Abstract
The cohesin complex is a chromosomal component required for sister chromatid cohesion that is conserved from yeast to man. The similarly conserved Nipped-B protein is needed for cohesin to bind to chromosomes. In higher organisms, Nipped-B and cohesin regulate gene expression and development by unknown mechanisms. Using chromatin immunoprecipitation, we find that Nipped-B and cohesin bind to the same sites throughout the entire non-repetitive Drosophila genome. They preferentially bind transcribed regions and overlap with RNA polymerase II. This contrasts sharply with yeast, where cohesin binds almost exclusively between genes. Differences in cohesin and Nipped-B binding between Drosophila cell lines often correlate with differences in gene expression. For example, cohesin and Nipped-B bind the Abd-B homeobox gene in cells in which it is transcribed, but not in cells in which it is silenced. They bind to the Abd-B transcription unit and downstream regulatory region and thus could regulate both transcriptional elongation and activation. We posit that transcription facilitates cohesin binding, perhaps by unfolding chromatin, and that Nipped-B then regulates gene expression by controlling cohesin dynamics. These mechanisms are likely involved in the etiology of Cornelia de Lange syndrome, in which mutation of one copy of the NIPBL gene encoding the human Nipped-B ortholog causes diverse structural and mental birth defects.
Introduction
Development of higher organisms requires tissue-specific activation and silencing of genes. Tissue-specific regulation is often mediated by sequences located several kilobases away from a gene and the combined actions of transcriptional activators, silencing proteins, and factors that modify chromatin structure. Studies in Drosophila reveal that chromosomal proteins required for sister chromatid cohesion also play critical roles in control of gene expression during development. The Drosophila Nipped-B protein was discovered in a screen for factors that facilitate expression of the cut homeobox gene in the developing wing margin that is driven by a distant transcriptional enhancer located more than 80 kb upstream of the transcription start site (Rollins et al. 1999). Nipped-B is essential, and homozygous Nipped-B mutants die as second instar larvae, while heterozygous Nipped-B mutations decrease expression of the cut and Ultrabithorax (Ubx) genes. These two genes and some unknown developmental processes are exquisitely sensitive to Nipped-B dosage: heterozygous Nipped-B null mutations reduce Nipped-B messenger RNA (mRNA) levels by only 25% and a 50% reduction induced by RNAi is lethal (Rollins et al. 2004).
Homozygous Nipped-B mutants show sister chromatid cohesion defects just before death as the maternally provided Nipped-B wanes (Rollins et al. 2004). Studies on Nipped-B orthologs in other organisms indicate that these defects result from a failure of the cohesin protein complex that mediates cohesion to bind to chromosomes (Arumugam et al. 2003; Ciosk et al. 2000; Gillespie and Hirano 2004; Seitan et al. 2006; Takahashi et al. 2004; Tomonaga et al. 2000; Watrin et al. 2006).
Cohesin binds to chromosomes throughout interphase when gene expression occurs. It contains four subunits, Smc1, Smc3, Rad21, and Stromalin (SA), which form a ring-like structure (reviewed in Hirano 2006; Huang et al. 2005; Losada 2007; Nasmyth and Haering 2005). In most organisms, cohesin is loaded along chromosomes during telophase and is removed from the arms at the subsequent prophase. A leading idea is that cohesin mediates cohesion by encircling both sister chromatids (Ivanov and Nasmyth 2005, 2007), although other mechanisms may occur at some locations (Chang et al. 2005; Dubey and Gartenberg 2007).
Heterozygous Nipped-B mutants do not show cohesion defects, indicating that their effects on gene expression are unlikely to be caused by a significant reduction in binding of cohesin to chromosomes. Changes in cohesin dosage, however, also affect cut expression, suggesting that Nipped-B's role in gene expression involves its ability to regulate cohesin binding. Although Nipped-B and cohesin are both needed for sister chromatid cohesion, they have opposite effects on cut expression. Reducing cohesin dosage increases cut expression in the developing wing margin while reducing Nipped-B decreases expression (Rollins et al. 1999; Rollins et al. 2004; Dorsett et al. 2005). This gave rise to the idea that cohesin binds to cut and inhibits expression, possibly by interfering with enhancer–promoter communication, and that Nipped-B maintains a dynamic cohesin-binding equilibrium to alleviate these effects (Dorsett 2004). Consistent with this idea, cohesin binds directly to cut regulatory sequences in cultured cells and to the cut locus in salivary gland chromosomes (Dorsett et al. 2005).
Cornelia de Lange syndrome (CdLS) is caused by heterozygous loss-of-function mutations in the Nipped-B-Like (NIPBL) ortholog of Nipped-B and, in a few cases, by viable missense mutations in the Smc1A or Smc3 cohesin subunit genes (Deardorff et al. 2007; Krantz et al. 2004; Musio et al. 2006; Tonkin et al. 2004). CdLS patients display slow growth, mental retardation, and defects in limbs and organs (Dorsett 2007; Jackson et al. 1993; Strachan 2005). Most do not show cohesion defects (Kaur et al. 2005; Vrouwe et al. 2007), suggesting that the diverse developmental deficits are caused by gene expression changes similar to those in Drosophila. The similar effects of reduced NIPBL activity and cohesin subunit missense mutations on human development in the absence of obvious effects on sister chromatid cohesion further suggest that Nipped-B/NIPBL are likely to dynamically regulate cohesin.
Another potential link between the effects of sister chromatid cohesion factors on development and effects on gene expression is provided by the finding that mice homozygous for a knockout of the Pds5B gene show developmental deficits reminiscent of some that occur in CdLS patients (Zhang et al. 2007). The Pds5 protein, which is also conserved from fungi to man, interacts with cohesin and plays roles in establishment and/or maintenance of sister chromatid cohesion (Dorsett et al. 2005; Hartman et al. 2000; Losada et al. 2005; Panizza et al. 2000; Stead et al. 2003; Sumara et al. 2007; Tanaka et al. 2001). In mammals, there are two Pds5 proteins, and the mice lacking Pds5B that show developmental abnormalities do not have cohesion defects. In Drosophila, there is a single pds5 gene, and heterozygous pds5 mutations alter cut gene expression without the effects on cohesion seen in homozygous mutants (Dorsett et al. 2005), suggesting that changes in gene expression also likely underlie the effects of Pds5B on mouse development.
The binding of cohesin and the Scc2 ortholog of Nipped-B have been mapped genome-wide in the yeast Saccharomyces cerevisiae (Glynn et al. 2004; Lengronne et al. 2004). Cohesin binds almost exclusively between genes in yeast, and most binding sites are between convergent transcription units. Coupled with the finding that Scc2 does not colocalize with cohesin, this led to the idea that cohesin loads onto chromosomes at Scc2 binding sites and then is pushed to the ends of genes by RNA polymerase (Glynn et al. 2004; Lengronne et al. 2004).
The intergenic localization of cohesin in yeast, where it rarely overlaps regulatory sequences, and the lack of co-localization with Scc2, which is inconsistent with dynamic control by Scc2, are incompatible with the models for how Nipped-B/NIPBL and cohesin regulate Drosophila gene expression. The yeast genome, however, is much more compact than that of higher eukaryotes, with smaller intergenic regions, few introns, and rare long-range regulation. Thus, the mechanisms that determine the location of cohesin binding sites are likely to differ in higher organisms. We mapped the Nipped-B and cohesin binding sites in the entire non-repetitive Drosophila genome to gain insights into how they interact with genes. Strikingly, we find that in contrast their orthologs in yeast, Nipped-B and cohesin colocalize and bind preferentially, but not exclusively, to active transcription units.
