Abstract
Previous studies demonstrated substantial variability of the location of primary visual cortex (V1) in stereotaxic coordinates when linear volume-based registration is used to match volumetric image intensities (Amunts et al., 2000). However, other qualitative reports of V1 location (Smith, 1904; Stensaas et al., 1974; Rademacher et al., 1993) suggested a consistent relationship between V1 and the surrounding cortical folds. Here, the relationship between folds and the location of V1 is quantified using surface-based analysis to generate a probabilistic atlas of human V1. High-resolution (about 200 μm) magnetic resonance imaging (MRI) at 7 T of ex vivo human cerebral hemispheres allowed identification of the full area via the stria of Gennari: a myeloarchitectonic feature specific to V1. Separate, whole-brain scans were acquired using MRI at 1.5 T to allow segmentation and mesh reconstruction of the cortical gray matter. For each individual, V1 was manually identified in the high-resolution volume and projected onto the cortical surface. Surface-based intersubject registration (Fischl et al., 1999b) was performed to align the primary cortical folds of individual hemispheres to those of a reference template representing the average folding pattern. An atlas of V1 location was constructed by computing the probability of V1 inclusion for each cortical location in the template space. This probabilistic atlas of V1 exhibits low prediction error compared to previous V1 probabilistic atlases built in volumetric coordinates. The increased predictability observed under surface-based registration suggests that the location of V1 is more accurately predicted by the cortical folds than by the shape of the brain embedded in the volume of the skull. In addition, the high quality of this atlas provides direct evidence that surface-based intersubject registration methods are superior to volume-based methods at superimposing functional areas of cortex, and therefore are better suited to support multi-subject averaging for functional imaging experiments targeting the cerebral cortex.
1 Introduction
The cerebral cortex can be parcellated into many distinct architectonic areas based on laminar differences in neuron and myelin density (Bolton, 1900; Smith, 1904; Campbell, 1905; Brodmann, 1909; Vogt, 1911; Flechsig, 1920; von Economo and Koskinas, 1925). These architectonic areas are thought to serve distinct functional roles, and thus a substantial amount of work has concentrated on locating areas and characterizing their features.
Directly determining the precise location of architectonic areas in a living human is not currently possible. However, observing macroscopic geometric features of the cerebral cortex in an individual is straightforward using standard structural MRI techniques. The goal of probabilistic atlases is to predict the location of cortical areas, which are difficult to image, from the easily imaged cortical geometry. In this context a probabilistic atlas refers to a map of the location of a cerebral cortical area relative to geometric features of the brain. For atlases of cortical areas to be useful, their location must be predictable from brain geometry, and the methods used to create and apply the atlas must effectively capture the relationship between areas and geometric features of the brain.
Recently, methods have been developed to generate probabilistic atlases of cortical areas in a stereotaxic coordinate system by matching the intensities of volumetric brain images via linear transformations (Talairach and Tournoux, 1988) or nonlinear transformations (Roland et al., 1994; Schormann and Zilles, 1998). These volume-based intersubject registration techniques were used to build a probabilistic atlas of human primary visual cortex (V1) (Amunts et al., 2000), as well as a number of other cortical areas (Amunts et al., 1999; Geyer et al., 1999; Grefkes et al., 2001; Morosan et al., 2001; Rademacher et al., 2001b,a; Eickhoff et al., 2007; Amunts et al., 2005; Wilms et al., 2005; Caspers et al., 2006). These studies represent the only probabilistic atlases of cortical areas delineated anatomically. Unfortunately, these atlases exhibit substantial intersubject variability, which indicates a substantial probability of error when predicting area location. This suggests that the geometric features of cortex used to produce the atlases are poor predictors of area location.
Because the cerebral cortex has the topology of a two-dimensional sheet, it is natural to represent cortical geometry in terms of its many sulci and gyri rather than in terms of a three-dimensional volume. Historically, cortical folds have been used as a qualitative method for describing cortical geometry, and in the last two decades several methods for quantitative surface-based analysis have been developed (Schwartz and Merker, 1986; Boissonnat, 1988; Dale and Sereno, 1993; Davatzikos and Bryan, 1996; Van Essen and Drury, 1997; Dale et al., 1999; Fischl et al., 1999a; Andrade et al., 2001; Goebel et al., 2006).
Previous studies investigating the relationship between primary cortical folds and primary sensory areas have found a consistent co-location (Sanides, 1970; Welker, 1990; Rademacher et al., 1993)—especially for V1. Smith (1904) performed an early study of the relationship between V1 and the cortical folds. He claimed that location of the stria of Gennari, a stripe of highly myelinated tissue particular to layer IV of V1 (Zeki, 1970; Boyd and Matsubara, 2005), was a reliable predictor of the location of the calcarine sulcus. Later studies (Putnam, 1926; Stensaas et al., 1974; Rademacher et al., 1993; Zilles et al., 1997; Hasnain et al., 2001) supported this general conclusion, but also reported intersubject variability in the location of the V1 border in relation to nearby gyri and sulci. Thus, previous studies indicate a gross consistency between the location of cortical folds and that of V1, but the lack of quantitative methods for analyzing this relationship dictates that only very general conclusions can be made about the strength of this relationship, and comparison of the results between studies is difficult. A goal of the work presented here was to measure the error in the predicted location of V1 when the primary cortical folds are used to align cortical geometry.
Probabilistic atlases are created and applied using intersubject registration techniques (Talairach and Tournoux, 1988; Roland et al., 1994; Friston et al., 1995; Fischl et al., 1999b; Van Essen, 2005), which provide a method for alignment based on cortical geometry. Thus far, atlases of V1 have been created using volume-based intersubject registration methods and have reported substantial variability in the location of cortical areas. However, because folds were not used in registration, this previous work has not addressed the relationship between the cortical folds and cortical areas.
Surface-based intersubject registration methods, which have been developed to aid in analysis and visualization of functional imaging data (Drury et al., 1996; Sereno et al., 1996; Thompson and Toga, 1996; Davatzikos, 1997; Fischl et al., 1999b), use the gross cortical folding pattern to derive a correspondence between the cortical surfaces of individuals. The goal of registration is to align common functional areas across subjects, and thus these methods have been developed and used under the assumption that alignment of cortical folds results in alignment of areas. Previous studies have suggested that aligning the folds effectively aligns areas (Fischl et al., 1999b), but no direct validation has been reported.
