Abstract
Drosophila Nipped-B is an essential protein that has multiple functions. It facilitates expression of homeobox genes and is also required for sister chromatid cohesion. Nipped-B is conserved from yeast to man, and its orthologs also play roles in deoxyribonucleic acid repair and meiosis. Mutation of the human ortholog, Nipped-B-Like (NIPBL), causes Cornelia de Lange syndrome (CdLS), associated with multiple developmental defects. The Nipped-B protein family is required for the cohesin complex that mediates sister chromatid cohesion to bind to chromosomes. A key question, therefore, is whether the Nipped-B family regulates gene expression, meiosis, and development by controlling cohesin. To gain insights into Nipped-B's functions, we compared the effects of several Nipped-B mutations on gene expression, sister chromatid cohesion, and meiosis. We also examined association of Nipped-B and cohesin with somatic and meiotic chromosomes by immunostaining. Missense Nipped-B alleles affecting the same HEAT repeat motifs as CdLS-causing NIPBL mutations have intermediate effects on both gene expression and mitotic chromatid cohesion, linking these two functions and the role of NIPBL in human development. Nipped-B colocalizes extensively with cohesin on chromosomes in both somatic and meiotic cells and is present in soluble complexes with cohesin subunits in nuclear extracts. In meiosis, Nipped-B also colocalizes with the synaptonemal complex and contributes to maintenance of meiotic chromosome cores. These results support the idea that direct regulation of cohesin function underlies the diverse functions of Nipped-B and its orthologs.
Introduction
Drosophila Nipped-B was discovered in a genetic screen for factors that facilitate long-range transcriptional activation of the cut and Ultrabithorax (Ubx) homeobox genes by enhancers positioned some 80 and 50 kbp away from the gene promoters (Rollins et al. 1999, 2004). Nipped-B is essential, and homozygous Nipped-B mutants die as larvae. Heterozygous Nipped-B null mutants, which show only a 25 to 30% reduction in Nipped-B expression, are viable but have reduced cut and Ubx expression during imaginal disk development.
Nipped-B encodes a member of a highly conserved protein family, which includes Scc2 and Mis4 of S. cerevisiae and S. pombe, discovered in screens for factors that control sister chromatid cohesion (Furuya et al. 1998; Michaelis et al. 1997), C. cinereus Rad9 identified in a screen for factors required for deoxyribonucleic acid (DNA) repair and meiosis (Valentine et al. 1995), and human Nipped-B-Like (NIPBL/delangin), mutated in Cornelia de Lange syndrome (CdLS; Krantz et al. 2004; Tonkin et al. 2004). Nipped-B family proteins contain seven HEAT repeats implicated in protein–protein interactions (Neuwald and Hirano 2000), and NIPBL missense mutations in all seven cause CdLS (Gillis et al. 2004; Miyake et al. 2005; Deardorff and Krantz, personal communication).
Homozygous Drosophila Nipped-B mutants display defects in sister chromatid cohesion before they die, and depletion of vertebrate Nipped-B homologs in vitro or in cultured cells cause cohesion defects, showing that all are functional orthologs of Scc2 and Mis4 (Gillespie and Hirano 2004; Rollins et al. 2004; Seitan et al. 2006; Takahashi et al. 2004; Watrin et al. 2006). Scc2 interacts with the Scc4 protein, which is also required for sister chromatid cohesion (Ciosk et al. 2000). Weakly conserved Scc4 homologs in S. pombe, C. elegans, Drosophila, and vertebrates interact with Nipped-B and its orthologs and function in sister chromatid cohesion and development (Bernard et al. 2006; Seitan et al. 2006; Watrin et al. 2006).
The Nipped-B family is required for the cohesin protein complex that mediates sister chromatid cohesion to bind to chromosomes, which explains their role in cohesion (Arumugam et al. 2003; Ciosk et al. 2000; Gillespie and Hirano 2004; Seitan et al. 2006; Takahashi et al. 2004; Tomonaga et al. 2000; Watrin et al. 2006). Cohesin consists of a heterodimer of the Smc1 and Smc3 structural maintenance of chromosome proteins, and two other proteins, the α-kleisin Rad21 (Mcd1/Scc1) and Stromalin (SA/Scc3; reviewed in Hirano 2006; Nasmyth and Haering 2005). The Rad21 termini interact with the head domains of Smc1 and Smc3, forming a ring-like structure. Many organisms contain meiosis-specific forms of some cohesin subunits, including Rec8 (Rad21 homolog) in yeast, C. elegans, and mammals and Smc1β and Stag3 (SA homolog) in mammals (reviewed in Revenkova and Jessberger 2006). The situation regarding meiosis-specific cohesin subunits is less clear in Drosophila, which lacks an obvious Rec8 homolog.
Cohesin is required but not sufficient for sister chromatid cohesion. Its ring-like structure is the basis of the current cohesion models, in which cohesin encircles two sister chromatids or cohesin rings bound to two sisters interlock with each other (Hirano 2006; Huang et al. 2005; Losada 2007; Nasmyth and Haering 2005). In some cases, a cohesin ring that encircles one sister may interact with proteins bound to the other sister to establish cohesion (Chang et al. 2005). During interphase, cohesin binds numerous sites along chromosome arms and more densely to pericentric heterochromatin, where it is critical for cohesion at metaphase.
Like the Nipped-B family, cohesin also has roles besides sister chromatid cohesion in meiosis, DNA repair, gene expression, and human development (reviewed in Dorsett 2007; Nasmyth and Haering 2005; Hagstrom and Meyer 2003; Hirano 2006; Revenkova and Jessberger 2006). Thus, a central question is whether regulation of cohesin also explains the functions of the Nipped-B family proteins in gene expression and meiosis. In this paper, we determine the effects of several Nipped-B mutations on gene expression, sister chromatid cohesion, and meiosis and the localization of Nipped-B and cohesin on somatic and meiotic chromosomes. Combined, the findings link Nipped-B's diverse roles to the regulation of cohesin activity.
Materials and methods
Sequencing Nipped-B alleles
Total ribonucleic acid (RNA) was isolated from wild-type and homozygous Nipped-B mutant second instar larvae using Trizol (Invitrogen) and reverse transcribed using SuperScript III (Invitrogen) and random hexamer primers. Overlapping segments of Nipped-B complementary DNAs (cDNAs) approximately 800 bp in length were amplified by polymerase chain reaction (PCR) and sequenced directly using the amplification primers (Retrogen). Sequence assembly and mutation analysis was performed using CodonCode Aligner software (CodonCode). For the N-terminal region, which showed significant alternative splicing, the PCR products were cloned into plasmid vectors, and several were sequenced. Primer sequences are available upon request.
Nipped-B, Pds5, and Rad21 antibodies
A His6-Nipped-B protein fusion containing Nipped-B residues 1 to 409 (GenBank AF114160) was expressed in E. coli using the pMCSG-7 vector (Stols et al. 2002) and purified under denaturing conditions. Insoluble purified protein was washed and suspended in phosphate-buffered saline (PBS) and used to immunize a guinea pig and a rabbit (Pocono Rabbit Farm and Laboratory, Canadensis, PA). A His6-Pds5 protein containing Pds5 residues encoded by exons 6, 7, and 8 of the Pds5 (CG17509) messenger RNA (mRNA) was prepared in the same manner and used to immunize a rabbit, and a His6-Rad21 fusion containing Rad21 residues 1–350 was used to immunize both a rabbit and a guinea pig.
Antibody specificities were confirmed by immunostaining and Western blots. In Western blots of cultured cell extracts, both Rad21 antisera recognized the same protein slightly larger than the predicted size for Rad21 as previously reported (Vass et al. 2003). The Rad21 protein was coprecipitated by SA, Smc1, and Nipped-B antisera (see “Results”) and was reduced in Rad21 RNA interference (RNAi)-treated cells. In some extracts, two major Rad21 bands were observed. All preimmune sera showed only low-level background immunostaining, and the Nipped-B, Pds5, and Rad21 antisera show staining that colocalizes with SA and Smc1 on polytene chromosomes (see “Results”). Homozygous pds5e3 mutants live to the third instar, and the maternal mRNA is virtually absent by the second instar stage (Dorsett et al. 2005). The salivary gland chromosomes from homozygous pds5e3 null mutants show staining for cohesin (Dorsett et al. 2005), but as expected, they show only background staining with the Pds5 antisera. By Western blots of whole-cell and nuclear extracts of cultured cells, whole-cell extracts of imaginal disks, adult ovaries, and embryos, the guinea pig Nipped-B antibody recognizes one major band close to the expected size (237 kDa) that is reduced in levels in embryos from heterozygous mutant mothers and cultured cells treated with anti-Nipped-B RNAi (Supplementary Fig. 1). The rabbit Nipped-B antisera precipitates all Nipped-B from nuclear extracts, as shown by guinea pig Nipped-B antibody Western blots of the precipitates and loss of the Nipped-B protein in the postimmunoprecipitation supernatant (Supplementary Fig. 1).