Materials and methods
Chromatin immunoprecipitation was performed as described before (Dorsett et al. 2005; Schwartz et al. 2006). Nipped-B, SA, and Smc1 antisera are described elsewhere (Dorsett et al. 2005; Gause et al. 2007). RNA polymerase II (PolII) antibody was purchased from Babco (MMS-126R). For controls, we precipitated with Smc1 preimmune serum or rabbit IgG or used input chromatin. Hybridization of probes prepared from the immune precipitated or input chromatin to tiled microarrays (Affymetrix no. 511262) was performed according to the manufacturer's directions. At least two independent precipitations using different chromatin preparations were used to probe separate microarrays for each protein. All experiments used at least two control hybridizations.
Trimmed mean log2 IP/control ratios for microarray features were calculated from the IP and control hybridization intensities using sliding 675-bp windows with the TiMAT programs (http://bdtnp.lbl.gov/TiMAT/TiMAT2/index.html). TiMAT was also used to predict binding peaks and regions at 1 and 25% false discovery rates. Data was viewed with the Affymetrix browser (www.affymetrix.com/support/developer/tools/download_igb.affx) and the April 2004 Drosophila annotated genome (Celniker et al. 2002; Berkeley Drosophila Genome Project, personal communication). The R statistical environment (R Foundation for Statistical Computing, Vienna, 2007; ISBN 3-900051-07-0; www.R-project.org) was used to calculate correlation coefficients, plot log2 IP/control values for microarray features and identify genes that differentially bind PolII, cohesin, and Nipped-B.
Results
Nipped-B and cohesin colocalize genome-wide
We mapped binding sites for Nipped-B, the Smc1 and SA cohesin subunits, and RNA polymerase II (PolII) in the entire non-repetitive genome of Drosophila using chromatin immunoprecipitation and hybridization of the precipitated DNA to tiled microarrays (ChIP-chip), as described previously for Polycomb group (PcG) proteins (Schwartz et al. 2006). We used cultured cells instead of whole organisms because cell lines should have less binding site heterogeneity. Three lines were used to look for differences in cohesin binding patterns. Two lines, the Sg4 subline (Schwartz et al. 2006) of Schneider line 2 and the Kc167 subline of Kc cells (Echalier and Ohanessian 1970), are embryonic in origin, and the ML-DmBG3 line (BG3; Ui et al. 1994) is derived from third instar central nervous system. The Affymetrix tiled microarray contains some 3×106 25-nt oligonucleotide features every 35 bp or so. Sliding windows of 675 bp were used to generate trimmed mean log2 IP/control ratios for the features, and statistical algorithms were used to predict binding peaks and regions at 1 and 25% false discovery rates.
We examined binding of Nipped-B, Smc1, and SA in Sg4 cells, Nipped-B and Smc1 in BG3 cells, and Smc1 in Kc cells. The genome-wide pattern for cohesin binding is very similar but not identical in all three cell lines, as shown by the maps for Smc1 binding in Fig. 1. In general, the cohesin-binding regions range from a few kilobases up to 100 kb or so in width, with large gaps between them. Most of the gaps are several kilobases in size but can extend to hundreds of kilobases. The largest gaps are about a megabase in size, such as the region around 15 Mb on chromosome 2L map and the region around 1 Mb on chromosome 2R (Fig. 1). Most of the larger gaps are in regions with low gene density.
Fig. 1.
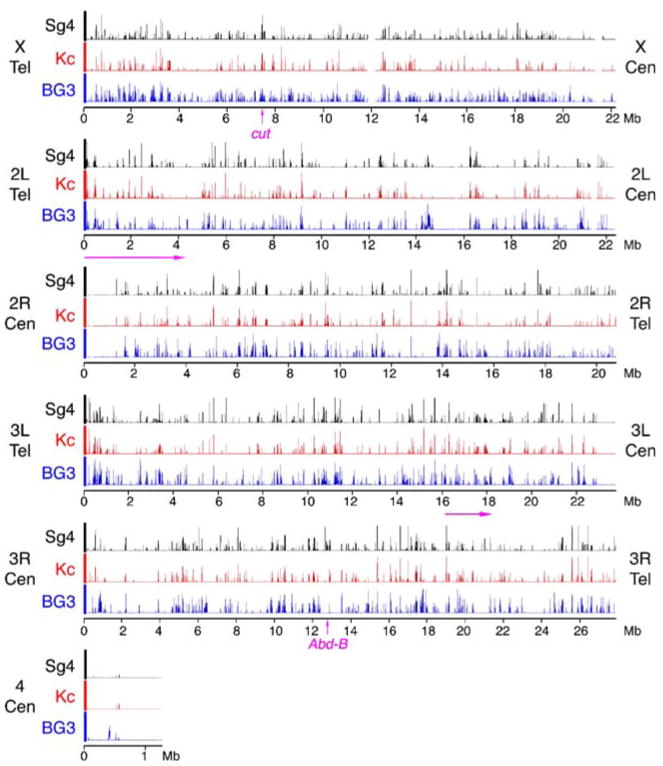
Binding of the Smc1 cohesin subunit to the non-repetitive genome in Sg4, Kc, and ML-DmBG3 (BG3) Drosophila cells as determined by chromatin immunoprecipitation. The three tracks for each chromosome arm plot the trimmed mean log2 IP/control ratios for Smc1 for Sg4 (black), Kc (red) and BG3 (blue) cells on a scale from 0.5 to 2.5. The positions of the telomeres (Tel) and centromeres (Cen) are indicated for each arm. The positions of the 2-Mb region of chromosome arm 3L shown in Fig. 2, and the 4.25 Mb region plotted in Fig. 3 are indicated with arrows underneath the map, and the positions of the cut and Abd-B genes shown in detail in other figures are indicated on the X and 3R maps. These data show that the cohesin binding patterns are very similar but not identical between the three cell lines, and that with the exception of the small chromosome 4, cohesin binds many regions along the chromosome arms. There are also large cohesin-free regions extending up to a megabase or so in size, including those around 15 Mb on the map of the chromosome 2L arm and 1 Mb on the 2R map. Many large cohesin-poor regions have low gene density
The microarrays lack repetitive sequences, so the centromeres and telomeres are not included in the data. While the non-repetitive regions flanking the telomeres shows some cohesin binding on all chromosomes, the centromere-flanking regions on chromosome arms 2R and 3L are cohesin-poor (Fig. 1). Chromosome 4, which is only a little over a megabase in size and contains substantial interspersed repetitive sequences, is also cohesin-poor (Fig. 1). These results do not mean, however, that repetitive sequences are generally cohesin-poor. By immunostaining, cohesin binds the pericentric heterochromatin of both mitotic and meiotic chromosomes in Drosophila (Warren et al. 2000; Valdeolmillos et al. 2004; Khetani and Bickel 2007; Gause et al. 2007).
By visual inspection, the patterns for Nipped-B and cohesin binding are nearly identical. Figure 2 shows a 2-Mb region of chromosome 3L that illustrates several typical features. The Nipped-B, SA, and Smc1 binding patterns in Sg4 cells are very similar, as are the Nipped-B and Smc1 patterns in BG3 cells. Nipped-B and cohesin bind to regions that range in size from a kilobase or so to more than 60 kb in length. Near the middle of the region of chromosome 3L shown in Fig. 2, there are two long cohesin-free regions, each some 200 kb in size, separated by a small cohesin peak. Much of this cohesin-poor region has a low gene density.
Fig. 2.