Surface-based analysis tools have been adopted mainly for use in functional imaging studies on live human subjects, and have not seen wide use in postmortem studies where histological analysis is possible. Because observation of the precise location of cortical areas has up to now required staining and slicing of the cortex postmortem—a process that complicates surface reconstruction—surface-based analysis has not been applied to studies investigating cortical areas. Recently, methods for imaging ex vivo human cortical hemispheres at high-resolution using MRI techniques (Augustinack et al., 2005; Hinds et al., 2005) have allowed investigation of microanatomical cortical features in intact hemispheres. A goal of the work presented here was to apply existing surface-based registration techniques to high-resolution structural MRI data of ex vivo human cortex.
The application of surface-based intersubject registration to the location of V1 provides a probabilistic atlas of V1, which here refers to a computational method for predicting the location of V1 in living subjects based on their particular pattern of cortical folds. The amount of error in the atlas prediction indicates the effectiveness of the registration method at creating overlap of common functional areas among a group of subjects. Because registration methods are used to enable intersubject averaging in functional or morphometric imaging experiments, the amount of overlap dictates the benefit gained by pooling data from corresponding cortical areas across subjects. Most methods for intersubject averaging use cortical geometry to compute correspondence between individual brains (Fischl et al., 1999b; Van Essen, 2005), thus assuming a particular relationship between geometry and areas that has not yet been directly established. The results of this work establish a stable relationship between the cortical folds and areas. This relationship suggests both that the location of the folds are functionally significant and that surface-based intersubject registration methods are effective tools increasing statistical power via multisubject averaging in functional imaging studies.
2 Materials and methods
2.1 Magnetic resonance imaging
Twenty whole, formalin-fixed ex vivo human hemispheres (ten right hemispheres and ten left hemispheres) were obtained under an Institutional Review Board-approved protocol through the autopsy service of the Massachusetts General Hospital and with the cooperation of the Massachusetts Alzheimer Disease Research Center, the Massachusetts Eye and Ear Infirmary, and the Center for Neuroimaging of Aging and Neurode-generative Disease. Five of the right hemispheres were from individuals with no diagnosed neurological disease, while one was from an individual diagnosed with Alzheimer’s Disease (AD), three were from individuals diagnosed with Huntington’s Disease (HD), and one hemisphere was from an individual for which no information regarding neurological disease history was available. Five of the left hemispheres were from individuals with no diagnosed neurological disease, while two were from individuals diagnosed with AD, two were from individuals with HD, and no information was available for two individuals.
2.1.1 Imaging microstructure
The stria of Gennari is a stripe of myelinated tissue confined to layer IVCα of V1 (Boyd and Matsubara, 2005). The stria is a highly reliable anatomical feature delineating V1 (Bolton, 1900; Henschen, 1920; Polyak, 1933; Clark, 1941) that has been imaged using MRI both in vivo (Clark et al., 1992; Barbier et al., 2002; Walters et al., 2003; Bridge et al., 2005) and ex vivo (Barbier et al., 2002; Fatterpekar et al., 2002; Hinds et al., 2005). This feature is one of only a few histological features that can be investigated using current MRI technology, which dictates that V1 is one of the very few cortical areas that can be investigated in whole-brain samples today.
High-resolution MRI scans of the occipital lobe of each hemisphere were acquired using a head-only 7 T MRI system based on either a Siemens Sonata or Avanto platform (Siemens Medical Solutions, Erlangen, Germany). The imaging volume covered approximately the posterior third of the hemisphere, containing V1 and surrounding tissue.
The optimal pulse sequence parameters for stria of Gennari contrast were taken to be the same as that for gray and white matter contrast, and were determined by synthesizing T1-weighted, T2*-weighted, and proton density-weighted images from a volume acquired using a three-dimensional, multi-echo FLASH pulse sequence (Fischl et al., 2004), then analyzing the contrast-to-noise ratio between gray and white matter per unit time. Fixation of the samples induced shortening of the difference in T1 relaxation time between gray and white matter as previously described (Tovi and Ericsson, 1992; Augustinack et al., 2005), resulting in MRI volumes that were predominantly T2*-weighted.
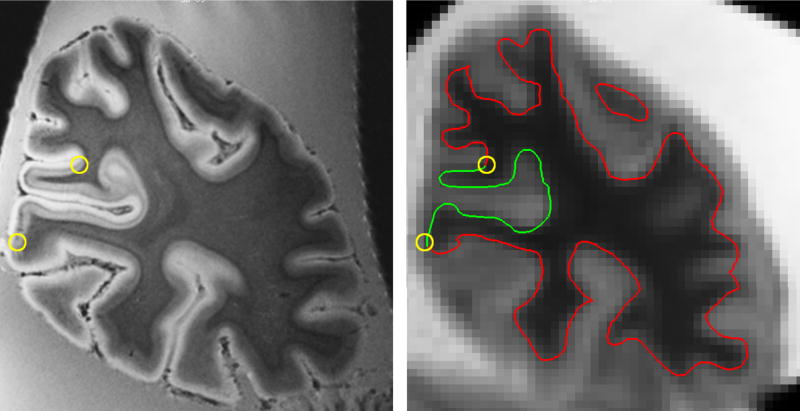
The voxel size varied from 170 to 250 μm, isotropic. All acquisitions were collected with an echo time of 20 ms, a repetition time of 40 ms, a flip angle of 30° and a bandwidth of 30 Hz per pixel. The low pixel bandwidth of this pulse sequence boosts the SNR within the small voxels, but increases imaging distortions. MR distortions for similar scans were found to be small compared to the thickness of cortex. Depending on the properties of the particular RF coil and the size of the FOV, the number of acquisitions required for reliable identification of the stria of Gennari varied. All RF coils used in this study were custom designed and built (Wald et al., 2005). The specific MR parameters used for each hemisphere are shown in Table 1 and a representative coronal slice showing the stria of Gennari is shown in Figure 1(b).
Table 1.