Western blots
Samples were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) using 6% gels and electroblotted to Immobilon membrane (Millipore). Nipped-B was detected using a 1:2,000 dilution of the guinea pig Nipped-B antiserum, followed by 1:7,500 dilution of donkey horse radish peroxidase-anti-guinea pig secondary antibody (Jackson Laboratories). Similar primary antibody dilutions of 1:2,000 to 1:5,000 were used for SA, Smc1, Rad21, and Pds5. The SA and Smc1 antisera have been described previously (Dorsett et al. 2005). Proteins were detected by chemiluminescence (Western Lightening, Perkin Elmer) and autoradiography. For embryo Western blots, 100 0- to 7-h-old embryos were crushed in 20 μl of PBS (pH 7.0) containing 0.2% Nonidet P40 (NP-40), 0.5 mM dithiothreitol (DTT), and protease inhibitors (1 mM benzamidine, 1 μg per ml pepstatin A, 1 μg per ml antipain, 0.5 mM phenymethylsulfonyl fluoride [PMSF]). Seven microliters of 4× SDS-PAGE loading dye was added to each sample, and the mixture was heated at 100°C for 10 min. Extracts of ovaries and imaginal disks were made using the same buffer. Whole-cell extracts of cultured cells were made using radioimmunoprecipitation assay (RIPA) buffer, and nuclear extracts were made as described below.
Effects of Nipped-B mutations on ctK and su(Hw)e2 bx34e mutant phenotypes
Males with wild-type or balanced mutant alleles of Nipped-B were crossed to ctK females at 25 and 27°C, and the wing margin nicks in the male progeny heterozygous for the Nipped-B allele being tested were quantified as previously described (Gause et al. 2001; Rollins et al. 2004). Nipped-B*/+; su(Hw)e2 bx34e flies were generated by crosses, and the bithorax phenotypes were quantified as previously described (Morcillo et al. 1996). Bonferroni–Dunn post-hoc analysis of variance tests were performed on phenotype data using Statview software (SAS Institute).
Neuroblast squashes
Precocious sister chromatid separation was scored in second instar larval neuroblasts from Oregon R wild-type and various Nipped-B mutants heteroallelic with Nipped-B02047 using colchicine and hypotonic treatments as previously described (Rollins et al. 2004). Data from the mutant groups were compared to each other and Oregon R wild-type using Fisher's exact test (Fisher 1922; http://www.matforsk.no/ola/fisher.htm).
Immunostaining
Salivary gland polytene chromosomes were prepared and immunostained as previously described (Dorsett et al. 2005) using a 1:100 or 1:200 dilution of primary antiserum and a 1:200 dilution of secondary antibodies that do not show cross-species reactivity (Jackson Laboratories). The SA and Smc1 antisera used for staining polytene chromosomes were described previously (Dorsett et al. 2005).
Intact ovaries whole-mount preparations were fixed and stained as described previously (Webber et al. 2004). Z-stacks were collected and deconvolved using the Improvision Volocity software. Meiotic chromosome spreads were prepared and immunostained as described previously (Khetani and Bickel 2007). For all experiments with meiotic cells, a mixture of guinea pig affinity-purified antibodies raised against Smc1 and Smc3 peptides (Khetani and Bickel 2007) was utilized to visualize cohesin.
Meiotic chromosome segregation
Nipped-B407/CyO or Nipped-B10E/+ females were crossed to attached X˄Y, v f B males and scored for Bar+ female and Bar male exceptional progeny. Transmission of the J21A minichromosome was measured as previously described (Murphy and Karpen 1995; Dobie et al. 2001). X˄Y; ry506 males were crossed to Nipped-B*/+; ry506; J21A, ry+ females, and the transmission frequency was calculated by dividing the number of ry+ female progeny by the total number of female progeny.
Immunoprecipitation of cohesin and Nipped-B from the nuclear extract
Nuclear extract was prepared from 100 ml of Kc cells grown to a density of approximately 5 × 106 cells per ml. Cells were collected by centrifugation at 300 × g for 5 min at 4°C, suspended in 15 ml of phosphate buffered saline (PBS, pH 7.0), and collected by centrifugation at 300 × g for 5 min at 4°C. The washed cells were suspended in 15 ml ice-cold hypotonic buffer (10 mM hydroxyethyl piperazineethanesulfonic acid (hepes)–KOH pH 7.9, 50 mM KCl, 1.5 mM DTT, 1 mM benzamidine, 1 μg per ml pepstatin A, 1 μg per ml antipain, 0.5 mM PMSF) and collected by centrifugation at 300 × g for 5 min at 4°C. The cell pellet was suspended in 5 vol of hypotonic buffer, incubated on ice for 10 min, and homogenized with 30 strokes in a ground glass homogenizer. Nuclei were collected by centrifugation at 2,000 × g for 5 min at 4°C. The nuclei were suspended in 5 vol of hypotonic buffer and collected by centrifugation at 2,000 × g for 5 min at 4°C. The nuclei were suspended in 2 vol of DNaseI digestion buffer (50 mM hepes–KOH pH 7.9, 100 mM KCl, 2.5 mM MgCl2, 1 mM MnCl2, 1 mM DTT, 0.2% [v/v] NP-40, 1 mM benzamidine, 1 μg per ml pepstatin A, 1 μg per ml antipain, 0.5 mM PMSF) and incubated on ice for 2 min. The suspended nuclei were treated with 20 μg of DNaseI (Sigma D-08876, 10 Kunitz units) at 25°C for 15 min. Insoluble material was removed by centrifugation at 100,000 × g for 60 min at 4°C, and after adjusting to 10% (v/v) glycerol, the supernatant was flash frozen in dry ice-ethanol and stored at −80°C.
For immunoprecipitation, 200 μg of nuclear extract was precleared by incubation with 20 μl of protein-A agarose beads (prewashed in DNaseI digestion buffer) for 1 h at 4°C, and beads were removed by centrifugation. The precleared extract was incubated with 10 μl of the indicated preimmune or immune serum and incubated for 4 hr at 4°C with gentle mixing. Immune complexes were collected by incubating with 20 μl of prewashed protein A-agarose beads for 1 h at 4°C and then collecting the beads by centrifugation for 15 s in an Eppendorf centrifuge. The beads were washed five times with 0.5 ml of DNaseI digestion buffer containing 0.4% NP-40 and then twice with DNaseI digestion buffer. The beads were boiled in SDS-PAGE dye for 5 min, and the samples were separated by SDS-PAGE in 6% gels. Western blots were prepared and probed for cohesin subunits, Nipped-B, and Pds5 as described above.
Results
Identification of Nipped-B mutations
We reasoned that if some of Nipped-B's functions do not involve cohesin, then it might be possible to find Nipped-B separation-of-function mutations. For example, a mutation could cause recessive cohesion defects but not have dominant effects on gene expression. To investigate this possibility, we characterized several Nipped-B mutations that were induced in an isogenic cn bw chromosome by ethylmethane sulfonate (EMS) mutagenesis (Myster et al. 2004). To identify the mutations and distinguish them from polymorphisms, we sequenced Nipped-B transcripts from an isogenic cn bw stock kept in our laboratory, several EMS-induced mutant alleles, and compared them to the previously reported Nipped-B cDNA sequence (Rollins et al. 1999).
The sequences revealed substantial amino acid coding variation in wild-type Nipped-B alleles, including differences in the number of residues (Supplementary Fig. 2). In addition to the wild-type polymorphisms, we found significant alternative splicing at the N terminus, some of which alters the encoded protein (Supplementary Figs. 3 and 4).