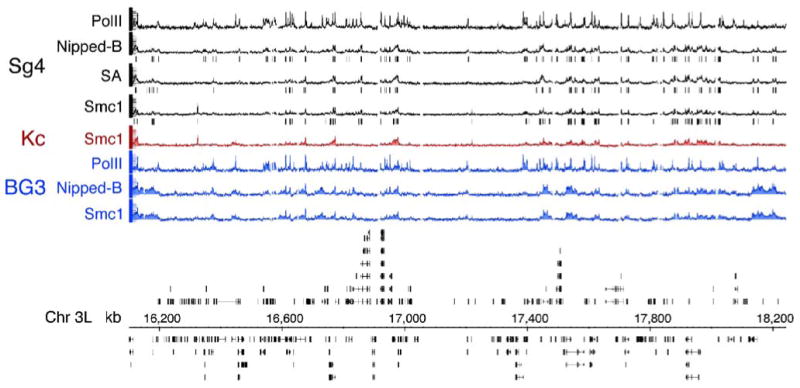
Binding of Nipped-B, cohesin subunits, and PolII to a 2 Mb region of chromosome 3L. This region was chosen to illustrate typical features of cohesin and Nipped-B binding patterns seen throughout the genome. The eight tracks at the top graph the trimmed mean log2 IP/control ratios for the microarray features on a scale of −0.5 to 3.0. The top four tracks (black) show RNA polymerase II (PolII), Nipped-B, and SA and Smc1 cohesin subunit binding in Sg4 cells. The red track shows Smc1 binding in Kc cells, and the three blue tracks show the PolII, Nipped-B, and Smc1 binding in BG3 cells. The vertical lines underneath the Sg4 Nipped-B, SA, and Smc1 tracks indicate microarray features predicted by TiMAT analysis to be binding peaks with a 1% false discovery rate. The annotated map of the chromosome 3L region (April 2004 release of the Drosophila melanogaster genome; Celniker et al. 2002; Berkeley Drosophila Genome Project, personal communication) is shown below the ChIP tracks. Black boxes indicate exons and lines indicate introns. The key features to note are that Nipped-B binding is virtually identical to that of the SA and Smc1 cohesin subunits, that the cohesin/Nipped-B binding patterns are very similar but not identical between the three cells lines, and that cohesin binds large regions ranging in size from a kilobase or so to more than 60 kb in length. There are also large regions, such as the 400 kb gene-poor domain near the middle of the graph that are nearly devoid of cohesin and Nipped-B
The peaks of Nipped-B and cohesin subunit binding sites predicted with a 1% false discovery rate are marked by vertical lines for Sg4 cells in Fig. 2. The predicted peaks are very similar for Nipped-B, SA, and Smc1, providing evidence for co-localization of Nipped-B and cohesin. Co-localization is also indicated by other analysis methods. Comparing the trimmed mean log2 IP/control values, the genome-wide correlation coefficient for Nipped-B and SA binding in Sg4 cells is 0.88, the Nipped-B-Smc1 correlation is 0.75, and the SA-Smc1 correlation is 0.71 (Table 1). In BG3 cells, the Nipped-B-Smc1 correlation is 0.92. To control for the possibility that systematic low-level signals might inflate the correlation, we compared Nipped-B in Sg4 cells to the randomly chosen Knirps protein binding in embryos measured using the same method (X.Y.L, M.D.B, unpublished). This gives a correlation of 0.11, indicating that systematic low-level signals do not make a significant contribution (Table 1).
Table 1.
Genome-wide correlation coefficients for protein binding
| Proteina | Nipped-B-Sg4 | SA-Sg4 | Smc1-BG3 | Smc1-Kc | PolII-BG3 |
|---|---|---|---|---|---|
| Smc1-Sg4 | 0.75b | 0.71 | 0.47 | 0.59 | − |
| SA-Sg4 | 0.88 | − | − | − | − |
| Nipped-B-BG3 | 0.56 | − | 0.92 | − | 0.51 |
| Smc1-BG3 | − | − | − | 0.55 | − |
| PolII-Sg4 | 0.62 | − | − | − | − |
| H3K27Me3-Sg4c | −0.30 | − | − | − | − |
| Knirps-embryod | 0.11 | − | − | − | − |
Indicates the protein and cell type: Sg4, Kc, BG3 (ML-DmBG3), or embryo
Correlation coefficients were calculated using the trimmed mean log2 IP/control ratio values for the microarray features.
Histone H3 lysine 27 trimethylation (Schwartz et al. 2006)
X.Y. Li and M.D. Biggin, unpublished
Plots of the Nipped-B vs SA or Smc1 trimmed mean log2 values provide evidence that the Nipped-B and cohesin peaks closely align with each by showing that individual array oligos have similar enrichment for Nipped-B and cohesin. Figure 3 shows these plots for the first 4.25 Mb of chromosome 2L in Sg4 and BG3 cells. Plots of other regions for both Sg4 and BG3 cells are very similar. A few Smc1 sites in Sg4 cells do not correlate with SA or Nipped-B (Fig. 3). We do not know if these are authentic or sites for a protein that cross-reacts with the Smc1 antibodies. However, the high correlation between Nipped-B and cohesin and between cohesin subunits indicates that the vast majority of sites are authentic and that Nipped-B and cohesin bind the same sites.
Fig. 3.
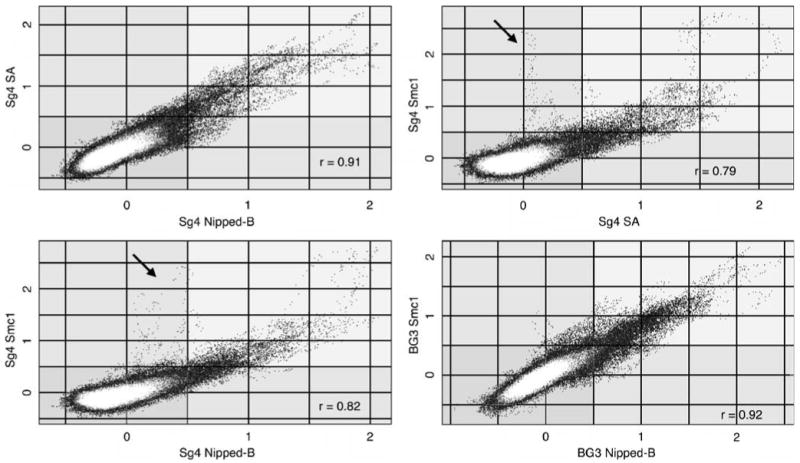
Colocalization of Nipped-B and cohesin subunit binding sites. The plots compare enrichment values for the SA and Smc1 cohesin subunits and Nipped-B at individual microarray features. The trimmed mean log2 IP/control values of 1×105 microarray features for chromosome 2L extending from nt 5,522–4,254,929 (Fig. 1, chromosome 2L, 0.055 to 4.25 Mb) for the indicated proteins and cells are plotted against each other. Plots of similar-sized regions across the genome are very similar. The correlation coefficients (r) for the plotted region are given, which are similar to those calculated for the entire non-repetitive genome (Table 1). Nipped-B, SA, and Smc1 values for individual microarray features correlate well with each other except for a few Smc1-positive features that are low for SA and Nipped-B in Sg4 cells (arrows), indicating that the Nipped-B and cohesin subunit peaks closely align with each other. The white masses centered close to log2 values of 0 for both proteins represent the majority of features that have low binding for both proteins.
Genome-wide correlation coefficients and plots indicate that, although the similarities are predominant, there are also significant differences in cohesin binding between Sg4, Kc, and BG3 cells. The genome-wide correlation for Smc1 binding between Sg4 and Kc cells is 0.59, and although plots reveal similar enrichment for many microarray features between the two cells indicating that many peaks occur in the same positions in the different cell types, there are also features that differ in binding (Fig. 4; Table 1). Similar correlation coefficients and plots are obtained when Nipped-B binding is compared between Sg4 and BG3 cells, or when Smc1 binding is compared between Sg4 and BG3 cells and between BG3 and Kc cells, indicating that there are similar differences in cohesin binding between all three cells lines (Table 1; Fig. 4). As described below, many of these binding differences occur within genes. An example of a difference in cohesin binding between Sg4 and BG3 cells is shown in Supplementary Fig. 1, and several differences are catalogued in Supplementary Table 1.
Fig. 4.
