conducted a similar study on enucleated eyes with melanoma to learn if PCR technology using cells aspirated from these tumors correlated with complementary studies involving the whole tumor specimen.
ACKNOWLEDGMENTS
Funding/Support: None.
Financial Disclosures: None.
DR. TIM STOUT
With no pertinent financial issues. I do not know how, as you mentioned, you would get perhaps ten or 20 cells and then perform an amplification step. This is just as a technical comment. I do not understand how that amplification step was performed, but perhaps this involved PCR based amplification. A recent study showed chromosomal variation, and for many years we have assumed that if you did a shotgun PCR on genomic DNA you would equally amplify the entire DNA present. The study showed that there is a lot of variability from chromosome to chromosome in terms of how efficient that process amplifies the DNA. I know your paper specifically involves chromosome 3, but one of the other markers was chromosome 8. I remember chromosome 8 as having DNA which, for unknown reasons, was preferentially less amplified than others in shotgun PCR approach. This is just a word of caution.
DR. JAMES J. AUGSBURGER
No financial conflict. According to Bill Harbour and coworkers, approximately 15% of the cases they have studied by both karyotyping and gene expression profiling have had isodisomy 3. These cases would not show up on microsatellite analysis. This suggests that it may be too early to celebrate the favorable survival prognosis of those patients who you interpreted as having disomy 3 in your study.
DR. DOUBLAS A. JABS
Obviously the ultimate goal of this investigation is to study and to determine if this method is clinically useful and whether or not it would have predictive value in enucleated specimens. No clinical data were included, but do you have any clinical information on the outcome of the patients who did not have the chromosomal 3 abnormalities?
DR. CAROL L. SHIELDS
I would like to thank all of you for your comments. Regarding Dr. Robertson, he mentioned that we have a procedure that should be safe, not costly, and provide useful information. I think we adequately explained that. This is safe and we do not have many complications. We have never seen seeding of a tumor in association with a needle biopsy tract in the more than 700 procedures that we have performed for cytogenetic studies or for cytology. One report in the literature documents seeding through a needle tract in which surgeons had used infusion through a 25 gauge needle. Perhaps the intraocular pressure associated with the infusion might promote some new cells to pass through the needle tract, but we have never seen that. He asked questions about the ten markers. I am not a geneticist, but a clinician like most of you and work with a genetics team. I basically obtain the cells and hand them over to a genetics team. As I understand it, from studying the ten markers they can identify in their microsatellite assay if any are missing, such as one marker or three markers. If some are absent, then we call that “partial monosomy”. If all ten markers are missing we call that “complete monosomy 3”. In this analysis anybody who had partial or complete monosomy 3 was included in our monosomy 3 group. Future studies will be published from the genetics side and help explain what partial monosomy 3 means. Future studies will examine the implications regarding prognosis of patients with partial or complete monosomy 3. The question that everyone asks about needle biopsy relates to sampling error. Are we obtaining cells that represent what is in the tumor? You know, in all fields of medicine needle biopsies are performed. For example, when you do a needle biopsy of a prostate tumor, a breast tumor, a thyroid tumor, or an eye tumor, the same question arises: are you getting representative tissue? We know that melanomas are heterogeneous, but there have been three good published studies on the correlation of genetic composition of uveal melanoma obtained with needle biopsy and the gross specimen.” Sisley did it, Damato performed it, and I mentioned in my talk that Naus et al from the Netherlands found good representation in the needle biopsy specimen compared to the gross specimen. Although there may be a risk of sampling error, I believe that we are getting representative tissue. Others have asked questions regarding the clinical usefulness of this investigation. I think it is too early in course of needle biopsy for genetics evaluation to predict clinical outcomes, as it has only been around since November 2005. With melanoma, as you all know, we needed at least 10 years of follow-up to correlate our findings with prognosis. Dr. Stout asked about amplification of the cells. We did use PCR and I know there are inherent problems with PCR. Lastly Dr. Augsburger mentioned the question of isodisomy and, as I am not a geneticist, I really cannot accurately answer that question. I am sure that Dr. Ganguly, a PhD who does our genetics can provide the answer.
ACKNOWLEDGMENTS
Funding/Support: This study was supported by the Retina Research Foundation Award of the Retina Society, Capetown, South Africa (C.L.S.); the Paul Kayser International Award of Merit in Retina Research, Houston Texas (J.A.S.); a donation from Michael, Bruce, and Ellen Ratner, New York, New York (J.A.S., C.L.S.); Mellon Charitable Giving from the Martha W. Rogers Charitable Trust, Philadelphia, Pennsylvania (C.L.S.); the LuEsther Mertz Retina Research Foundation, New York, New York (C.L.S.); and the Eye Tumor Research Foundation, Philadelphia (C.L.S., J.A.S.). Financial Disclosures: None.
Author Contributions: Dr Carol L. Shields has had full access to all of the data in this study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Other Acknowledgments: Statistical analysis was provided by Rishita Nutheti, International Centre for Advancement of Rural Eye Care, L. V. Prasad Eye Institute, Hyderabad, India.