We also identified sequence changes unique to each of nine EMS-induced Nipped-B mutant alleles (Fig. 1). Three are missense mutations (Nipped-BNC39, Nipped-BNC71; Nipped-BNC78), two of which (NC71, NC78) occur in HEAT repeats. Each missense mutation affects a residue conserved in human NIPBL, and all are close to conserved residues altered by CdLS-causing missense NIPBL mutations (Fig. 2; Gillis et al. 2004; Miyake et al. 2005; Schoumans et al. 2007). Two other Nipped-B mutations, Nipped-BNC7 and Nipped-BNC59, truncate the encoded protein in the C-terminal region after the HEAT repeats, and four, Nipped-BNC6, Nipped-BNC23, Nipped-BNC41, and Nipped-BNC77, truncate the protein in the N-terminal region before the HEAT repeats. All the mutations are single-nucleotide changes, except Nipped-BNC77, which is a short duplication that creates a stop codon and a new splice donor site. All the mutations affect residues encoded by all splicing variants and thus are unlikely to produce any wild-type Nipped-B isoforms.
Fig. 1.
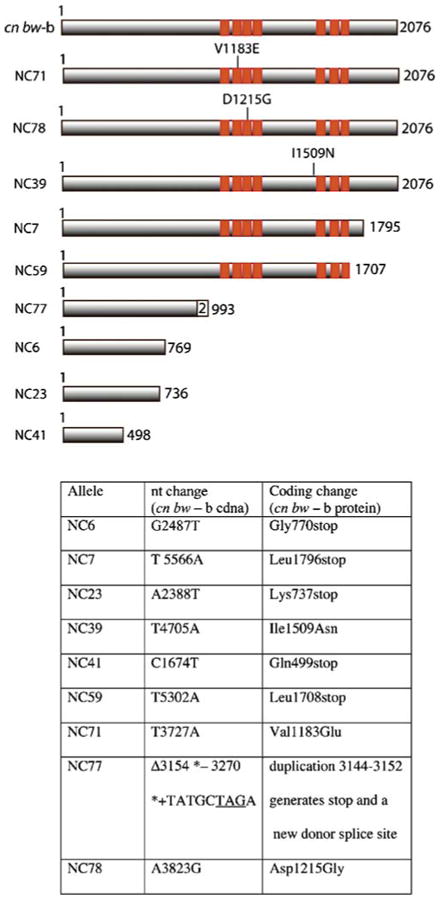
Nipped-B mutations and predicted mutant proteins. The diagrams at the top indicate the predicted protein products of the sequenced Nipped-B mutant alleles, and the table at the bottom gives the changes in nucleotide sequence from the parental wild-type cn bw sequence and the associated change in protein coding. Orange boxes show the positions of the HEAT repeats
Fig. 2.

Alignment of Nipped-B missense mutations to CdLS-causing NIPBL mutations. Nipped-BNC71 and Nipped-BNC78 alter conserved residues in HEAT repeats 2 and 3, respectively, and are close to conserved residues altered in NIPBL missense mutants. Nipped-BNC39 affects a conserved residue between HEAT repeats 4 and 5 that is immediately adjacent to a conserved residue altered by a NIPBL missense mutation
All the EMS-induced mutations, including the three missense alleles, are recessive lethal at the second to third instar molt, the same as null alleles (Rollins et al. 1999). We tried to detect the truncated proteins by Western blot using antibodies generated against the N-terminal region of the protein in early embryos, which show the highest levels of Nipped-B protein because of maternal loading (Rollins et al. 1999). Only very minor amounts of Nipped-B protein migrating at the predicted smaller sizes, which could potentially be the truncated proteins, were detected in embryos from Nipped-BNC7/+ and Nipped-BNC59/+ mothers (Supplementary Fig. 1). The full-length maternal protein is reduced in level, and we deduce, therefore, that the truncated forms of the protein are unstable. Because the truncation alleles are virtually equivalent to null alleles, it cannot be determined if the C-terminal residues are critical for specific Nipped-B functions.
Dominant effects of Nipped-B mutations on cut and Ultrabithorax phenotypes
To determine if any of the EMS-induced mutations separate the gene expression and cohesion functions of Nipped-B, we measured their effects on cut and Ubx phenotypes and on sister chromatid cohesion. Nipped-B loss-of-function alleles generated by γ-rays, including Nipped-B407, Nipped-B292.1, and T(2;3)Nipped-B359.1, dominantly enhance the mutant phenotypes of some cut gene alleles and reduce expression of a wild-type cut gene (Rollins et al. 1999, 2004). Similarly, Nipped-B loss-of-function alleles dominantly increase the severity of the bx34e allele of Ubx when it is partially suppressed by the su(Hw)e2 mutation (Rollins et al. 1999). Homozygous Nipped-B mutations and heteroallelic combinations with the Nipped-B02047 P insertion allele cause sister chromatid cohesion defects before death (Rollins et al. 2004).
To measure effects of the EMS-induced Nipped-B mutations on cut expression, we used the wing-nicking phenotype caused by the ctK mutation. The ctK allele is an insertion of a gypsy transposon that has fewer potential Su(Hw) insulator protein-binding sites. It causes partial loss of the adult wing margin by partially blocking the remote wing margin enhancer that drives cut expression during wing margin development. Decreases in ctK expression further reduce the amount of wing margin as quantified by counting the number of nicks or gaps in the bristle row along the adult wing margin (Dorsett et al. 2005; Gause et al. 2001; Rollins et al. 2004). Null Nipped-B alleles and in vivo Nipped-B RNAi increase the wing nicking displayed by ctK (Rollins et al. 2004).
All the EMS-induced Nipped-B alleles dominantly increased the ctK wing nicking (Fig. 3). The two missense mutations affecting HEAT repeats, Nipped-BNC71 and Nipped-BNC78, exhibited a significantly weaker effect than the other alleles, while the other missense allele, Nipped-BNC39, had effects similar to the truncations. One of the C-terminal truncation alleles, Nipped-BNC59, reproducibly had stronger effects on the ctK phenotype than the others, but we cannot rule out the possibility that there are other mutations on this chromosome that enhance the effect. It is also possible that small amounts of the truncated protein are present during wing margin development, and that this truncated protein might be antimorphic.
Fig. 3.

Effects of Nipped-B mutations on su(Hw)e2 bx34e and ctK mutant phenotypes. The dominant effects of the indicated Nipped-B alleles on the bithorax phenotype displayed by su(Hw)e2 bx34e males and the cut wing phenotype displayed by ctK males were quantified as described in the text. The error bars on the bithorax phenotype data are standard errors. The cut wing phenotype data are presented as box plots with the vertical lines indicating the 10th, 25th, 50th (median), 75th, and 90th percentiles for each genotype. The circles are data points that fall outside the 10th and 90th percentiles. Asterisks indicate phenotypes that differ significantly from the wild-type control (cn bw-a) by the Bonferroni–Dunn post-hoc test
All the EMS-induced Nipped-B alleles dominantly enhanced the bithorax phenotype of su(Hw)e2 bx34e, similar to the effects of null Nipped-B alleles (Rollins et al. 1999). Although the Nipped-BNC71 and Nipped-BNC78 missense mutations had the smallest effects, they were only slightly less than those caused by the other mutant alleles, suggesting that Ubx expression is more sensitive to reduced Nipped-B activity than is cut (Fig. 3). When the alleles are broken into three categories, missense, truncations after the HEAT repeats (C region truncations), and truncations before the HEAT repeats (N region truncations), the missense mutants as a group induce a significantly weaker bithorax phenotype than either of the truncation classes (5.0 vs 5.4 and 5.5). Thus, as with cut, the missense mutants as a class have weaker effects on Ubx expression than do the truncation classes.
Recessive effects of Nipped-B mutations on sister chromatid cohesion
We compared the effects of the Nipped-B alleles on sister chromatid cohesion by examining neuroblast metaphase nuclei for precocious sister chromatid separation (PSCS). To avoid effects of other potential recessive mutations, we tested heteroallelic combinations of each allele with the Nipped-B02047 P element insertion (Rollins et al. 1999, 2004).
Table 1 shows the number of metaphases examined and the number of examples of PSCS detected for each mutant, and Fig. 4 shows some examples of the PSCS phenotype. Because the Nipped-B alleles are recessive lethal before the molt to third instar and the mitotic index in the mutants is low, it is difficult to obtain enough metaphases to accurately measure the frequency of PSCS for each individual allele. To make statistical comparisons, therefore, the mutants were categorized into missense, C-terminal region truncation, and N-terminal region truncation classes as described above. All classes increase the frequency of PSCS compared to the wild-type control. The missense alleles, Nipped-BNC39, Nipped-BNC71, and Nipped-BNC78, individually and as a class gave a significantly lower frequency of PSCS (∼30%) than did the truncation classes, which were similar to each other (59 and 62%) and the frequency reported for Nipped-B null alleles (61%, Rollins et al. 2004). Using Fisher's exact test, which is applicable when there are low sample numbers (Fisher 1922), the missense mutant class is significantly different from wild-type and both truncation classes, while the two truncation classes are not different from each other (Table 1). Thus, the missense mutants have weaker effects on both gene expression and sister chromatid cohesion than do truncation alleles, and as expected, both classes of truncation alleles have effects on gene expression and sister chromatid cohesion similar to those of null alleles. We conclude, therefore, that none of the nine EMS-generated alleles, including the three missense alleles, separate the gene expression and sister chromatid cohesion functions of Nipped-B.