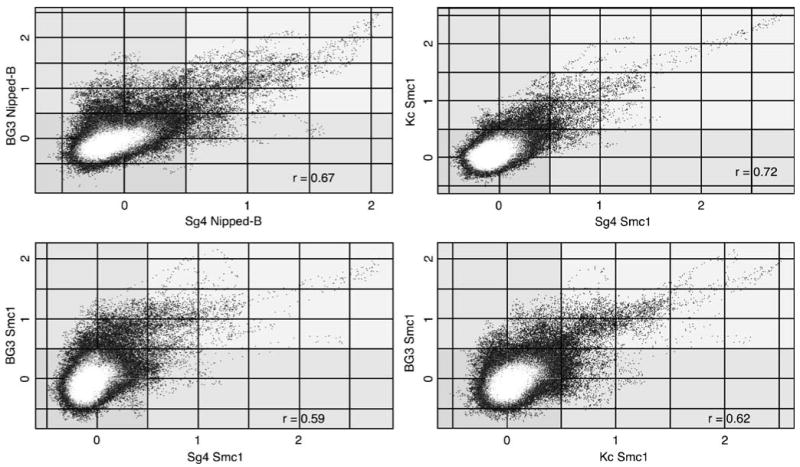
Similarity of cohesin binding in different cell lines. The Smc1 (Sg4, Kc, and BG3) or Nipped-B (Sg4 and BG3) trimmed mean log2 IP/control values of individual microarray features for different cell lines are plotted against each other. The plots cover the same region used in Fig. 2 (chromosome 2L nt 5,522–4,254,929), and other regions show very similar results. The correlation coefficients (r) for the plotted regions are similar to those for the entire genome (Table 1). In all cases, the similarities in cohesin binding between cell lines predominate, with many features showing similar values between the two cell types being compared, indicating that that the peaks occur in precisely the same positions in the two cell lines. There are also features that have significant values in one cell type and not the other, indicating that there are also peaks specific to each cell type
Nipped-B and cohesin bind transcribed regions
Nipped-B and cohesin localization in Drosophila contrasts sharply with that in S. cerevisiae. Cohesin binds every 10 kb or so in yeast, and the peaks are generally less than a few kilobases in width. The yeast Scc2 ortholog of Nipped-B binds different sites than cohesin, and almost all cohesin binds between genes (Glynn et al. 2004; Lengronne et al. 2004). As shown above, however, Drosophila Nipped-B colocalizes with cohesin, and there are large cohesin-binding and cohesin-free regions that extend for several kilobases.
Another key difference is that in contrast to the almost completely intergenic localization of cohesin in yeast, cohesin binds to many transcription units in Drosophila. We looked closely at the cut gene because it is regulated by Nipped-B and cohesin in vivo, and we have previously mapped cohesin binding to the upstream control region by conventional ChIP (Dorsett et al. 2005). The Smc1 binding between the wing margin enhancer and the cut transcription start site in Kc cells by ChIP-chip is virtually identical to that previously mapped by conventional ChIP, with relatively narrow peaks a few kilobases wide located 0.5 and 4 kb upstream of the transcription start site (marked by asterisks in Fig. 5) and additional distal peaks between the wing margin enhancer and promoter. The binding is very similar but not identical in Sg4 cells (Fig. 5). In both Kc and Sg4 cells, there are also multiple peaks of cohesin binding in the cut transcription unit. Cohesin also binds cut in BG3 cells, but in both the upstream regulatory region and transcription unit, the binding is more extensive, such that a 180-kb region starting upstream of the distal wing margin enhancer extending to the 3′ end of the transcription unit is bound by cohesin and Nipped-B (Fig. 5). In contrast, the 25-kb wide cohesin binding region located 10 kb downstream of cut, like most sites in the genome, is very similar in all three cell lines, indicating that the increased cohesin and Nipped-B binding to cut in BG3 cells is authentic.
Fig. 5.
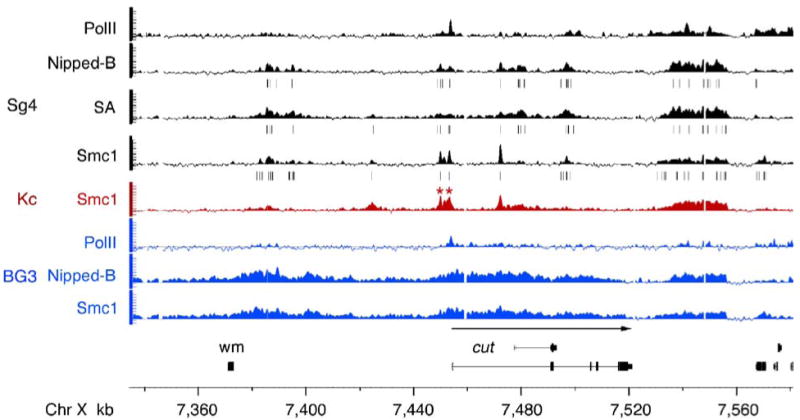
Binding of Nipped-B and cohesin to the cut gene regulatory region and transcription unit. Tracks above the chromosome map show cohesin subunit, Nipped-B, and PolII binding as trimmed mean log2 IP/control values (scale −0.5 to 3) for Sg4 (black), Kc (red) and BG3 (blue) cells. The peaks predicted with a 1% false discovery rate for Nipped-B, SA, and Smc1 in Sg4 cells are indicated with vertical lines underneath the tracks. The extent and direction of cut transcription is indicated with a left-to-right arrow above the cut transcript maps, and the distal wing margin enhancer is indicated by a box (wm). Nipped-B and cohesin bind to the upstream regulatory region and cut transcription unit in all three cell lines, but the binding is more extensive in BG3 cells. The promoter proximal peaks 0.5 and 4 kb upstream of the transcription start site in Kc cells (asterisks) occur precisely in the same positions as previously mapped by conventional ChIP experiments (Dorsett et al. 2005). PolII is found predominantly at the promoter in both Sg4 and BG3 cells, indicating that the difference in cohesin binding between the two cell types is unlikely to reflect a substantial difference in transcription
cut is one of 369 genes in the entire non-repetitive genome that binds cohesin within the transcription unit in all three cell lines (Supplementary Table 1). This group includes the Act5C actin gene, indicating that high transcription does not prevent cohesin binding (Supplementary Table 1; Supplementary Fig. 1). Thus, it is unlikely that in Drosophila, as proposed to explain cohesin localization in yeast (Glynn et al. 2004; Lengronne et al. 2004), RNA polymerase pushes cohesin to the ends of genes.
Binding of Nipped-B and cohesin to transcription units prompted us to compare their localization relative to RNA polymerase II (PolII). Genome-wide, the Nipped-B vs PolII correlation is 0.62 in Sg4 cells and 0.51 in BG3 cells (Table 1). The antibody we used detects PolII with a hypophosphorylated C terminal domain, which generally localizes at promoters, while Nipped-B usually binds extended regions. Plots reveal extensive overlap in PolII and Nipped-B in both Sg4 and BG3 cells but less direct correlation at individual features than between cohesin and Nipped-B (Fig. 6). Many features have low Nipped-B values and high PolII values and vice versa, meaning that the cohesin and PolII peaks usually do not precisely align with each other. The substantial overlap with PolII in both cell lines, however, indicates that Nipped-B and cohesin bind many transcriptionally active regions.
Fig. 6.