MRI parameters used for each ex vivo hemisphere.
| hemi | acquisitions | time | voxel size (mm3) | FOV (mm2) |
|---|---|---|---|---|
| RH1 | 25 | 35.9 h | 0.17 ×0.17 ×0.17 | 76.00 × 87.00 |
| RH2 | 10 | 15.2 h | 0.18 ×0.18 ×0.18 | 57.50 × 138.00 |
| RH3 | 9 | 11.3 h | 0.20 ×0.20 ×0.20 | 71.00 × 90.00 |
| RH4 | 6 | 11.5 h | 0.20 ×0.20 ×0.20 | 84.00 × 90.00 |
| RH5 | 3 | 3.3 h | 0.25 ×0.25 ×0.25 | 72.00 × 176.00 |
| RH6 | 12 | 15.1 h | 0.20 × 0.20 × 0.20 | 100.00 × 75.00 |
| RH7 | 5 | 10.5 h | 0.20 × 0.20 × 0.20 | 102.00 × 102.00 |
| RH8 | 3 | 12.2 h | 0.18 × 0.18 × 0.18 | 115.00 × 115.00 |
| RH9 | 4 | 11.8 h | 0.18 × 0.18 × 0.18 | 161.00 × 92.00 |
| RH10 | 9 | 14.7 h | 0.18 × 0.18 × 0.18 | 69.43 × 81.00 |
| LH1 | 4 | 9.4 h | 0.20 × 0.20 × 0.20 | 76.80 × 128.00 |
| LH2 | 8 | 18.3 h | 0.20 × 0.20 × 0.20 | 89.00 × 115.00 |
| LH3 | 7 | 14.0 h | 0.20 × 0.20 × 0.20 | 77.00 × 90.00 |
| LH4 | 6 | 9.8 h | 0.18 × 0.18 × 0.18 | 69.43 × 81.00 |
| LH5 | 12 | 18.0 h | 0.20 × 0.20 × 0.20 | 77.00 × 96.00 |
| LH6 | 11 | 13.8 h | 0.20 × 0.20 × 0.20 | 100.00 × 75.00 |
| LH7 | 5 | 10.5 h | 0.20 × 0.20 × 0.20 | 102.00 × 102.00 |
| LH8 | 4 | 11.8 h | 0.18 × 0.18 × 0.18 | 115.00 × 115.00 |
| LH9 | 3 | 12.2 h | 0.18 × 0.18 × 0.18 | 115.00 × 115.00 |
| LH10 | 6 | 11.5 h | 0.20 × 0.20 × 0.20 | 104.00 × 85.00 |
Figure 1.
The same coronal slice through volumes acquired at high and standard resolution. The high-resolution acquisition allowed delineation of V1 via the stria of Gennari, which is visible as a dark stripe in layer IV of the calcarine sulcus in the left panel. The manually located V1/V2 boundary is shown in both panels as the center of the yellow circles. The standard-resolution acquisition allowed reconstruction of a whole-brain surface mesh, which is superimposed on the slice shown in the right panel with portions of the surface that are identified as V1 shown in green.
Ex vivo hemispheres were chosen over in vivo subjects for imaging the stria in this study because the entire extent of V1 is reliably identifiable ex vivo. However, some of the samples used in this study had a known history of neurological disease. Subject information, including history of disease, is shown in Table 2. Both AD and HD are known to cause changes in cortical thickness (Frisoni, 1996; De Leon et al., 1997; Jack et al., 1997; Vonsattel and DiFiglia, 1998; Halliday et al., 1998), but no study has demonstrated a change in the location of area boundaries due to these diseases. A discussion of the effect of including individuals with history of neurological disease is located in the Supplementary Materials.
Table 2.
Information about the individual hemispheres used in this study.
| hemi | age | gender | PMI | cause of death | neurological disease |
|---|---|---|---|---|---|
| RH1 | M | 80 | – | heart failure | Alzheimer’s |
| RH2 | – | – | – | – | Huntington’s |
| RH3 | – | – | – | – | Huntington’s |
| RH4 | – | – | – | – | Huntington’s |
| RH5 | M | 67 | 12 | – | Normal |
| RH6 | F | 50 | 16 | – | Normal |
| RH7 | F | 75 | 15 | pancreatic cancer | Normal |
| RH8 | M | 81 | 24 | heart failure | Normal |
| RH9 | – | – | – | – | – |
| RH10 | – | – | – | – | Normal |
| LH1 | M | 80 | – | pneumonia | Alzheimer’s |
| LH2 | – | – | – | – | – |
| LH3 | – | – | – | – | – |
| LH4 | – | – | – | – | Huntington’s |
| LH5 | – | – | – | – | Huntington’s |
| LH6 | M | 79 | 24 | pneumonia | Normal |
| LH7 | F | 75 | 15 | pancreatic cancer | Normal |
| LH8 | M | – | 7 | – | Normal |
| LH9 | M | 81 | 24 | heart failure | Normal |
| LH10 | – | – | – | – | Normal |
2.1.2 Whole-brain structural imaging
The same hemispheres that were scanned to image V1 were scanned at standard resolution for gray and white matter tissue over the whole brain using a whole-body 1.5 T Siemens Sonata or Avanto MRI system. The high-bandwidth, multi-echo FLASH pulse sequence presented by Fischl et al. (2004) was used to provide low distortion images with optimal contrast between tissue types. The resulting image volumes had an isotropic voxel size of 1 mm, with a TR of 20 ms and bandwidth of 650 Hz per pixel. Eight echos were acquired during each repetition, and volumes were acquired at each of six flip angles ranging from 5° to 30° in steps of 5°. The FOV was 192 mm × 256 mm × 96 mm if the only one hemisphere was imaged, but was 192 mm × 256 mm × 144 mm if a pair of hemispheres was imaged simultaneously.
2.2 Gray matter segmentation and reconstruction
To allow surface-based analysis of the cortex, a two-dimensional mesh representation of the white and gray matter interface was generated from the whole-brain MRI data. The standard resolution volumes were processed using the FreeSurfer software package (Dale et al., 1999; Fischl et al., 1999a). The cortical white matter was segmented for each hemisphere, and this segmentation was used to build a mesh representation of the interface between the gray and white matter by tiling the voxel faces on this interface with a surface mesh. This voxel interface surface was then smoothed respecting the intensities in the original volume, resulting in a mesh representation of the entire cortical surface.