Table 1.
Precocious sister chromatid separation in Nipped-B mutants
| Nipped-B allelea | Number of metaphases | Number of PSCSb | Frequency of PSCS | Fisher exact test P valuec vs | ||
|---|---|---|---|---|---|---|
| Missense | C-region truncations | N-region truncations | ||||
| Wild type | ||||||
| Oregon R | 30 | 1 | 0.03 | 5.2×10−3 | 1.5×10−6 | 3.4×10−9 |
| Missense | ||||||
| NC39 | 14 | 5 | 0.36 | |||
| NC71 | 17 | 4 | 0.24 | |||
| NC78 | 12 | 4 | 0.33 | |||
| Total | 43 | 13 | 0.30 | 2.5×10−2 | 6.5×10−4 | |
| C region truncations | ||||||
| NC7 | 6 | 3 | 0.50 | |||
| NC59 | 31 | 19 | 0.61 | |||
| Total | 37 | 22 | 0.59 | 0.55 | ||
| N region truncations | ||||||
| NC6 | 34 | 21 | 0.62 | |||
| NC23 | 17 | 10 | 0.59 | |||
| NC41 | 22 | 14 | 0.64 | |||
| NC77 | 8 | 6 | 0.75 | |||
| Total | 81 | 51 | 0.63 | |||
All alleles were tested as heterozygotes with Nipped-B02047.
Metaphases with one or more examples of sister chromatid separation
Fisher's exact test (Fisher 1922) calculates the probability (P value) that two populations are same using small sample sizes. In this table, we compared the wild-type sample vs the totals for the missense mutant, the C-region truncation, and N-region truncation groups and the totals for each of the mutant groups to each other. All the comparisons indicate that the groups are significantly different from each other (low P values), except for the comparison of the C and N truncation mutant groups.
Fig. 4.
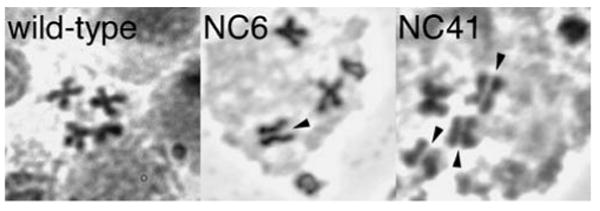
Examples of precocious sister chromatid separation (PSCS) in Nipped-B mutants. The panels show an example wild-type second instar neuroblast metaphase spread and examples of PSCS in Nipped-BNC6/Nipped-B02047 and Nipped-BNC41/Nipped-B02047 second instar metaphase spreads. Arrowheads indicate instances of PSCS. For Table 1, a metaphase was scored as having PSCS if one or more chromosomes show PSCS
Nipped-B, cohesin, and Pds5 colocalize on salivary polytene chromosomes
If Nipped-B's roles in sister chromatid cohesion, gene expression, and meiosis all involve regulating cohesin function, we would expect Nipped-B and cohesin to colocalize on chromosomes. To test this, we double immunostained salivary gland polytene for Nipped-B and cohesin. In all the many nuclei examined, Nipped-B staining was completely coincident with staining for the Smc1 (Fig. 5) and SA cohesin subunits (not shown). There was a general correlation in the intensity of staining—Regions that stained strongly for Nipped-B also stained strongly for cohesin. Similar to the results for Nipped-B, we found that the Pds5 protein, which is required for establishment and/or maintenance of cohesion in multiple organisms (Dorsett et al. 2005; Hartman et al. 2000; Losada et al. 2005; Panizza et al. 2000; Tanaka et al. 2001), also colocalizes with cohesin on polytene chromosomes (Fig. 5). Preimmune serum did not show staining, and the secondary antibodies used for the coimmunostaining experiments do not show species cross-reactivity. Homozygous pds5e3 null mutant polytene chromosomes, which stain for cohesin (Dorsett et al. 2005), do not stain for Pds5.
Fig. 5.

Immunostaining of salivary gland polytene chromosomes for Nipped-B, Smc1, Pds5, and HP1 proteins. The upper panels show double immunostaining of Oregon R wild-type salivary gland chromosomes for Nipped-B and the Smc1 cohesin subunits. The upper left panel shows a whole nucleus, and the right shows a higher magnification view of the tip of the X chromosome from the same nucleus. The Nipped-B and Smc1 proteins colocalize, and there is a strong correlation in Nipped-B and Smc1 staining intensity. The bottom right panel shows double immunostaining of Oregon R wild-type chromosomes for Pds5 and Smc1, which also colocalize almost completely. The lower left panel shows double immunostaining for Nipped-B and HP1, which do not colocalize. The bright HP1 staining is the heterochromatic chromocenter. White scale bars in all panels indicate 5 μm
It has been proposed that in addition to regulation of cohesin, interactions between the NIPBL ortholog of Nipped-B with heterochromatin protein HP1 might be functionally important for Nipped-B's human ortholog (Lechner et al. 2005). In S. pombe, interactions between the Swi6 HP1 ortholog and cohesin are important for sister chromatid cohesion (Bernard et al. 2001; Nonaka et al. 2002). In contrast to the colocalization of Nipped-B with cohesin, however, double-immunostaining experiments indicate that Nipped-B does not colocalize with HP1 on chromosomes (Fig. 5). The brightest regions of HP1 staining, which include the chromocenter and region 31B on the left arm of chromosome 2, stain weakly if at all for Nipped-B. A few bright Nipped-B bands stain weakly for HP1, but most do not. These results indicate that significant HP1–Nipped-B interactions do not occur on chromosomes. We cannot exclude the possibility that interactions with HP1 mask the epitopes recognized by the Nipped-B antibodies, but if this is the case, we deduce that such sites would lack cohesin, given the complete colocalization of Nipped-B and cohesin.
It is possible that HP1-Nipped-B interactions occur only when the proteins are not associated with chromosomes. Nevertheless, the phenotypes of HP1 mutants indicate that if such interactions do occur, they are not critical for Nipped-B and cohesin function. It has been reported that polytene chromosomes appear relatively normal in HP1 [Su(var)2–5] mutants and cohesion defects do not occur in any of the several tissues examined (Fanti et al. 1998). Polytene chromosomes that lack cohesin, on the other hand, show a significant change in morphology (Dorsett et al. 2005).
Nipped-B colocalizes with cohesin and the synaptonemal complex during female meiosis
There is evidence in yeast and vertebrates that meiosis-specific forms of cohesin are involved in formation of the synaptonemal complex (SC; reviewed in Revenkova and Jessberger 2006), and the mushroom Rad9 adherin mutant starts but fails to complete SC assembly (Seitz et al. 1996; Cummings et al. 2002). Rad9 mutants also show defective cohesion. Combined, these observations suggest that Nipped-B might also facilitate SC formation and/or maintenance by regulating cohesin function.
We immunostained Drosophila oocyte chromosome spreads with anti-Nipped-B, and observed that it colocalizes extensively with the Smc1 and Smc3 cohesin subunits in a thread-like pattern along the arms of meiotic chromosomes (Fig. 6). It also colocalizes with the Ord protein, which is required for meiotic cohesion in both sexes and SC maintenance in oocytes (Bickel et al. 1997; Khetani and Bickel 2007; Webber et al. 2004), and with C(3)G, a transverse filament protein that resides within the central element of the SC (Anderson et al. 2005). These data argue that Nipped-B may play a direct role in regulating meiotic cohesion as well as SC structure and function.
Fig. 6.