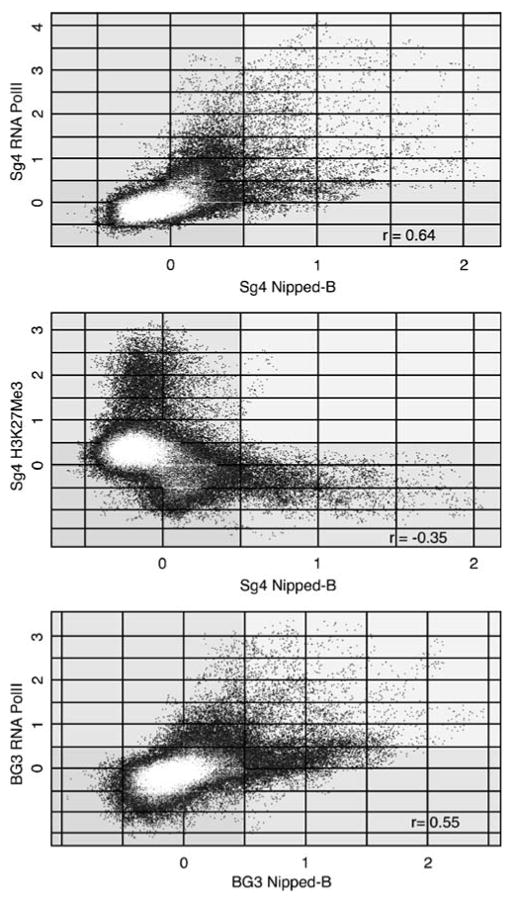
Overlap of Nipped-B and RNA polymerase II (PolII) binding and lack of Nipped-B binding to regions enriched in histone H3 lysine 27 trimethylation (H3K27Me3). The Nipped-B trimmed mean log2 IP/control values for the individual microarray features are plotted against those for PolII or H3K27Me3. The same 4.25 Mb region of chromosome 2L used for Figs. 2 and 3 is plotted for each comparison, but other regions of the genome show a nearly identical pattern. The top two panels compare Nipped-B to PolII and H3K27Me3 in Sg4 cells, and the bottom panel compares Nipped-B and PolII in BG3 cells. The correlation coefficients (r) for the plotted region are similar to those calculated for the entire non-repetitive genome (Table 1). The plots show that many sequences are enriched by both Nipped-B and PolII immunoprecipitation, but there is less direct correlation in enrichment values at individual features than between Nipped-B and cohesin subunits (Fig. 2), indicating that although Nipped-B and PolII binding overlap, the peaks usually do not align with each other. The middle plot shows that there is essentially no Nipped-B binding to regions with high levels of H3K27Me3
Indeed, detailed analysis shows that cohesin preferentially binds to active genes. We defined active genes as those that bind PolII at the transcription start site and also have the histone H3 lysine 4 trimethylation (H3K4Me3) transcriptional elongation mark (reviewed by Shilatifard 2006; Y.B.S., T.G.K., V.P., unpublished) close to the promoter. Using twofold enrichment or greater for both as the criteria, there are 5,954 active genes in Sg4 cells and 423 of these overlap SA-binding regions with a twofold enrichment or greater. In contrast, 9,711 genes lack both PolII and H3K4Me3 and only 32 of these overlap SA-binding regions. Thus, by these criteria, active genes are more than 20-fold more likely to bind cohesin than are silent genes.
Genes are also more likely to have PolII downstream of the promoter when they bind cohesin. Thirty-five genes in the non-repetitive genome bind Nipped-B and cohesin in their transcribed regions in Sg4 but not in BG3 cells, and 80 bind cohesin in BG3 and not in Sg4 cells (Supplementary Table 1; see Supplementary Fig. 1 for an example). Thirty-three of the 35 genes that bind cohesin in Sg4 and not in BG3 cells bind PolII in Sg4 cells, and PolII is also present more than a kilobase downstream of the promoter in 26 (74%) of these. In contrast, while 16 of these genes bind PolII in BG3 cells, only 2 (6%) have PolII downstream of the transcription start site. Similar results are seen for the 80 genes that bind cohesin in BG3 and not in Sg4 cells: 45 out of 80 (56%) have downstream PolII in BG3 cells, and only 2 (3%) have downstream PolII in Sg4 cells (Supplementary Table 1). Thus, cohesin is more likely to bind a gene when it is actively transcribed.
As expected from the above analysis, Nipped-B and cohesin bind less to intergenic sequences than to genes, and within genes, they usually bind 5′ untranslated regions (UTRs) and introns. These trends were quantified by determining the fraction of the Nipped-B and SA peaks predicted with a 25% false discovery rate for Sg4 cells that occur in intergenic sequences, introns, exons, and 5′ and 3′ UTRs (Supplementary Fig. 2). The results were normalized to the percent of the genome that consists of these features to calculate the binding preferences (Table 2). For example, 40 to 50% of Nipped-B and SA cohesin peaks occur in introns, which comprise about a third of the non-repetitive genome (Supplementary Fig. 2). Thus, SA and Nipped-B bind slightly more to introns than expected if they bound at random, and the calculated preferences are 1.45 and 1.33, respectively (Table 2). About a third of Nipped-B and SA peaks occur in intergenic sequences and thus bind slightly less than expected on a random basis with preferences of 0.81 and 0.74 (Table 2; Supplementary Fig. 2). They occur six- to eightfold more than expected on a random basis in 5′ UTRs and much less than expected in coding sequences (preference ratios of 0.27 and 0.18) and 3′ UTRs (ratios of 0.3 and 0.2). This analysis was performed using a peak prediction algorithm, so this does not mean that cohesin does not bind to coding sequences, but that the binding to coding exons is significantly lower on average than to the flanking introns.
Table 2.
SA cohesin subunit and Nipped-B binding preferences for genome features
| Genome feature | Fraction of genome | Binding preference ratioa | |
|---|---|---|---|
| SA | Nipped-B | ||
| Intergenic | 0.41 | 0.74 | 0.81 |
| Intron | 0.33 | 1.45 | 1.32 |
| Coding | 0.17 | 0.27 | 0.18 |
| 3′ UTR | 0.03 | 0.34 | 0.2 |
| 5′ UTR | 0.02 | 6.0 | 8.3 |
Ratios greater than 1 indicate that binding is more than expected on a random basis, and values less than 1 indicate less binding. These values were calculated from data in Supplementary Fig. 1 by dividing the fraction of the top-ranked half of the peaks predicted at a 25% false discovery rate occurring in a genome feature by the fraction of the non-repetitive genome (column 2) containing that feature.
The preference for 5′ UTRs correlates with frequent occurrence of cohesin and PolII peaks at transcription start sites (see Fig. 5 and Supplementary Fig. 1 for examples). Averaging the enrichment for Nipped-B and SA from 10 kb upstream to 10 kb downstream of the transcription start site for all genes that overlap cohesin-binding regions in Sg4 cells demonstrates a peak in enrichment for both at transcription start sites (Supplementary Fig. 3). The curve is skewed towards the transcribed region, which likely explains the higher occurrence of predicted cohesin peaks in introns relative to intergenic regions but not the much lower occurrence in coding sequences. The reason for the low preference of cohesin for coding sequences is unknown. It is also unknown what portions of the intergenic regions bound by cohesin might be transcribed or are regulatory sequences, such as those upstream of cut (Fig. 5).
Nipped-B and cohesin binding correlates with Abd-B expression
Nipped-B facilitates expression of the Ubx gene of the bithorax complex (BX-C) in vivo (Rollins et al. 1999). Thus, we closely examined binding of Nipped-B and cohesin to the BX-C, which also contains abd-A and Abd-B. In Sg4 cells, Abd-B is expressed, but PcG proteins silence Ubx and abd-A, such that they are expressed at a 300-fold lower level than Abd-B (Schwartz et al. 2006). Nipped-B, cohesin, and PolII bind the transcribed Abd-B gene but not the silent Ubx or abd-A genes, with the exception of Smc1-only sites near abd-A (Fig. 7). PolII, cohesin, and Nipped-B bind the same 75-kb region starting near the upstream Abd-B promoter, extending past the 3′ end of Abd-B through the iab-7 enhancer region, ending at the Fab-7 boundary (Fig. 7). This region is flanked on both sides by histone H3 lysine 27 trimethylation (H3K27Me3) domains associated with PcG silencing (Fig. 7; Kahn et al. 2006; Schwartz et al. 2006). The lack of cohesin binding to the silent Ubx and abd-A genes suggested that cohesin might bind Abd-B only when it is expressed. Indeed, cohesin does not bind Abd-B in Kc or BG3 cells, in which Abd-B is silent (Fig. 7).