The surface representation of the white and gray matter interface was used instead of a pial surface because the differences in the spatial profile of MR image intensities external to gray matter between in vivo and ex vivo scans affect the accuracy of the detection of the gray matter-pia interface in scans of ex vivo hemispheres using methods developed for in vivo scans. In particular, the formalin used to fix the hemispheres can exhibit similar image intensities to gray matter, which affects the accuracy of pia identification without affecting identification of the gray and white matter interface.
2.3 Identifying V1
The boundary of V1 was identified manually in the high-resolution images as the terminus of the stria of Gennari. To determine the location of V1 on the cortical surface, the V1 boundary for each subject was projected from the high-resolution volume to the whole-brain volume. First, the high-resolution and whole-brain images for each individual were rigidly co-registered. Next, each point on the boundary of V1 was projected onto the nearest vertex on the cortical surface mesh. Finally, a post-processing step ensured that V1 was a closed polygon. This was accomplished by identifying and eliminating discontinuities in the V1 boundary, which were defined as boundary vertices that have no neighboring surface vertex on the V1 boundary. Discontinuities were corrected by labeling all vertices on the shortest path (computed via the Dijkstra shortest path algorithm (Dijkstra, 1959)) between the vertex on one side of a discontinuity and the surface vertex to which the adjacent V1 boundary point projected. The accuracy of the resulting V1 surfaces were verified by inspection of the alignment between the surface labels of V1 and the high-resolution volume. Errors in the surface projection were corrected manually. The complete set of ten left and ten right hemisphere V1 surfaces are shown in the Supplementary Materials.
2.4 Intersubject registration
To align cortical geometry across subjects, intersubject registration was computed using the method presented by Fischl et al. (1999b). For each hemisphere, an inflated cortical surface mesh was created from the surface representing the interface between white and gray matter by numerically integrating a cost function consisting of a spring force that smooths the surface and a term that limits metric distortion (Fischl et al., 1999a). The mean curvature of the inflated surface was computed at each vertex using neighboring vertices in a radius of 10 mm. The pattern of mean curvature computed over these large neighborhoods can be used to aid initial registration because it reflects mainly the large-scale features of the inflated surface such as the occipital, temporal, and frontal poles, and the dorsal-medial, ventro-medial, and ventro-lateral margins of the inflated cortex.
During inflation, vertices move toward the centroid of their nearest neighbors. Thus, vertices in deep folds will move outward while vertices at the crowns of gyri will move inward. The amount and direction of the overall movement of a vertex represents its sulcal depth (Fischl et al., 1999a; Van Essen, 2005), which can be computed by integrating the distance that a vertex moves along its surface normal during the inflation process. This quantity, called average convexity (Fischl et al., 1999a), effectively captures the geometry of the primary folds due to the consideration of the total vertex movement. The secondary and tertiary folds are less prominent (Bailey and von Bonin, 1951; Chi et al., 1977) and therefore they are smoothed out of the inflated surface more quickly than primary folds. Therefore primary folds contribute more to the convexity because they determine the direction of a vertex normal for a greater number of inflation steps. In contrast, primary, secondary, and tertiary folds are equally represented in the pattern of mean curvature. Therefore, average convexity was used as the main feature representing the geometry of the cortical folds during registration. To generate the two-dimensional coordinate system required to register surface meshes, the cortex of each individual was mapped to the surface of a sphere with minimal distortion as described by Fischl et al. (1999a).
Intersubject registration was performed independently for the groups of left and right hemispheres. First, the pattern of curvature and convexity on the spherical surface of one subject was arbitrarily chosen as an initial target folding pattern, or initial template, and the surface geometry of all other subjects was registered to the geometry of that subject using Automated Spherical Warping (Fischl et al., 1999b). This initial alignment was used to create a surface template that represented the primary folding pattern of all subjects by averaging the convexity in the template space over all individuals. All subjects were then registered to this averaged template.
The process of template generation by averaging the registered convexity among subjects followed by registration to the average template represents an iteration of the template generation process. The number of iterations of template generation that produces optimal V1 alignment is unknown, so several iterations of template generation were performed, and alignment quality was computed for each. The number of productive iterations was determined by identifying the number of iterations after which V1 alignment quality did not consistently increase.
The surface-based registration method of Fischl et al. (1999b) registers the vertices of one surface with a template by iteratively displacing vertices along the spherical surface in the direction that reduces the cost function
| (1) |
where Jp represents mismatch in the folding pattern, JA and Jd represent area and distance distortion, respectively, between the original surface and the registered spherical surface, and JT represents topological defects introduced by the fluid morph. The term
defines similarity in the cortical geometry, which includes the large-scale curvature of the inflated cortical surface through Ji and the average convexity through Jf, which represents the primary folds. The term
where H− is the negative Heaviside step function, penalizes topological defects in the morph independent of the strength of area distortion penalization. For a detailed description of the cost function and the registration procedure, see Fischl et al. (1999a,b).
The result of surface registration is a correspondence between vertices of the spherical surface for each individual to that of the template, which provides a registration of the original cortical surface of each individual in a common surface-based reference space. For visualization purposes an average cortical surface representation was created for the groups of left and right hemispheres from the surfaces of the individual ex vivo hemispheres using the surface averaging technique provided by FreeSurfer, which uses the spherical projection of each individual surface to provide a reference space for averaging individual geometry and curvature patterns.
2.5 Predicting V1 from the folds
The degree to which the location of V1 can be predicted from the pattern of cortical folds was measured by computing probability maps for V1 in the template space. After registration, V1 labels for each individual were mapped into the template space using vertex correspondence between the spherical and registered surfaces. The probability of lying in V1 was then computed for each vertex in the template by averaging the registered V1 label for that vertex across individuals.
Atlasing gives a spatial profile of V1 predictability and provides a method for predicting V1 location in novel subjects, but directly comparing atlases produced by studies employing different imaging and registration methods is difficult. Similarity measures serve as an indicator of alignment quality and allow comparison of atlases across studies without applying each atlas to a common data set. Thus, these measures can be used to compare the alignment quality for individual atlases. Here, three similarity measures were computed: mean kernel size, percent overlap, and the Jaccard similarity coefficient. The mean kernel size similarity measure captures the amount of spatial smoothing required to match individual and averaged V1 labels. It is computed by finding the standard deviation of the Gaussian kernel that, when convolved with the label of an individual, produces the best match between an individual V1 label and the population average. The mean kernel size Kσ (in mm) is computed as
where N is the number of individuals and σk is the width (standard deviation) of the Gaussian kernel for individual k. In essence, the V1 label of each individual is treated as a step function that is smoothed until the spread at the edges of the label match that of the group average, so the mean kernel size is a measure of the average spread of V1 around its boundary in a population. Smaller mean kernel sizes indicate better alignment.