Immunostaining of female meiotic chromosome spreads for Nipped-B, C(3)G, Smc1/3, Ord, and histone H3 variant, CID. The panels show that Nipped-B colocalizes extensively along the arms with the Smc1 and Smc3 cohesin subunits, the SC component C(3)G and the cohesion protein Ord. Little or no Nipped-B signal is detected at centromeric regions (arrows), as confirmed by staining for the centromere-specific histone H3 variant, CID (bottom panels)
Although Nipped-B exhibits robust localization along the arms of meiotic chromosomes, centromeric Nipped-B staining was surprisingly weak or nonexistent (Fig. 6). In contrast to Nipped-B, bright staining for the cohesion proteins Ord and Smc1/Smc3 is visible in centromeric heterochromatin, which is identified by bright diamidinophenylindole staining and immunostaining for the CID centromere-specific histone (Fig. 6; Khetani and Bickel 2007; Webber et al. 2004). These data suggest that Nipped-B may not associate strongly with meiotic centromeres and are consistent with the lack of Nipped-B signal at the centromeres of polytene chromosomes. However, we cannot rule out the possibility that the epitopes recognized by Nipped-B antibodies are masked within pericentric heterochromatin.
Nipped-B mutations cause early chromosome core dissolution
Nipped-B homozygotes die before meiosis can be observed, but Nipped-B loss-of-function mutations have a dominant effect on meiotic chromosome morphology. Female meiosis starts at the tip of each ovariole of the ovary in the germarium, which contains four developmental regions (1, 2A, 2B, 3). Germline mitotic divisions take place in region 1, and prophase I initiates in region 2A, followed by formation of the SC. Even after the oocyte exits the germarium, the SC is normally maintained throughout pachytene until stage 6 of oogenesis.
In 108 out of the 112 ovarioles examined from Nipped-B10E heterozygotes, the thread-like C(3)G and Smc1/3 staining starts to fragment in germarial region 3 or stage 2 oocytes (Fig. 7). Fragmentation does not occur this early in the wild type. Nipped-B10E is a loss-of-function allele generated by excision of the Nipped-B02047 P element insertion, and similar defects were also observed for females heterozygous for the γ-ray-induced Nipped-B407, Nipped-B292.1, and T(2;3)Nipped-B359.1 alleles.
Fig. 7.

Dominant effect of the Nipped-B10E mutation on chromosome cores and synaptonemal complex during female meiosis. Meiotic chromosomes at the indicated stages of meiosis and oogenesis were visualized by staining for Smc1/3 and C(3)G protein in wild-type and Nipped-B10E heterozygous mutant ovarioles (whole-mount preparation). Nipped-B10E is a null-like allele generated by P element excision. The developmental stages (germarium regions 2A, 2B, 3, and stage 2 of oogenesis) are organized in temporal order from the top down. During wild-type prophase I, the synaptonemal complex first forms in region 2A of the germarium, becomes restricted to the oocyte by region 3, and remains stable until stage 6 of oogenesis. In heterozygous Nipped-B mutants, formation of the chromosome cores in region 2A appears to occur normally, but the cores fragment (lose their linear structure) in germarium region 3 to stage 2 of oogenesis, which is substantially earlier than in the wild type. Despite the early fragmentation, Smc1/3 staining is retained at the centromeres. The bright Smc1/3 spots in the stage 2 oocyte also stain for CID, a centromere-specific histone H3 variant (Fig. 8). Temporal progression of fragmentation defects was quantified in 42 Nipped-B10E/+ ovarioles and revealed that more than 50% of region 3 and more than 90% of stage 2 oocytes exhibit extensive fragmentation of chromosome cores. Similar results were obtained with other Nipped-B alleles (see text). The wild type never shows chromosome disintegration at these early stages
Although fragmentation of cohesin chromosome cores is severe in stage 2 Nipped-B mutant oocytes, Smc1 and Smc3 proteins remain associated with meiotic centromeres (Figs. 7 and 8). The bright Smc1/3 spots in the Nipped-B mutant stage 2 oocytes in Fig. 7 also stain for the centromere-specific histone, CID (Fig. 8). Combined with the lack of Nipped-B staining at centromeres, retention of normal Smc1 and Smc3 staining at centromeres in Nipped-B mutants suggest that Nipped-B may not modulate the behavior of cohesin within pericentric heterochromatin.
Fig. 8.

Colocalization of Smc1/3 and CID in Nipped-B mutant meiotic chromosomes. The upper panels show the staining of the same stage 2 oocyte in Fig. 7 for Smc1/3 and C(3)G in color, and the lower left panel shows the staining for the CID centromere-specific histone. The lower right panel is a merge of the Smc1/3 and CID staining, showing that the bright Smc1/3 spots occur at centromeres
Early SC breakdown in Nipped-B mutants does not affect meiotic chromosome segregation
In homozygous ord mutants, Smc1 and Smc3 fail to accumulate at centromeres, and the Smc1 and Smc3 threadlike staining along chromosome arms is already disrupted in germarium region 2B, which is earlier than in the heterozygous Nipped-B mutants (Khetani and Bickel 2007). The ord mutants also show elevated levels of meiotic nondisjunction (Bickel et al. 1997). We tested if the changes in SC structure caused by heterozygous Nipped-B mutants affect chromosome segregation and found no significant meiotic nondisjunction (Table 2; 0 of 1,303 total progeny from Nipped-B mutant females, 1 of 1,342 progeny from Nipped-B mutant males).
Table 2.
Meiotic chromosome segregation in heterozygous Nipped-B mutants
| Parent | Na | Nondisjunctionb | J21A transmissionc |
|---|---|---|---|
| ♀ Nipped-B407/+ | 636 | 0 | nad |
| ♀ Nipped-B292.1/+ | 667 | 0 | na |
| ♂ Nipped-B407/+ | 635 | 0 | na |
| ♂ Nipped-B10E/+ | 431 | 0.0023 | na |
| ♂ Nipped-B292.1/+ | 150 | 0 | na |
| ♂ Nipped-B359.1/+ | 126 | 0 | na |
| ♀ Nipped-B+/+; ry506; J21A, ry+ | 1441 | na | 0.24±0.03e |
| ♀ Nipped-B407/+; ry506; J21A, ry+ | 1315 | na | 0.29±0.03 |
| ♀ Nipped-B292.1/+; ry506; J21A, ry+ | 1479 | na | 0.33±0.03 |
Number of progeny scored
Frequency of exceptional progeny; see text for assay
Frequency of J21A presence in female progeny; see text for assay. The female parents in the seventh row were heterozygous for the parental chromosome (Nipped-B+) used to generate the Nipped-B407 and Nipped-B292.1 mutations (Rollins et al. 1999).
not applicable
95% confidence interval
A more sensitive test also failed to detect effects on meiotic segregation in heterozygous Nipped-B mutants. Maternal transmission of the J21A minichromosome with a weak centromere is roughly half that expected for a normal chromosome (Murphy and Karpen 1995). This reduced transmission is very sensitive to changes in the dosage of genes involved in chromosome segregation (Dobie et al. 2001). For example, heterozygous wings-apart-like (wapl) mutations, which decrease mitotic chromatid separation by reducing prophase removal of cohesin from chromosome arms, decrease J21A transmission in female meiosis (Dobie et al. 2001; Gandhi et al. 2006; Kueng et al. 2006; Verni et al. 2000). Females heterozygous for the parental chromosome in which the Nipped-B407 and Nipped-B292.1 mutations were induced (Rollins et al. 1999) transmitted J21A at a frequency of 0.24 (348 of 1,441; Table 2). J21A was transmitted at a frequency of 0.29 (384 of 1,315) by females heterozygous for Nipped-B407, and a frequency of 0.33 (493 of 1,479) by Nipped-B292.1/+ females (Table 2). The control and Nipped-B mutant J21A transmission frequencies all fall within the normal range (0.22 to 0.37) and are close to the wild-type average of 0.27 (Dobie et al. 2001). Thus, centromeric cohesion does not appear to be altered in heterozygous Nipped-B mutant females, and the early SC dissolution does not alter chromosome segregation during meiosis I.
Nipped-B is in complexes with the Rad21 and SA cohesin subunits
The colocalization of Nipped-B with cohesin along polytene chromosomes and meiotic chromosome cores and the effects of Nipped-B mutations on mitotic sister chromatid cohesion and meiotic chromosome core structure suggest that Nipped-B is directly involved in regulating association of cohesin with chromosomes and its organization on chromosomes. To determine if Nipped-B forms complexes with cohesin, we conducted immunoprecipitation experiments with nuclear extracts of Kc-cultured cells. We prepared extracts using moderate ionic strength and DNaseI digestion to release chromosome-bound cohesin and maximize the probability of detecting Nipped-B–cohesin complexes.
As expected, immunoprecipitation with anti-Smc1 brings down high levels of the SA and Rad21 cohesin subunits, and precipitation with anti-SA coprecipitates high amounts of Smc1 and Rad21 (Fig. 9). In contrast, none of the cohesin subunit precipitations brought down detectable amounts of Nipped-B. Precipitation of Nipped-B, however, brought down small but detectable amounts of SA and Rad21 but not Smc1 (Fig. 9). Because the amounts were small, we repeated the experiment nine times with different nuclear extracts and a different cell line (MLDmD8) and always detected SA and Rad21 but not Smc1.