Fig. 7.
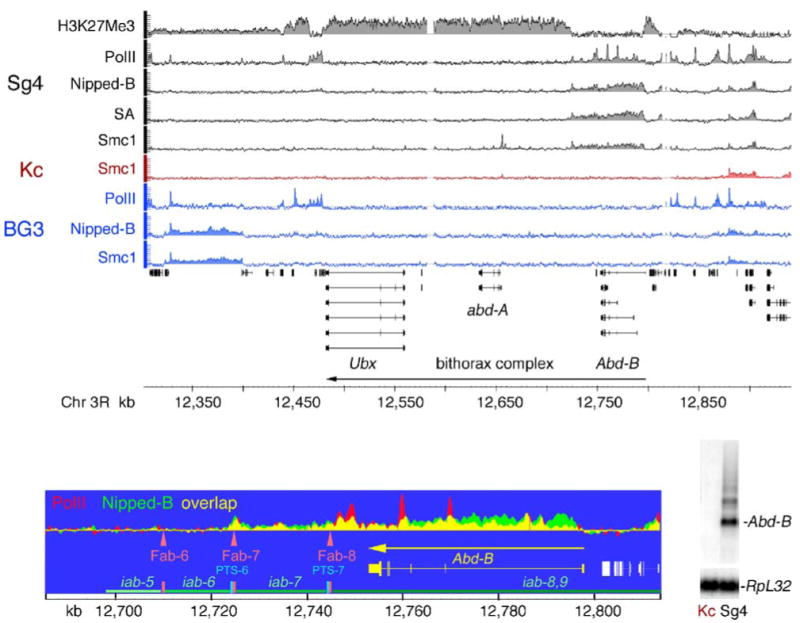
Binding of Nipped-B and cohesin to the active Abd-B gene in Sg4 cells. The tracks show histone H3 lysine 27 trimethylation (H3K27Me3), PolII, Nipped-B, SA, and Smc1 localization in the bithorax complex (BX-C) for Sg4 cells (black and gray), Smc1 binding in Kc cells (red), and PolII, Nipped-B, and Smc1 binding in BG3 cells (blue). Trimmed mean log2 IP/control values are plotted on a scale from −0.5 to 3. The H3K27Me3 data is from Schwartz et al. 2006. The direction of transcription for the BX-C is indicated with an arrow. An expanded map of Abd-B showing the regulatory region (Akbari et al. 2006; Maeda and Karch 2006) with enhancers (iab, green horizontal lines), boundary elements (Fab, orange vertical bars and arrowheads) and promoter-targeting sequences (PTS, cyan vertical bars) with Nipped-B and PolII binding overlaid on each other illustrates the coincidence of some Nipped-B and PolII peaks. At the lower right is a Northern blot showing Abd-B transcripts in Sg4 cells and their absence in Kc cells. The blot was reprobed for RpL32 as a loading control. Ubx and abd-A transcripts are at least 300-fold lower than Abd-B transcripts in Sg4 cells (Schwartz et al. 2006). Nipped-B and cohesin bind to the Abd-B transcription unit and downstream regulatory region in Sg4 cells where Abd-B is active but not in Kc or BG3 cells where it is silent. The Ubx and abd-A genes, which are actively silenced by Polycomb group proteins in Sg4 cells, as indicated by high H3K27Me3 (Schwartz et al. 2006), do not bind Nipped-B and cohesin in any of the cell lines, although Ubx is regulated by Nipped-B in vivo (Rollins et al. 1999)
Cohesin is also not found in other silenced regions, such as the entire Antennapedia complex in Sg4, BG3, and Kc cells (Fig. 1, chromosome arm 3R, 2.48–2.72 Mb). Like the silenced region of the BX-C, H3K27Me3 also coats the Antennapedia complex in Sg4 cells (Kahn et al. 2006; Schwartz et al. 2006). The cut locus is also a PcG target in Sg4 cells (Schwartz et al. 2006; Schwartz and Pirrotta 2007), although this does not prevent binding of PolII to the cut promoter (Fig. 5). It is possible that the expansion of Nipped-B and cohesin binding to cut in BG3 cells relative to Sg4 cells may reflect loss of PcG silencing of cut in BG3 cells, although this does not result in gene activation and increased PolII binding (Fig. 5). Significantly, Nipped-B binding anti-correlates with H3K27Me3 in Sg4 cells, with a genome-wide correlation coefficient of −0.30 (Table 1). This negative correlation is also illustrated by a plot of the trimmed mean log2 IP/control values for the microarray features, which reveals very little or no overlap in Nipped-B binding and H3K27Me3 (Fig. 6).
Discussion
Cohesin localization differs between yeast and Drosophila
The studies reported in this paper represent the first large-scale mapping of cohesin binding to a metazoan genome. Cohesin and the Scc2 ortholog of Nipped-B have been mapped genome-wide in S. cerevisiae, where it was found that nearly all cohesin binds between genes, about every 10 kb or so, and that Scc2 and cohesin do not colocalize (Glynn et al. 2004; Lengronne et al. 2004). Cohesin actually does colocalize with Scc2 in an scc2 temperature-sensitive mutant at the nonpermissive temperature, and then translocates away from Scc2 when shifted to the permissive temperature, supporting the idea that cohesin loads at Scc2 sites (Lengronne et al. 2004). Because the majority of cohesin binding sites in yeast are between convergent transcription units, it has also been proposed that RNA polymerase then pushes cohesin to the ends of genes.
In sharp contrast to yeast, we find that Drosophila Nipped-B and cohesin colocalize throughout the non-repetitive genome and preferentially bind to actively transcribed regions. Thus, in Drosophila, PolII does not push cohesin to the ends of genes, and it is possible for Nipped-B to dynamically regulate cohesin binding instead of just loading it onto chromosomes. The cohesin binding regions in Drosophila are much larger on average than in yeast, extending from a few kilobases up to 100 kb or so in length, and cohesin-free regions can extend from several kilobases in size up to a megabase or so.
The reasons for the differences in cohesin localization between yeast and Drosophila are unknown, but multiple speculative possibilities can be considered. One is that, in Drosophila, transcription might be needed in many cases to provide a 10-nm chromatin fiber that fits into the 35-nm internal diameter of the cohesin ring (Anderson et al. 2002), while in yeast, much of the chromosome already has an accessible structure. For instance, the H1 linker histone that helps form higher order chromatin structures is likely present at most nucleosomes in metazoan organisms, while in yeast, the related Hho1 linker histone is present at low levels and does not globally regulate chromatin structure or gene expression (Freidkin and Katcoff 2001). It is also feasible that in yeast, which has a small compact genome, the positions of cohesin binding sites have been evolutionarily optimized to avoid interference with transcription. It is also worth noting that in Drosophila, cohesin peaks occur three- to eightfold less frequently in coding sequences than in intergenic sequences or introns. In yeast, where most genes lack introns, similar preferences would favor binding to intergenic sequences. It is unclear why cohesin prefers noncoding over coding sequences in Drosophila, but it is possible that differences in DNA sequence or binding of other proteins could be critical factors.