Average percent overlap, as used by Nieto-Castanon et al. (2003) and Hinds et al. (2005), is computed as
for R ∈ {2, . . .,N} V1 surfaces in each combination, where is the number of combinations of the N V1 surfaces, āj is the average area of the R surfaces in combination j, and ∩ai represents the area of the spatial intersection across the V1 surfaces in combination j, indexed by i ∈ {1, . . .,R}. The average percent overlap is an estimate of the ratio between the area common to all V1 maps to the average area of V1. Therefore, it represents the amount of overlap that is expected on average when registering a new subject to the atlas. Larger percent overlaps indicate better alignment.
The Jaccard similarity coefficient SJ (Jaccard, 1908) is the ratio between the spatial intersection and the union over all V1 estimates:
SJ is commonly used as a similarity measure that represents the expected worst case alignment of a new subject to the atlas. Larger Jaccard similarity coefficients indicate better alignment.
In addition, the percent blurring statistic used by Fischl et al. (1999b) to measure functionally defined V1 spread is reported for comparison with the results of previous studies. It is computed as
where Pb is the percent blurring, ā is the average area of V1, and ∪ak is the spatial union of all V1 estimates. The percent blurring is the ratio of the area of the total spread of V1 to the mean area. Lower percent blurring indicates better alignment.
To compare alignment quality between atlases when the individual subjects used in the atlas generation process are not available, the cumulative probability was computed directly from the atlas probabilities. Cumulative V1 probability C(p) is the proportion of the atlas that exhibits probability of less than or equal to probability p, computed here by approximating the cumulative density as a sum
where Gu is the proportion of the atlas exhibiting V1 probability u. The steps of u are determined by the probability values possible given the number of subjects used to build the atlas.
2.6 Optimizing surface registration
The terms JA and Jd in (1) represent area and distance distortion, respectively, between the original surface and the registered spherical surface. The amount of metric distortion allowed during registration of the folds is controlled by the parameters λA and λd. For details regarding the computation of JA and Jd see Fischl et al. (1999a). These quantities are metric properties of the original and deformed surfaces, which for computational reasons are estimated in a neighborhood of each vertex individually instead of considering the global metric structure of the surface. It has been demonstrated that preserving local metric structure is effective at preserving global metric structure when using the FreeSurfer surface flattening method (Balasubramanian et al., 2006), a process similar to the local geometric surface deformations performed during the fluid morph.
When the surface registration method of Fischl et al. (1999b) was developed, they proposed values for JA and Jd based on the behavior of surface geometry under other surface morphing processes such as inflation and spherical projection. Because at that time no direct method for evaluating the alignment quality of cortical areas was available, the parameter values producing optimal alignment were left as an empirical question. Here, the location of V1 on the cortical surface provides a method for evaluating registration quality under various parameter values.
To determine the amount of distortion that should be allowed to optimize V1 registration, the relevant parameters were varied (λA ∈ {0.0,0.1,0.4,1.0,2.5,5.0} and λd ∈ {0.1,0.4,1.0,2.5,5.0,10.0} for 36 distinct parameter value pairs) and the quality of alignment was evaluated in the resulting registrations. Because each of the similarity measures indicates a different relevant property of the atlas, the mean kernel size, Jaccard similarity coefficient, and percent overlap were combined to obtain a single measure of V1 alignment quality. Alignment quality was computed by rank ordering the parameter value pairs with respect to each of the three similarity measures, then averaging the rank over all measures. This represents the best compromise between similarity measures that yields good alignment based on all measures. Optimization was performed at each successive iteration of the template generation process, choosing the registration that produced the best V1 alignment to serve as the template for the next iteration.
2.7 Alignment quality of the calcarine sulcus and V1
The calcarine sulcus is the feature of the cortical folding pattern that is nearest to V1, and thus its alignment has the greatest effect on the alignment of V1. However, the optimal registration parameter search procedure outlined above does not directly address whether good calcarine sulcus registration leads to good V1 registration. To address this question, calcarine alignment quality was independently measured and compared with V1 alignment quality.
The calcarine sulcus was identified as the region of positive average convexity just anterior of the occipital pole on the medial surface. In all hemispheres, the calcarine sulcus joins the parieto-occipital sulcus. The calcarine sulcus was determined to end at a line drawn parallel to the parieto-occipital sulcus beginning at the cuneal point. In several of the hemispheres the calcarine sulcus was interrupted by a gyrus approximately one-third of the way from the occipital pole to the cuneal point. In these cases, the two islands of positive average convexity were taken as the calcarine sulcus.
A probabilistic atlas of the calcarine was generated using the same methods as for the V1 probabilistic atlas for the same parameter value pairs. Similarity measures were computed for each pair, and the average rank of each pair was taken to be the calcarine alignment quality, as for V1. To address the relationship between calcarine and V1 alignment quality, a simple linear regression was performed to find the correlation between the average rank for each feature over parameter value pair.
3 Results
3.1 Iterations of template generation
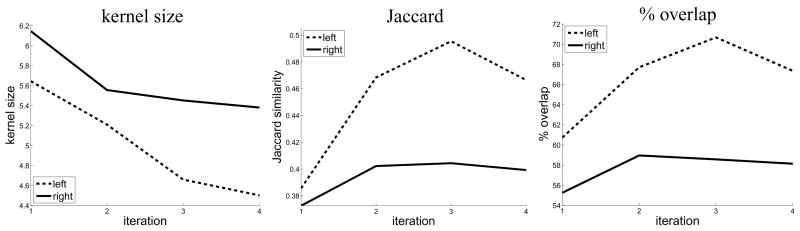
To determine the number of productive iterations of template generation, V1 alignment quality was computed for between one and four iterations. Figure 2 shows the relationship between alignment quality and iteration number. Alignment quality increases for the first and second iterations of template generation for each hemisphere and similarity measure and no benefit consistent over all similarity measures is gained after the third iteration. Therefore, in the following V1 alignment was considered final after iteration three.