Fig. 9.
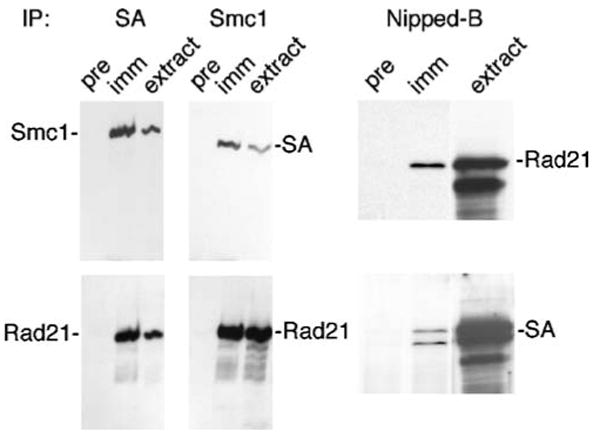
Immunoprecipitation of cohesin subunits and Nipped-B from Kc cell nuclear extracts. The serum used for precipitation is indicated above each Western blot. For each blot, pre indicates a preimmune serum control precipitation, imm indicates immune serum precipitation, and extract is a control lane containing input nuclear extract. The left panels show that precipitation with anti-SA or anti-Smc1 precipitates significant amounts of other cohesin subunits. We were unable to detect Nipped-B or Pds5 in these precipitates in multiple experiments. The panels on the right show that precipitation of Nipped-B coprecipitates small amounts of Rad21 and SA. In all nine repeated Nipped-B immunoprecipitation experiments with different extract preparations, we detected Rad21 and SA but were unable to detect Smc1. The rabbit anti-Nipped-B in these experiments precipitates all Nipped-B from the extract (Supplementary Fig. 1). In some experiments, such as those on the right, we detected multiple forms of Rad21 or SA but do not know if these represent protein modifications or partial proteolysis
A Rad21–SA subcomplex is present in Drosophila cultured cells (Vass et al. 2003), and the immunoprecipitation results thus suggest that there are small amounts of complexes that contain Nipped-B and the Rad21–SA subcomplex in nuclear extracts. These complexes likely do not contain Smc1, although we cannot rule out the possibility that the anti-Nipped-B antibodies displace the Smc1–Smc3 heterodimer from the Rad21–SA subcomplex. Even so, these results, combined with the colocalization with cohesin on somatic and meiotic chromosomes, support the idea that Nipped-B is directly involved in regulating cohesin function. The apparent lack of stable association between Nipped-B and the complete cohesin complex suggests that such interactions are either less stable and more transient than those with the Rad21–SA subcomplex or that they may be stabilized by DNA.
Discussion
In this study, we examined the functions of Drosophila Nipped-B in gene expression, mitotic sister chromatid cohesion, and meiosis. The results support the idea that Nipped-B's diverse functions in somatic and meiotic cells all involve regulation of cohesin function.
Nipped-B HEAT repeat mutations link Nipped-B's roles in gene expression and sister chromatid cohesion
Relative to null alleles, the missense Nipped-BNC71 and Nipped-BNC78 mutations that alter HEAT repeats have weaker effects on both gene expression and sister chromatid cohesion. This agrees with prior studies suggesting that Nipped-B's role in gene expression is to regulate cohesin chromosome binding. The prior studies showed that Nipped-B facilitates activation of the cut gene by a distant transcriptional enhancer and that cohesin has an opposite effect to Nipped-B on cut expression (Rollins et al. 1999; Dorsett et al. 2005; Rollins et al. 2004). They also showed that cohesin binds to cut in multiple cell types and that a unique pds5 mutation that reduces binding of cohesin to chromosomes increases cut expression (Dorsett et al. 2005). Combined, these findings and the genetic linkage between Nipped-B's roles in gene expression and sister chromatid cohesion reported here support the hypothesis that Nipped-B facilitates cut expression by alleviating negative effects of cohesin (Dorsett 2004, 2007). This model suggests that cohesin binding to cut inhibits expression and by maintaining a dynamic cohesin chromosome-binding equilibrium, Nipped-B facilitates cohesin removal or relocation.
Based on this model, we posit that the Nipped-BNC71 and Nipped-BNC78 missense mutations in the HEAT repeats are hypomorphic and reduce but do not abolish the ability of Nipped-B to control cohesin binding to chromosomes. In this scenario, the homozygous HEAT repeat mutants show milder cohesion defects than null alleles because loading of cohesin onto chromosomes is only partially reduced, and the heterozygous mutants have weaker effects on gene expression because they only slightly alter the cohesin chromosome-binding equilibrium.
The Nipped-BNC71 and Nipped-BNC78 missense mutations provide the first evidence that some of the HEAT repeats are important for gene expression and sister chromatid cohesion, and they also link Nipped-B's role in gene expression to the function of NIPBL and cohesin in human development. There are CdLS-causing missense mutations in all seven HEAT repeats of NIPBL, and Nipped-BNC71 and Nipped-BNC78 affect HEAT repeats 2 and 3, respectively (Gillis et al. 2004; Miyake et al. 2005; Deardorff and Krantz, personal communication). Indeed, Nipped-BNC71 and Nipped-BNC78 both affect conserved residues adjacent or very close to conserved residues affected by CdLS-causing mutations, and thus the CdLS mutations are likely to have dominant effects on gene expression similar to Nipped-BNC71 and Nipped-BNC78. The structural similarity between Nipped-BNC71 and Nipped-BNC78 and the NIPBL mutations provides a link to cohesin in human development because like the NIPBL HEAT repeat mutations, missense mutations in the Smc1 and Smc3 cohesin subunits also cause CdLS (Deardorff et al. 2007; Musio et al. 2006).
Does Nipped-B directly regulate cohesin function?
The functional connections between cohesin and the Nipped-B protein family in sister chromatid cohesion, gene expression, and development that have been revealed genetically likely reflect direct regulation of cohesin function by Nipped-B. This is supported by the finding that Nipped-B colocalizes with cohesin on polytene and meiotic chromosomes and the presence of soluble complexes containing Nipped-B and the Rad21 and SA cohesin subunits in cell nuclear extracts. We do not know why soluble complexes containing Nipped-B and whole cohesin were not detected, but it is possible that the antibodies used for immunoprecipitation disrupt these interactions, that the epitopes are masked in such complexes, that such interactions are transitory, or that they are stable only when Nipped-B and cohesin are both bound to DNA.
In S. cerevisiae, cohesin does not colocalize with the Scc2 ortholog of Nipped-B on chromosomes (Lengronne et al. 2004). Nevertheless, purification of yeast Scc2–Scc4 complex brings along small amounts of cohesin subunits, and thus it has also been proposed that the Scc2–Scc4 complex also directly regulates binding of cohesin to chromosomes (Arumugam et al. 2003). It is suggested that cohesin loads at Scc2-binding sites and translocates away (Lengronne et al. 2004). In contrast, chromatin immunoprecipitation experiments using cultured Drosophila cells reveals that Nipped-B colocalizes with cohesin in the entire nonrepetitive genome, confirming that their colocalization is not unique to polytene and meiotic chromosomes (Misulovin et al., submitted for publication). Thus, the separate localization of the Nipped-B/Scc2 cohesin loader and cohesin may be unique to yeast chromosomes.
Does Nipped-B regulate cohesin function at centromeres?
Our results raise the possibility that Nipped-B is not critical for the function of cohesin at centromeres. Cohesin binds densely to pericentric heterochromatin, and cohesin at centromeres is protected in both mitotic and meiotic cells by a member of the Shugoshin/Mei-S332 protein family (Kitajima et al. 2004; McGuinness et al. 2005; Salic et al. 2004; Tang et al. 1998, 2004). Although Nipped-B completely colocalizes with Smc1 and Smc3 along meiotic chromosome cores, we did not see Nipped-B staining around the centromeres, where cohesin staining is the strongest. We also did also not detect significant staining for Nipped-B at the centromeres of polytene chromosomes, but this might be explained by the under-replication of pericentric heterochromatin in these cells. Consistent with this idea, we also did not see strong cohesin staining at polytene centromeres. Nipped-B, however, also does not colocalize with the HP1 heterochromatin protein, whose Swi6 ortholog in S. pombe is required for cohesion and interacts with cohesin (Bernard et al. 2001; Nonaka et al. 2002), at positions that are not under-replicated. In cultured Drosophila cells, Rad21 and SA persist at the centromeres until the metaphase–anaphase transition (Valdeolmillos et al. 2004; Warren et al. 2000). We have immunostained metaphase chromosome spreads from Kc cells and can see Smc1 and SA that colocalizes with the CID centromere-specific histone at centromeres but have not been able to detect Nipped-B (Gause and Dorsett, unpublished). In this case, however, given the relatively weak signals, lack of staining elsewhere in the same nucleus, and the different fixation conditions needed to get good chromosome morphology, it is difficult to rule out the possibility that we cannot detect Nipped-B because of insufficient sensitivity.