In yeast, cohesin binds more densely around centromeres (Blat and Kleckner 1999; Megee et al. 1999; Tanaka et al. 1999; Glynn et al. 2004; Lengronne et al. 2004; Weber et al. 2004). In Drosophila, the centromeres are in heterochromatin that consists largely of repetitive sequences. Thus, the studies reported in this paper provide no information regarding the binding of cohesin or Nipped-B binding to centromeres. By immunostaining, cohesin binds to both mitotic and meiotic centromeres in Drosophila (Warren et al. 2000; Valdeolmillos et al. 2004; Khetani and Bickel 2007; Gause et al. 2007). Immunostaining with the same Nipped-B antibody used in the experiments reported in this paper indicates that Nipped-B colocalizes with cohesin along chromosome arms in both polytene and meiotic chromosomes, but not at centromeres in meiotic chromosomes (Gause et al. 2007). Thus, Nipped-B might not be involved in regulating association of cohesin with centromeres during meiosis.
Potential effects of cohesin and Nipped-B on gene expression
Based on effects of Nipped-B and cohesin on cut expression in vivo, it was originally proposed that cohesin binding to the cut regulatory region hinders enhancer–promoter interactions and that Nipped-B alleviates this effect by dynamic control of cohesin binding (Dorsett 2004). The finding that Nipped-B colocalizes with cohesin supports the idea that it dynamically regulates binding. The preferential association of cohesin with transcribed regions suggests additional mechanisms by which cohesin binding might affect transcription, and vice versa. As a general model, we envision that transcription facilitates cohesin binding and that the cohesin that binds affects subsequent transcription. Nipped-B then regulates these effects on transcription by dynamic control of cohesin binding or subunit interactions.
Features of the cohesin binding to the active Abd-B gene in Sg4 cells raise the possibility that in some cases, cohesin could interfere with both transcriptional elongation and activation. Some cohesin and PolII peaks coincide in both the Abd-B transcription unit and 3′ regulatory region, which contains intergenic transcription units likely involved in Abd-B regulation (Bae et al. 2002; Drewell et al. 2002). The cohesin in the regulatory region could hinder Abd-B activation by affecting this intergenic transcription. For instance, in the human β-globin gene, blocking intergenic transcription between the enhancer and promoter by insertion of a transcription terminator or an insulator reduces activation (Ling et al. 2004; Zhao and Dean 2004). Genes with distant regulatory elements, such as cut and Ubx, may be more sensitive to Nipped-B dosage because of combined effects on activation and elongation.
Cohesin might also have positive effects on gene expression in some cases. Although it is unknown if the effect is direct, reduction of Rad21 dosage decreases runx gene expression during early zebrafish development (Horsfield et al. 2007). Similarly, Smc1 homozygous mutant clones in the Drosophila mushroom body show reduced ecdysone receptor (EcR) gene expression (Oren Schuldiner and Liqun Luo, personal communication), and cohesin binds EcR in all three cell lines examined in this study. Our findings do not provide an obvious explanation for how cohesin could directly facilitate gene expression, except the possibility that it might help maintain the chromatin in an unfolded state that is more conducive to transcription. Another possibility is that, in specific cases, cohesin might contribute to chromatin boundary function to block the spread of silencing factors as it does at the HMR silent locus in yeast (Donze et al. 1999). There is a cohesin/Nipped-B peak at the Fab-7 boundary element flanking the active Abd-B domain in Sg4 cells, and thus we cannot rule out the possibility that cohesin plays a role in defining chromatin domains permissive for gene expression.
Effects of transcription on cohesin binding
The data indicate that cohesin and Nipped-B bind preferentially, but not exclusively, to active genes. As mentioned above, we speculate that transcription facilitates cohesin binding by unfolding chromatin to a 10-nm fiber that can be encircled by cohesin. Based on the anti-correlation with histone H3 lysine 27 trimethylation, it also appears likely that silencing, either by preventing transcription or through an independent effect on chromatin structure, inhibits cohesin binding.
Transcription is neither necessary nor sufficient for cohesin binding because some poorly expressed genes, such as cut, bind cohesin, and some active genes, such as SA, do not. In the case of cut, PolII binds primarily at the promoter in both Sg4 and BG3 cells. There is little downstream polymerase in the cut transcription unit in either cell type, yet there is substantially more cohesin binding to this region in BG3 cells. Thus, there must be additional factors besides transcription that regulate cohesin binding.
Implications for Cornelia de Lange syndrome
Association of cohesin and Nipped-B with many genes suggests that the diversity of CdLS phenotypes stems from effects on multiple genes. Many of the genes bound by cohesin in Drosophila cells encode evolutionarily conserved transcription factors and receptors that control limb, organ, peripheral, and central nervous system development (see Supplementary Table 1). These include the genes encoding the Notch receptor, its Serrate and Delta ligands and mastermind coactivator, the thickvein transforming growth factor beta (TGFβ) receptor and the Mad DNA-binding protein that mediates TGFβ signaling, the patched hedgehog receptor, the ecdysone steroid hormone receptor, and the epidermal growth factor receptor. Homeobox genes bound by cohesin include cut, Lim1, Distal-less (Dll), homeobrain (hbn), Abd-B, invected (inv), homothorax (hth), and C15, among others. There are also multiple zinc finger protein genes that bind cohesin, including the pannier (pnr) GATA1 ortholog and its interaction partner u-shaped (ush). In BG3 cells, the entire Enhancer of split gene complex encoding multiple bHLH transcription factors involved in nervous system development is bound by cohesin and Nipped-B.
The finding that cohesin binding to Abd-B correlates with Abd-B expression and the variation in cohesin binding between the three cell lines indicate that many other genes are also likely to bind cohesin in other cell types. Thus, identification of target genes that cause specific CdLS phenotypes will require mapping cohesin binding and gene expression patterns in affected tissues at critical stages of development. Because many genes are bound by cohesin in each cell type, we speculate that some of the individual patient phenotypes might stem from simultaneous effects on the expression of multiple genes.
Supplementary Material
The online version of this article (doi:10.1007/s00412-007-0129-1) contains Supplementary material, which is available to authorized users.
Acknowledgments
The authors thank Cheri van de Bunte and Joel Eissenberg for comments on the manuscript and Jumin Zhou for helpful discussions. Kc167 and ML-DmBG3 cells were obtained from the Drosophila Genomics Resource Center at Indiana University. This work was supported by NIH grants R01GM055683 (DD), R01GM070444 (MDB), P01HD052860 (DD, Project III Director; Ian Krantz, PI), and March of Dimes FY05-103 (DD). Work at Lawrence Berkeley National Laboratory was performed under Department of Energy contract DE-AC02-05CH11231. The microarray CEL files have been deposited with the NCBI GEO database under accession no. GSE9248. Processed ChIP data files are available upon request.
Footnotes
F. Uhlmann
Contributor Information
Ziva Misulovin, Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, Saint Louis, MO 63104, USA.
Yuri B. Schwartz, Department of Molecular Biology and Biochemistry, Rutgers University, Piscataway, NJ 08854, USA
Xiao-Yong Li, Berkeley Drosophila Transcription Network Project, Genomics Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Tatyana G. Kahn, Department of Molecular Biology and Biochemistry, Rutgers University, Piscataway, NJ 08854, USA
Maria Gause, Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, Saint Louis, MO 63104, USA.
Stewart MacArthur, Berkeley Drosophila Transcription Network Project, Genomics Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Justin C. Fay, Department of Genetics, Washington University School of Medicine, Saint Louis, MO 63108, USA
Michael B. Eisen, Center for Integrative Genomics, Department of Molecular and Cell Biology, University of California, Berkeley, CA, USA Berkeley Drosophila Transcription Network Project, Genomics Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA.
Vincenzo Pirrotta, Department of Molecular Biology and Biochemistry, Rutgers University, Piscataway, NJ 08854, USA.
Mark D. Biggin, Berkeley Drosophila Transcription Network Project, Genomics Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA
Dale Dorsett, Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, Saint Louis, MO 63104, USA, e-mail: dorsettd@slu.edu.