Figure 2.
The similarity measures at the optimal parameter values for each iteration of the template construction process with the dashed line indicating values for the left hemispheres and the solid line indicating right hemisphere values. Substantial increase in alignment quality is evident from the first to the third iterations, after which there is no consistent increase or decrease in alignment quality.
Table 3 shows the parameter values that produced the best V1 alignment for between one and four iterations of template generation. Notably, the optimal value for λd rises considerably between iterations one and two for both hemispheres. This indicates that more distance distortion should be introduced in the first iteration to create good V1 alignment.
Table 3.
The parameter values that produced the best V1 alignment at each iteration of template generation for each hemisphere. The average rank over similarity measures is shown for each parameter value pair.
| Left | Right | |||||
|---|---|---|---|---|---|---|
| iteration | λd | λA | rank | λd | λA | rank |
| 1 | 0.0 | 0.1 | 1.00 | 0.1 | 0.1 | 1.00 |
| 2 | 1.0 | 0.0 | 2.00 | 2.5 | 0.4 | 1.33 |
| 3 | 5.0 | 0.1 | 1.00 | 5.0 | 1.0 | 2.00 |
| 4 | 5.0 | 2.5 | 1.67 | 10.0 | 2.5 | 1.33 |
3.2 Optimal registration parameters
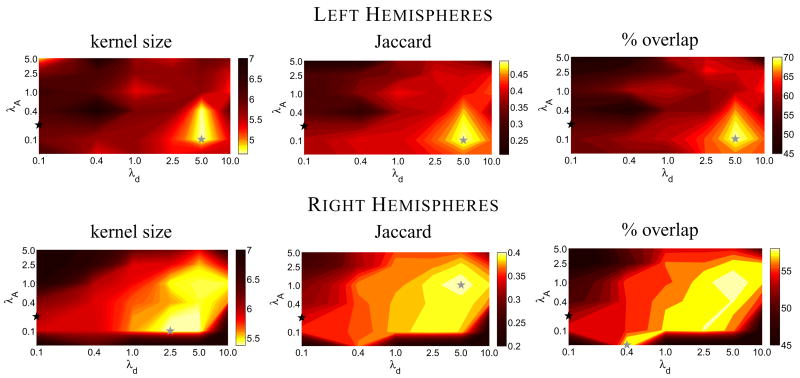
The quality of V1 alignment was examined for registrations performed for several iterations of template generation using a range of parameter values for both λd, which controls the amount of distance distortion introduced by the registration, and λA, which controls the area distortion. The average kernel size, Jaccard similarity coefficient, and the percent overlap of all subjects for both hemispheres after three iterations of template generation are shown in Figure 3. There is a gross consistency in the alignment quality across hemispheres and similarity measures, although the parameter value pair exhibiting the best alignment varies somewhat.
Figure 3.
Alignment quality of V1 computed for several values of λA and λd after three iterations of the template generation process. The top row represents left hemispheres and the bottom row represents right hemispheres. The average kernel size is shown in the first column. Lower sizes represent better V1 alignment, as indicated by the color bars. The Jaccard similarity coefficient for the same parameter values is shown in the second column, with higher values representing better alignment. The third column shows the percent overlap of V1 for all individuals Po(10). In all images, the location of the gray star indicates the parameter values that produced the best V1 alignment for that measure. The location of the black star indicates the parameter value pair commonly employed using this registration method.
Taking the lowest total sum ranked parameter value pair over the various similarity measures indicated that λA = 0.1 and λd = 5.0 (average rank 1.0) produces the optimal alignment of V1 for the left hemispheres and λA = 1.0 and λd = 5.0 (average rank 2.0) produces the optimal alignment of V1 for the right hemispheres. These values of λA are similar to the value of 0.2 that is routinely used in imaging studies using the FreeSurfer software package. However, the routinely used value of λd = 0.1 is much lower than the optimal values identified here, as shown in Table 4.
Table 4.
Summary of the results of the optimal parameter value search. The similarity measure value under the V1 alignment using the optimal parameter values is shown for each measure and hemisphere. In addition, the similarity measure value using the standard parameter values of λA = 0.2 and λd = 0.1 is shown for each hemisphere.
| Left | Right | |||
|---|---|---|---|---|
| similarity measure | optimal | standard | optimal | standard |
| kernel size | 5.28 | 6.12 | 5.55 | 7.06 |
| % blurring | 45.4 | 59.6 | 46.6 | 71.8 |
| Jaccard coef. | 0.47 | 0.33 | 0.40 | 0.27 |
| % overlap | 68.8 | 52.1 | 58.9 | 47.0 |
The similarity values shown in Table 4 indicate a much higher V1 alignment quality than that reported by previous studies (Amunts et al., 2000; Fischl et al., 1999b). This suggests that surface-based alignment via the cortical folds can produce high-quality alignment of the primary visual cortex.
To determine the amount of increased V1 alignment quality using the optimal parameter values, the standard values of λd = 0.1 and λA = 0.2 were used to register V1 to the surface template. The resulting values for the similarity measures are shown in Table 4. For the left hemispheres, V1 alignment quality increased using the optimal parameters by 29.8% for kernel size, 20.9% for percent blurring, 50.5% for Jaccard similarity, and 41.6% for percent overlap. For the right hemispheres, V1 alignment quality increased using the optimal parameters by 32.1% for kernel size, 50.6% for percent blurring, 36.2% for Jaccard similarity, and 17.7% for percent overlap.
In addition, a rigid surface-based registration was performed by matching the folding pattern between the individual and the template without the surface morphing step described in Section 2.4. This process only allows rotations of the spherical surface representation without introducing any further metric distortion. V1 alignment quality was lower without the morphing step, resulting in a percent overlap of 28.8% for the left hemispheres and 22.7% for the right hemispheres. The Jaccard similarity coefficients were 0.14 and 0.12, and the percent blurring was 104.2% and 98.7% for the left and right hemispheres. The percent overlap is low but still greater than for volumetric registration, suggesting that this rigid surface-based registration discounts some of the variability due to the cortical folds, but a lesser amount than observed when surface morphing is employed.