Although the lack of pericentric Nipped-B staining in meiotic chromosomes might be caused by epitope masking, other evidence indicates that Nipped-B function is also not critical at meiotic centromeres. In particular, the retention of Smc1 and Smc3 protein at the meiotic chromosome centromeres in heterozygous Nipped-B mutants and the normal meiotic transmission of the J21A minichromosome with a weak centromere indicate that centromeric cohesion is not altered, although the chromosome cores disintegrate early when the dosage of Nipped-B protein is reduced. This contrasts sharply with the chromosome nondisjunction and lack of cohesin binding to the meiotic centromeres in ord mutants (Khetani and Bickel 2007). Thus, the evidence suggests that either Nipped-B does not play a critical role in the function of cohesin at meiotic centromeres or that its role at centromeres is not as dosage sensitive as its function along the chromosome cores. Given the lack of Nipped-B staining, the colocalization of Ord and cohesin, and the lack of cohesin at centromeres in ord mutants, it is possible that Ord functionally substitutes for Nipped-B at meiotic centromeres.
Roles of Nipped-B in meiosis
Our data provide evidence for a functional link between the Nipped-B protein family and cohesin in meiosis. The C. coprinus Rad9 hypomorphic mutant, in addition to having meiotic cohesion defects, fails to complete SC formation, indicating that the Nipped-B family is involved in SC assembly (Cummings et al. 2002; Seitz et al. 1996). The findings that Nipped-B colocalizes with cohesin subunits along meiotic chromosome cores and that cores disassemble prematurely when Nipped-B dosage is reduced indicates that the Nipped-B family is also involved in the maintenance of SC structure.
The Nipped-B family functions in SC assembly and maintenance are both likely to involve cohesin. Cohesin colocalizes with SC proteins and is involved in homolog pairing, SC formation, and SC structure in diverse organisms (Bannister et al. 2004; Chan et al. 2003; Eijpe et al. 2000; Khetani and Bickel 2007; Klein et al. 1999; Pasierbek et al. 2001, 2003; Revenkova et al. 2004; Xu et al. 2005). The C(2)M protein, which contains kleisin motifs similar to those in Rad21 and which interacts with Smc3, is needed for SC formation and the coalescence of Smc1 and Smc3 into chromosome cores (Heidmann et al. 2004; Khetani and Bickel 2007; Manheim and McKim 2003). C(2)M, however, is not required for binding of the Smc1 and Smc3 cohesin subunits to either centromeres or chromosome arms (Khetani and Bickel 2007). Taken together, these data suggest a model in which Nipped-B and C(2)M collaborate in formation of the cohesin chromosome cores and that Nipped-B and Ord then cooperate to maintain this structure. Thus, the Nipped-B protein family plays a role beyond simply loading cohesin onto chromosomes and is involved directly in regulating the higher-order meiosis-specific organization of cohesin.
Supplementary Material
The online version of this article (doi:10.1007/s00412-007-0125-5) contains supplementary material, which is available to authorized users.
Acknowledgments
The authors thank Steve Myster and Mark Peifer for the EMS-generated Nipped-B mutants, Scott Hawley for C(3)G antibodies, and Gary Karpen for the J21A fly stocks. We also thank Patrick Morcillo for advice on statistical analysis and Ian Krantz and Matt Deardorff for information on human NIPBL mutations. This work was supported by NIH Grants R01 GM055683 (D.D.), R01 GM059354 (S.E.B.); P01 HD058260 (D.D., Project III Director), and March of Dimes FY05-103 (D.D.).
Contributor Information
Maria Gause, Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, Saint Louis, MO 63104, USA.
Hayley A. Webber, Department of Biological Sciences, Dartmouth College, Hanover, NH, USA
Ziva Misulovin, Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, Saint Louis, MO 63104, USA.
Gabe Haller, Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, Saint Louis, MO 63104, USA.
Robert A. Rollins, Wyeth Research, Pearl River, NY, USA
Joel C. Eissenberg, Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, Saint Louis, MO 63104, USA
Sharon E. Bickel, Department of Biological Sciences, Dartmouth College, Hanover, NH, USA
Dale Dorsett, Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, Saint Louis, MO 63104, USA, e-mail: dorsettd@slu.edu.
References
- Anderson LK, Royer SM, Page SL, McKim KS, Lai A, Lilly MA, Hawley RS. Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc Natl Acad Sci USA. 2005;102:4482–4487. doi: 10.1073/pnas.0500172102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K. ATP hydrolysis is required for cohesin's association with chromosomes. Curr Biol. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- Bannister LA, Reinholdt LG, Munroe RJ, Schimenti JC. Positional cloning and characterization of mouse mei8, a disrupted allele of the meiotic cohesin Rec8. Genesis. 2004;40:184–194. doi: 10.1002/gene.20085. [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Bernard P, Drogat J, Maure JF, Dheur S, Vaur S, Genier S, Javerzat JP. A screen for cohesion mutants uncovers Ssl3, the fission yeast counterpart of the cohesin loading factor Scc4. Curr Biol. 2006;16:875–881. doi: 10.1016/j.cub.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Bickel SE, Wyman DW, Orr-Weaver TL. Mutational analysis of the Drosophila sister-chromatid cohesion protein ORD and its role in the maintenance of centromeric cohesion. Genetics. 1997;146:1319–1331. doi: 10.1093/genetics/146.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RC, Chan A, Jeon M, Wu TF, Pasqualone D, Rougvie AE, Meyer BJ. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature. 2003;423:1002–1009. doi: 10.1038/nature01697. [DOI] [PubMed] [Google Scholar]
- Chang CR, Wu CS, Hom Y, Gartenberg MR. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 2005;19:3031–3042. doi: 10.1101/gad.1356305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Cummings WJ, Merino ST, Young KG, Li L, Johnson CW, Sierra EA, Zolan ME. The Coprinus cinereus adherin Rad9 functions in Mre11-dependent DNA repair, meiotic sister-chromatid cohesion, and meiotic homolog pairing. Proc Natl Acad Sci USA. 2002;99:14958–14963. doi: 10.1073/pnas.232316999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deardorff MA, Kaur M, Yaeger D, Rampuria A, Korolev S, Pie J, Gil-Rodriguez C, Arnedo M, Loeys B, Kline AD, Wilson M, Lillquist K, Siu V, Ramos FJ, Musio A, Jackson LS, Dorsett D, Krantz ID. Mutations in cohesin complex members Smc3 and Smc1A cause a mild variant of Cornelia de Lange syndrome with predominant mental retardation. Am J Hum Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobie KW, Kennedy CD, Velasco VM, McGrath TL, Weko J, Patterson RW, Karpen GH. Identification of chromosome inheritance modifiers in Drosophila melanogaster. Genetics. 2001;157:1623–1637. doi: 10.1093/genetics/157.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Adherin: key to the cohesin ring and cornelia de Lange syndrome. Curr Biol. 2004;14:R834–R836. doi: 10.1016/j.cub.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Dorsett D. Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma. 2007;116:1–13. doi: 10.1007/s00412-006-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development. 2005;132:4743–4753. doi: 10.1242/dev.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijpe M, Heyting C, Gross B, Jessberger R. Association of mammalian SMC1 and SMC3 proteins with meiotic chromosomes and synaptonemal complexes. J Cell Sci. 2000;113:673–682. doi: 10.1242/jcs.113.4.673. [DOI] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. J R Stat Soc. 1922;85:87–94. [Google Scholar]
- Furuya K, Takahashi K, Yanagida M. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev. 1998;12:3408–3418. doi: 10.1101/gad.12.21.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Morcillo P, Dorsett D. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol Cell Biol. 2001;21:4807–4817. doi: 10.1128/MCB.21.14.4807-4817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol. 2004;14:1598–1603. doi: 10.1016/j.cub.2004.07.053. [DOI] [PubMed] [Google Scholar]