References
- Akbari OS, Bousum A, Bae E, Drewell RA. Unraveling cis-regulatory mechanisms at the abdominal-A and Abdominal-B genes in the Drosophila bithorax complex. Dev Biol. 2006;293:294–304. doi: 10.1016/j.ydbio.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419–424. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K. ATP hydrolysis is required for cohesin's association with chromosomes. Curr Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- Bae E, Calhoun VC, Levine M, Lewis EB, Drewell RA. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc Natl Acad Sci U S A. 2002;99:16847–16852. doi: 10.1073/pnas.222671299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, Adams M, Champe M, Dugan SP, Frise E, Hodgson A, George RA, Hoskins RA, Laverty T, Muzny DM, Nelson CR, Pacleb JM, Park S, Pfeiffer BD, Richards S, Sodergren EJ, Svirskas R, Tabor PE, Wan K, Stapleton M, Sutton GG, Venter C, Weinstock G, Scherer SE, Myers EW, Gibbs RA, Rubin GM. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0079. RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CR, Wu CS, Hom Y, Gartenberg MR. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 2005;19:3031–3042. doi: 10.1101/gad.1356305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodriguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, Siu V, Ramos FJ, Musio A, Jackson LS, Dorsett D, Krantz ID. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Adherin: key to the cohesin ring and Cornelia de Lange syndrome. Curr Biol. 2004;14:R834–R836. doi: 10.1016/j.cub.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Dorsett D. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma. 2007;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development. 2005;132:4743–4753. doi: 10.1242/dev.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewell RA, Bae E, Burr J, Lewis EB. Transcription defines the embryonic domains of cis-regulatory activity at the Drosophila bithorax complex. Proc Natl Acad Sci USA. 2002;99:16853–16858. doi: 10.1073/pnas.222671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey RN, Gartenberg MR. A tDNA establishes cohesion of a neighboring silent chromatin domain. Genes Dev. 2007;21:2150–2160. doi: 10.1101/gad.1583807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echalier G, Ohanessian A. In vitro culture of Drosophila melanogaster embryonic cells. In Vitro. 1970;6:162–172. doi: 10.1007/BF02617759. [DOI] [PubMed] [Google Scholar]
- Freidkin I, Katcoff DJ. Specific distribution of the Saccharomyces cerevisiae linker histone homolog HHO1p in the chromatin. Nucleic Acids Res. 2001;29:4043–4051. doi: 10.1093/nar/29.19.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Webber HA, Misulovin Z, Haller G, Rollins RA, Eissenberg JC, Bickel SE, Dorsett D. Functional links between Drosophila Nipped-B and cohesin in somatic and meiotic cells. Chromosoma. 2007 Oct 2; doi: 10.1007/s00412-007-0125-5. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Horsfield JA, Anagnostou SH, Hu JK, Cho KH, Geisler R, Lieschke G, Crosier KE, Crosier PS. Cohesin-dependent regulation of Runx genes. Development. 2007;134:2639–2649. doi: 10.1242/dev.002485. [DOI] [PubMed] [Google Scholar]
- Huang CE, Milutinovich M, Koshland D. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Philos Trans R Soc Lond B Biol Sci. 2005;360:537–542. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K. A topological interaction between cohesin rings and a circular minichromosome. Cell. 2005;122:849–860. doi: 10.1016/j.cell.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Ivanov D, Nasmyth K. A physical assay for sister chromatid cohesion in vitro. Mol Cell. 2007;27:300–310. doi: 10.1016/j.molcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Jackson L, Kline AD, Barr M, Koch S. de Lange syndrome: a clinical review of 310 individuals. Am J Med Genet. 1993;47:940–946. doi: 10.1002/ajmg.1320470703. [DOI] [PubMed] [Google Scholar]
- Kahn TG, Schwartz YB, Dellino GI, Pirrotta V. Polycomb complexes and the propagation of the methylation mark at the Drosophila Ubx gene. J Biol Chem. 2006;281:29064–29075. doi: 10.1074/jbc.M605430200. [DOI] [PubMed] [Google Scholar]
- Kaur M, DeScipio C, McCallum J, Yaeger D, Devoto M, Jackson LG, Spinner NB, Krantz ID. Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am J Med Genet A. 2005;138:27–31. doi: 10.1002/ajmg.a.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetani RS, Bickel SE. Regulation of meiotic cohesion and chromosome core morphogenesis during pachytene in Drosophila oocytes. J Cell Sci. 2007;120:3123–3137. doi: 10.1242/jcs.009977. [DOI] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Ainol L, Zhang L, Yu X, Pi W, Tuan D. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J Biol Chem. 2004;279:51704–51713. doi: 10.1074/jbc.M404039200. [DOI] [PubMed] [Google Scholar]
- Losada A. Cohesin regulation: fashionable ways to wear a ring. Chromosoma. 2007;116:321–329. doi: 10.1007/s00412-007-0104-x. [DOI] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. The ABC of the BX-C: the bithorax complex explained. Development. 2006;133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- Megee PC, Mistrot C, Guacci V, Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila Nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, Rollins RA, Erdjument-Bromage H, Tempst P, Benard CY, Hekimi S, Newbury SF, Strachan T. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol. 2006;4:e242. doi: 10.1371/journal.pbio.0040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, Guacci V. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J Cell Biol. 2003;163:729–741. doi: 10.1083/jcb.200305080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan T. Cornelia de Lange Syndrome and the link between chromosomal function, DNA repair and developmental gene regulation. Curr Opin Genet Dev. 2005;15:258–264. doi: 10.1016/j.gde.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2007;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Cosma MP, Wirth K, Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hao Z, Kai M, Okayama H. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 2001;20:5779–5790. doi: 10.1093/emboj/20.20.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey SE, Uhlmann F, Nasmyth K, Yanagida M. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- Ui K, Nishihara S, Sakuma M, Togashi S, Ueda R, Miyata Y, Miyake T. Newly established cell lines from Drosophila larval CNS express neural specific characteristics. In Vitro Cell Dev Biol Anim. 1994;30A:209–216. doi: 10.1007/BF02632042. [DOI] [PubMed] [Google Scholar]
- Valdeolmillos A, Rufas JS, Suja JA, Vass S, Heck MM, Martinez AC, Barbero JL. Drosophila cohesins DSA1 and Drad21 persist and colocalize along the centromeric heterochromatin during mitosis. Biol Cell. 2004;96:457–462. doi: 10.1016/j.biolcel.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Vrouwe MG, Elghalbzouri-Maghrani E, Meijers M, Schouten P, Godthelp BC, Bhuiyan ZA, Redeker EJ, Mannens MM, Mullenders LH, Pastink A, Darroudi F. Increased DNA damage sensitivity of Cornelia de Lange syndrome cells: evidence for impaired recombinational repair. Hum Mol. 2007;16:1478–1487. doi: 10.1093/hmg/ddm098. [DOI] [PubMed] [Google Scholar]
- Warren WD, Steffensen S, Lin E, Coelho P, Loupart M, Cobbe N, Lee JY, McKay MJ, Orr-Weaver T, Heck MM, Sunkel CE. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr Biol. 2000;10:1463–1466. doi: 10.1016/s0960-9822(00)00806-x. [DOI] [PubMed] [Google Scholar]
- Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol. 2006;16:863–874. doi: 10.1016/j.cub.2006.03.049. [DOI] [PubMed] [Google Scholar]
- Weber SA, Gerton JL, Polancic JE, DeRisi JL, Koshland D, Megee PC. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2:E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Jain S, Song H, Fu M, Heuckeroth RO, Erlich JM, Jay PY, Milbrandt J. Mice lacking sister chromatid cohesion protein PDS5B exhibit developmental abnormalities reminiscent of Cornelia de Lange syndrome. Development. 2007;134:3191–201. doi: 10.1242/dev.005884. [DOI] [PubMed] [Google Scholar]
- Zhao H, Dean A. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 2004;32:4903–4919. doi: 10.1093/nar/gkh832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The online version of this article (doi:10.1007/s00412-007-0129-1) contains Supplementary material, which is available to authorized users.