3.3 V1 atlas
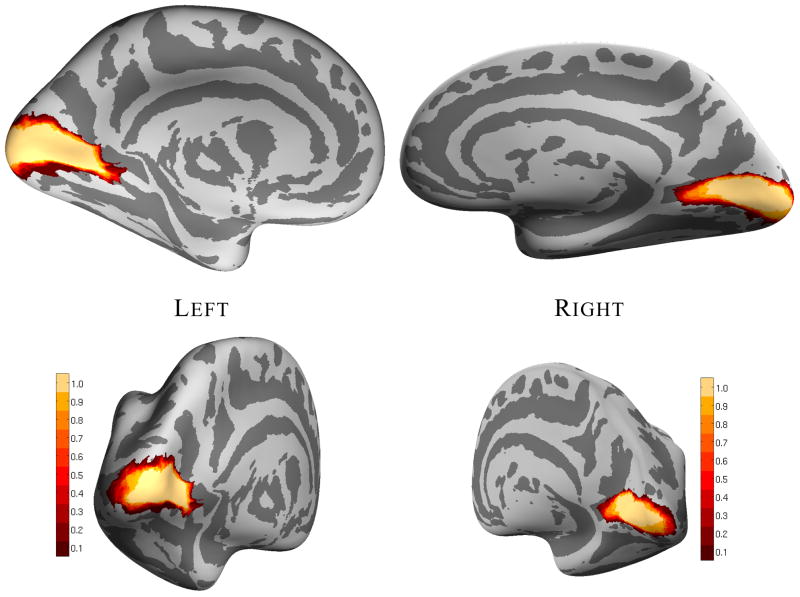
Spatial probability maps for V1 following intersubject registration are displayed on an average cortical surface representation in Figure 4. A large area of probability 1.0 of lying in V1 is evident along the fundus and walls of the calcarine sulcus for both the left and right hemispheres. A V1 probabilistic atlas exhibiting similar alignment quality was obtained via registration of the individual ex vivo hemispheres to a template generated from 40 healthy, living humans as shown in the Supplementary Material. This atlas has been made available as part of the FreeSurfer software package (http://surfer.nmr.mgh.harvard.edu/) for predicting the location of V1 from cortical surface representations of living subjects.
Figure 4.
A view of the medial surface of the inflated average left hemisphere cortical surface is shown on the top left, and an oblique posterior view is shown below it. A medial view and an oblique posterior view of the inflated average right hemisphere cortical surface is shown to the right. V1 probability is indicated by the color of the vertex, with yellow indicating high probability and red indicating low probability, as indicated by the color bars.
3.4 V1 similarity
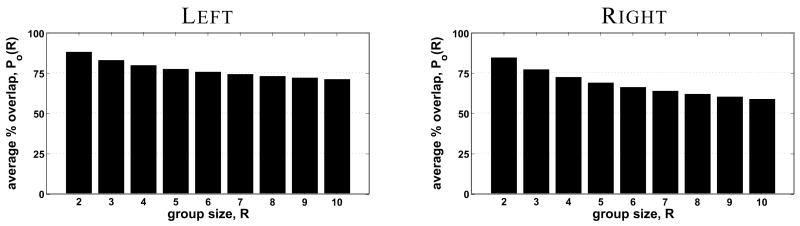
The average kernel size required to match individual and registered average labels was 4.7 mm for left hemisphere V1 and 5.4 mm for right hemisphere V1. The average percent overlap for all ten left hemispheres was 70.6%, while the ten right hemispheres exhibited a percent overlap of 58.5%. These overlaps are much greater than that observed by other V1 atlasing studies using volume-based registration (17.8% by Roland et al. (1997) and 0.13% by Amunts et al. (2000)). The average percent overlap for groups of R subjects computed using the optimal registration parameter values is shown in Figure 5. The percent blurring for V1 was 42.7% for the left hemispheres and 44.9% for the right hemispheres, lower than that observed by Fischl et al. (1999b) using the same registration methods to align functionally defined V1 (about 80%).
Figure 5.
Average percent overlap of V1 over all possible groups of R left hemispheres is shown on the left, and right hemispheres on the right.
3.5 Comparing volume and surface-based registration
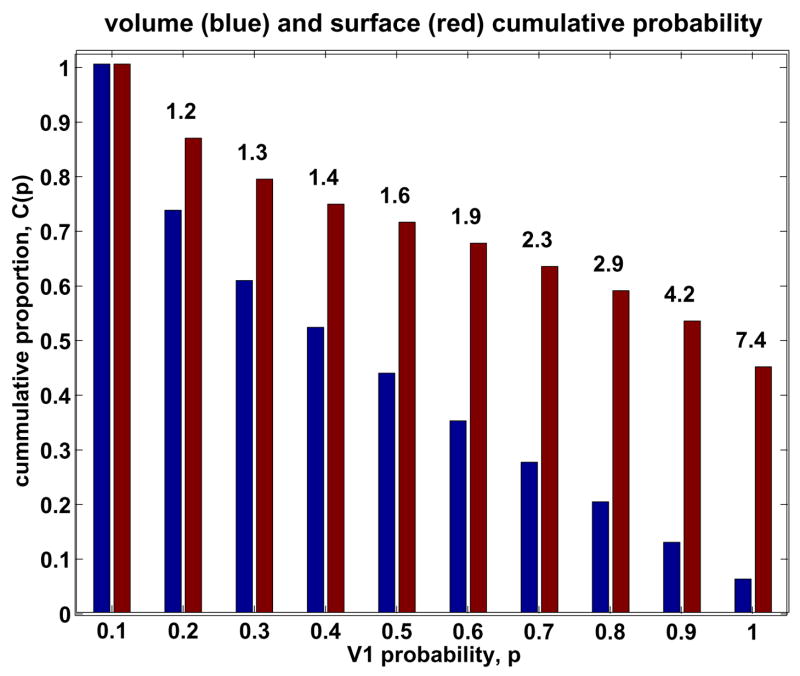
The atlas quality observed under surface-based registration was compared to that observed using nonlinear volume-based registration by examining the cumulative atlas probability for the atlas generated here and the publicly available V1 atlas (http://www.fz-juelich.de/inb/inb-3/spm_anatomy_toolbox) based on the work of Amunts et al. (2000). Figure 6 indicates that the atlas generated using surface-based registration has a substantially greater registration quality. In particular, the proportion of the atlas exhibiting high probability of V1 is substantially greater for surface-based registration.