REFERENCES
- 1.Weis E, Shah CP, Lajous M, Shields JA, Shields CL. The association between host susceptibility factors and uveal melanoma: a meta-analysis. Arch Ophthalmol. 2006;124:54–60. doi: 10.1001/archopht.124.1.54. [DOI] [PubMed] [Google Scholar]
- 2.Shah CP, Weis E, Lajous M, Shields JA, Chields CL. Intermitttent and chronic ultraviolet light exposure and uveal melanoma: a meta-analysis. Ophthalmology. 2005;112:1599–1607. doi: 10.1016/j.ophtha.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 3.Horsthemke B, Prescher G, Bornfeld N, Becher R. Loss of chromosome 3 alleles and multiplication of chromosome 8 alleles in uveal melanoma. Genes Chromosomes Cancer. 1992;4:217–221. doi: 10.1002/gcc.2870040305. [DOI] [PubMed] [Google Scholar]
- 4.Prescher G, Bornfeld N, Hirche H, Horsthemke B, Jockel KH, Becher R. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996;347:1222–1225. doi: 10.1016/s0140-6736(96)90736-9. [DOI] [PubMed] [Google Scholar]
- 5.Sisley K, Rennie IG, Cottam DW, Potter AM, Potter CW, Rees RC. Cytogenetic findings in six posterior uveal melanomas: involvement of chromosomes 3, 6, and 8. Genes Chromosomes Cancer. 1990;2:205–209. doi: 10.1002/gcc.2870020307. [DOI] [PubMed] [Google Scholar]
- 6.Sisley K, Cottam DW, Rennie IG, et al. Non-random abnormalities of chromosomes 3, 6, and 8 associated with posterior uveal melanoma. Genes Chromosomes Cancer. 1992;5:197–200. doi: 10.1002/gcc.2870050304. [DOI] [PubMed] [Google Scholar]
- 7.Sisley K, Rennie IG, Parsons MA, et al. Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes Chromosomes Cancer. 1997;19:22–28. doi: 10.1002/(sici)1098-2264(199705)19:1<22::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 8.Sisley K, Parsons MA, Garnham J, et al. Association of specific chromosome alteration with tumour phenotype in posterior uveal melanoma. Br J Cancer. 2000;82:330–338. doi: 10.1054/bjoc.1999.0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes S, Damato BE, Giddings I, Hiscott PS, Humphreys J, Houlston RS. Microarray comparative genomic hybridisation analysis of intraocular uveal melanomas identifies distinctive imbalances associated with loss of chromosome 3. Br J Cancer. 2005;93:1191–1196. doi: 10.1038/sj.bjc.6602834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harbour JW. Eye cancer: unique insights into oncogenesis: the Cogan Lecture. Invest Ophthalmol Vis Sci. 2006;47:1736–1745. doi: 10.1167/iovs.05-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ehlers JP, Harbour JW. Molecular pathobiology of uveal melanoma. Int Ophthalmol Clin. 2006;46:167–180. doi: 10.1097/01.iio.0000195855.31324.db. [DOI] [PubMed] [Google Scholar]
- 13.Midena E, Bonaldi L, Parrozzani R, Tebaldi E, Boccassini B, Vujosevic S. In vivo detection of monosomy 3 in eyes with medium-sized uveal melanoma using transscleral fine needle aspiration biopsy. Eur J Ophthalmol. 2006;16:422–425. doi: 10.1177/112067210601600310. [DOI] [PubMed] [Google Scholar]
- 14.Augsburger JJ, Shields JA. Fine needle aspiration biopsy of solid intraocular tumors. Trans Pa Acad Ophthalmol Otolaryngol. 1983;36:169–172. [PubMed] [Google Scholar]
- 15.Shields JA, Shields CL, Ehya H, Eagle RC, Jr, De Potter P. Fine-needle aspiration biopsy of suspected intraocular tumors. The 1992 Urwick Lecture. Ophthalmology. 1993;100:1677–1684. doi: 10.1016/s0161-6420(93)31418-1. [DOI] [PubMed] [Google Scholar]
- 16.Naus NC, Verhoeven AC, van Drunen E, et al. Detection of genetic prognostic markers in uveal melanoma biopsies using fluorescence in situ hybridization. Clin Cancer Res. 2002;8:534–539. [PubMed] [Google Scholar]
- 17.Sisley K, Nichols C, Parsons MA, Farr R, Rees RC, Rennie IG. Clinical applications of chromosome analysis, from fine needle aspiration biopsies, of posterior uveal melanomas. Eye. 1998;12(Pt 2):203–207. doi: 10.1038/eye.1998.48. [DOI] [PubMed] [Google Scholar]
- 18.Cross NA, Ganesh A, Parpia M, Murray AK, Rennie IG, Sisley K. Multiple locations on chromosome 3 are the targets of specific deletions in uveal melanoma. Eye. 2006;20:476–481. doi: 10.1038/sj.eye.6701906. [DOI] [PubMed] [Google Scholar]
- 19.Hausler T, Stang A, Anastassiou G, et al. Loss of heterozygosity of 1p in uveal melanomas with monosomy 3. Int J Cancer. 2005;116:909–913. doi: 10.1002/ijc.21086. [DOI] [PubMed] [Google Scholar]
- 20.White VA, McNeil BK, Horsman DE. Acquired homozygosity (isodisomy) of chromosome 3 in uveal melanoma. Cancer Genet Cytogenet. 1998;102:40–45. doi: 10.1016/s0165-4608(97)00290-2. [DOI] [PubMed] [Google Scholar]
- 21.Teixeira MR, Ribeiro FR, Eknaes M, et al. Genomic analysis of prostate carcinoma specimens obtained via ultrasound-guided needle biopsy may be of use in preoperative decision-making. Cancer. 2004;101:1786–1793. doi: 10.1002/cncr.20527. [DOI] [PubMed] [Google Scholar]
- 22.Hoffer FA, Chung T, Diller L, Kozakewich H, Fletcher JA, Shamberger RC. Percutaneous biopsy for prognostic testing of neuroblastoma. Radiology. 1996;200:213–216. doi: 10.1148/radiology.200.1.8657913. [DOI] [PubMed] [Google Scholar]