- Gillis LA, McCallum J, Kaur M, DeScipio C, Yaeger D, Mariani A, Kline AD, Li HH, Devoto M, Jackson LG, Krantz ID. NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype-phenotype correlations. Am J Hum Genet. 2004;75:610–623. doi: 10.1086/424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom KA, Meyer BJ. Condensin and cohesin: more than chromosome compactor and glue. Nat Rev Genet. 2003;4:520–534. doi: 10.1038/nrg1110. [DOI] [PubMed] [Google Scholar]
- Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesin and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann D, Horn S, Heidmann S, Schleiffer A, Nasmyth K, Lehner CF. The Drosophila meiotic kleisin C(2)M functions before the meiotic divisions. Chromosoma. 2004;113:177–187. doi: 10.1007/s00412-004-0305-5. [DOI] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Huang CE, Milutinovich M, Koshland D. Rings, bracelet or snaps: fashionable alternatives for Smc complexes. Phil Trans R Soc Lond B Biol Sci. 2005;360:537–542. doi: 10.1098/rstb.2004.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetani RS, Bickel SE. Regulation of meiotic cohesion and chromosome core morphogenesis during pachytene in Drosophila oocytes. J Cell Sci. 2007;120:3123–3137. doi: 10.1242/jcs.009977. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SB, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, Jukofsky L, Wasserman N, Bottani A, Morris CA, Nowaczyk MJ, Toriello H, Bamshad MJ, Carey JC, Rappaport E, Kawauchi S, Lander AD, Calof AL, Li HH, Devoto M, Jackson LG. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher FJ., 3rd The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem Biophys Res Commun. 2005;331:929–937. doi: 10.1016/j.bbrc.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A. Cohesin regulation: fashionable ways to wear a ring. Chromosoma. 2007;116:321–329. doi: 10.1007/s00412-007-0104-x. [DOI] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Hirano T. Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci. 2005;118:2133–2141. doi: 10.1242/jcs.02355. [DOI] [PubMed] [Google Scholar]
- Manheim EA, McKim KS. The synaptonemal complex component C(2)M regulates meiotic crossing over in Drosophila. Curr Biol. 2003;13:276–285. doi: 10.1016/s0960-9822(03)00050-2. [DOI] [PubMed] [Google Scholar]
- McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Miyake N, Visser R, Kinoshita A, Yoshiura K, Niikawa N, Kondoh T, Matsumoto N, Harada N, Okamoto N, Sonoda T, Naritomi K, Kaname T, Chinen Y, Tonoki H, Kurosawa K. Four novel NIPBL mutations in Japanese patients with Cornelia de Lange syndrome. Am J Med Genet A. 2005;135:103–105. doi: 10.1002/ajmg.a.30637. [DOI] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Dorsett D. Genes regulating the remote wing margin enhancer in the Drosophila cut locus. Genetics. 1996;144:1143–1154. doi: 10.1093/genetics/144.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Interactions between the nod+ kinesin-like gene and extracentromeric sequences are required for transmission of a Drosophila minichromosome. Cell. 1995;81:139–148. doi: 10.1016/0092-8674(95)90378-x. [DOI] [PubMed] [Google Scholar]
- Musio A, Selicorni A, Focarelli ML, Gervasini C, Milani D, Russo S, Vezzoni P, Larizza L. X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat Genet. 2006;38:528–530. doi: 10.1038/ng1779. [DOI] [PubMed] [Google Scholar]
- Myster SH, Wang F, Cavallo R, Christian W, Bhotika S, Anderson CT, Peifer M. Genetic and bioinformatic analysis of 41C and the 2R heterochromatin of Drosophila melanogaster: a window on the heterochromatin-euchromatin junction. Genetics. 2004;166:807–822. doi: 10.1534/genetics.166.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Hirano T. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 2000;10:1445–1452. doi: 10.1101/gr.147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Panizza S, Tanaka T, Hochwagen A, Eisenhaber F, Nasmyth K. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr Biol. 2000;10:1557–1564. doi: 10.1016/s0960-9822(00)00854-x. [DOI] [PubMed] [Google Scholar]
- Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek P, Fodermayr M, Jantsch V, Jantsch M, Schweizer D, Loidl J. The Caenorhabditis elegans SCC-3 homologue is required for meiotic synapsis and for proper chromosome disjunction in mitosis and meiosis. Exp Cell Res. 2003;289:245–255. doi: 10.1016/s0014-4827(03)00266-0. [DOI] [PubMed] [Google Scholar]
- Revenkova E, Jessberger R. Shaping meiotic prophase chromosomes: cohesins and synaptonemal complex proteins. Chromosoma. 2006;115:235–240. doi: 10.1007/s00412-006-0060-x. [DOI] [PubMed] [Google Scholar]
- Revenkova E, Eijpe M, Heyting C, Hodges CA, Hunt PA, Liebe B, Scherthan H, Jessberger R. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152:577–593. doi: 10.1093/genetics/152.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Schoumans J, Wincent J, Barbaro M, Djureinovic T, Maguire P, Forsberg L, Staaf J, Thuresson AC, Borg A, Nordgren A, Malm G, Anderlid BM. Comprehensive mutational analysis of a cohort of Swedish Cornelia de Lange syndrome patients. Eur J Hum Genet. 2007;15:143–149. doi: 10.1038/sj.ejhg.5201737. [DOI] [PubMed] [Google Scholar]
- Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, Rollins RA, Erdjument-Bromage H, Tempst P, Benard CY, Hekimi S, Newbury SF, Strachan T. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol. 2006;4:e242. doi: 10.1371/journal.pbio.0040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz LC, Tang K, Cummings WJ, Zolan ME. The rad9 gene of Coprinus cinereus encodes a proline-rich protein required for meiotic chromosome condensation and synapsis. Genetics. 1996;142:1105–1117. doi: 10.1093/genetics/142.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6:991–996. doi: 10.1038/ncb1177. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hao Z, Kai M, Okayama H. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 2001;20:5779–5790. doi: 10.1093/emboj/20.20.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TT, Bickel SE, Young LM, Orr-Weaver TL. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes Dev. 1998;12:3843–3856. doi: 10.1101/gad.12.24.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Sun Y, Harley SE, Zou H, Yu H. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci USA. 2004;101:18012–18017. doi: 10.1073/pnas.0408600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey SE, Uhlmann F, Nasmyth K, Yanagida M. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- Valdeolmillos A, Rufas JS, Suja JA, Vass S, Heck MM, Martinez AC, Barbero JL. Drosophila cohesins DSA1 and Drad21 persist and colocalize along the centromeric heterochromatin during mitosis. Biol Cell. 2004;96:457–462. doi: 10.1016/j.biolcel.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Valentine G, Wallace YJ, Turner FR, Zolan ME. Pathway analysis of radiation-sensitive meiotic mutants of Coprinus cinereus. Mol Gen Genet. 1995;247:169–179. doi: 10.1007/BF00705647. [DOI] [PubMed] [Google Scholar]
- Vass S, Cotterill S, Valdeolmillos AM, Barbero JL, Lin E, Warren WD, Heck MM. Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr Biol. 2003;13:208–218. doi: 10.1016/s0960-9822(03)00047-2. [DOI] [PubMed] [Google Scholar]
- Verni F, Gandhi R, Goldberg ML, Gatti M. Genetic and molecular analysis of wings apart-like (wapl), a gene controlling heterochromatin organization in Drosophila melanogaster. Genetics. 2000;154:1693–1710. doi: 10.1093/genetics/154.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WD, Steffensen S, Lin E, Coelho P, Loupart M, Cobbe N, Lee JY, McKay MJ, Orr-Weaver T, Heck MM, Sunkel CE. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr Biol. 2000;10:1463–1466. doi: 10.1016/s0960-9822(00)00806-x. [DOI] [PubMed] [Google Scholar]
- Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol. 2006;16:863–874. doi: 10.1016/j.cub.2006.03.049. [DOI] [PubMed] [Google Scholar]
- Webber HA, Howard L, Bickel SE. The cohesion protein ORD is required for homologue bias during meiotic recombination. J Cell Biol. 2004;164:819–829. doi: 10.1083/jcb.200310077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Beasley MD, Warren WD, van der Horst GT, McKay MJ. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev Cell. 2005;8:949–961. doi: 10.1016/j.devcel.2005.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The online version of this article (doi:10.1007/s00412-007-0125-5) contains supplementary material, which is available to authorized users.