Figure 6.
The cumulative V1 probability for the atlas presented here generated using surface-based registration (red) and an atlas generated using nonlinear volume-based registration (blue). The ratio of the cumulative probability using surface-based and volume-based registration is indicated above the bar for each probability.
3.6 Calcarine and V1 alignment quality
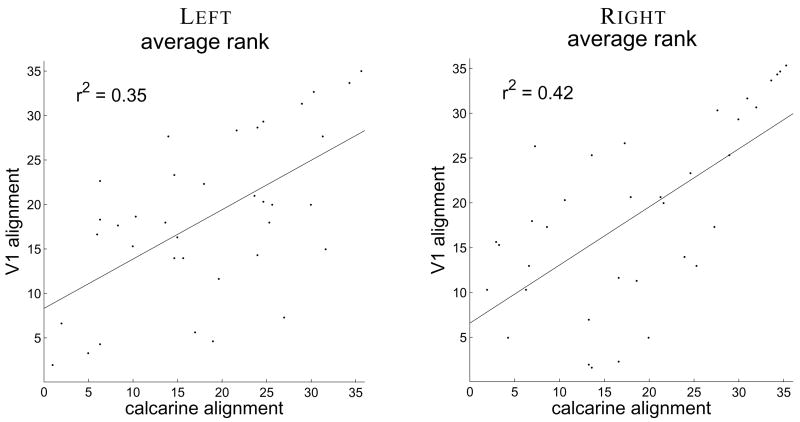
To examine the possibility that certain parameter value pairs resulted in good V1 alignment without necessarily better alignment of the folding pattern, the alignment quality of the calcarine sulcus was used to predict V1 alignment quality. The labels representing the calcarine sulcus were averaged in the registered space across individuals resulting in a probabilistic atlas of the calcarine sulcus (see Supplementary Materials). The similarity measures and average rank were computed for each parameter value pair. The predictability of V1 alignment quality from calcarine alignment quality was examined by computing the coefficients β of the simple linear regression model y = β0 + β1x by minimizing the residual sum of the squared error. Here y represents a vector of average ranks of V1 alignment over parameter values and x represents a vector of average ranks of calcarine alignment.
Figure 7 shows the linear model fit and the correlation coefficient for both hemispheres (r2 = 0.35 for the left hemispheres and r2 = 0.42 for the right hemispheres), indicating predictability of V1 alignment from calcarine alignment. Computing the F-statistic for the regression confirms that the correlation is not due to chance (p < 0.001 for both hemispheres). This quantitatively establishes a consistent relationship between V1 and the folding pattern. However, the regression accounts for only about 40% of the variance, indicating that there are additional factors involved in V1 alignment quality.
Figure 7.
Linear regression of V1 alignment quality average rank over parameter values with respect to calcarine alignment average rank for each group of hemispheres. The line corresponding to the coefficients β0 and β1 is drawn, and the r2 value for the regression is shown for each hemisphere to the top left.
4 Discussion
4.1 Predicting V1 from folds
The results of the work presented here indicate that the primary folds serve as a good predictor for the location of V1. The degree to which this is the case has thus far been overlooked due the lack of adequate quantitation in previous studies.
Stensaas et al. (1974) investigated the relationship of V1 to the location of the lips of the calcarine sulcus in an effort to determine the best placement for electrodes as part of a potential cortical visual prosthesis. They found that on average 67% of V1 was buried within the calcarine sulcus, and reported the amount of V1 outside the calcarine for exposed portions of cortex posterior (average 6%), dorsal (average 21%), and ventral (average 4%) to the sulcus. Standard deviations of the locations of these features were not reported, only ranges. The amount of exposed V1 ranged between 0% and 22% posterior to the calcarine, 9% and 36% dorsal, and 0% and 20% ventral, indicating a fairly substantial range of variation.
Rademacher et al. (1993) measured the position of V1 on the cortical surface relative to several salient landmarks of the folding pattern, and found on average 62% of V1 to be inside the calcarine sulcus. Although very little statistical analysis was presented, they did report that that 92% ± 7.0% (mean ± standard deviation) of V1 was posterior to the cuneal point. This finding complements the results of Stensaas et al. (1974) by providing an estimate of variability of anterior V1 with respect to a feature of the cortical folds.
While the results of Stensaas et al. (1974) and Rademacher et al. (1993) are consistent with those presented here, a rigorous comparison is not possible due to methodological differences. Stensaas et al. (1974) reported detailed values for V1 location relative only to the lips of the calcarine sulcus, while the surface-based registration method used here considers the entire mosaic of primary folds via average convexity and mean curvature. In addition, the lack of statistics prevents a true variability analysis of their data. Rademacher et al. (1993) did not report detailed values for V1 location except for the location of its anterior boundary relative to the cuneal point, making comparisons of their results with those of other studies difficult. In addition, they concentrated on measuring the degree to which folds serve as effective limiting landmarks beyond which V1 does not extend instead of reporting the spread around these landmarks. Thus, their work is effective as a guide to predicting where V1 is not, rather than where it is.
4.2 Predicting the location of other cortical areas
Much effort has been applied to the study of the relationship between cortical folds and architectonic cortical areas. In his classical work, Brodmann (1909) indicated that there was considerable variability in the relative location of areas and folds. However, other researchers have described a consistent relationship between particular cortical areas and folds (Smith, 1904; Sanides, 1970), leading to a widespread belief that certain folds predict the location of cortical areas (Welker, 1990; Rademacher et al., 1993).
Over the past decade, the Amunts and Zilles group has produced maps of many cytoarchitectonically delineated cortical areas in one group of ten ex vivo human brains (Amunts et al., 1999; Geyer et al., 1999; Amunts et al., 2000; Grefkes et al., 2001; Morosan et al., 2001; Rademacher et al., 2001b,a; Amunts et al., 2005; Wilms et al., 2005; Caspers et al., 2006; Eickhoff et al., 2007). The results of these studies allowed an overall analysis of the relationship between prominent cortical folds and the location of area boundaries (Zilles et al., 1997). It was concluded that folds are reliable predictors of area boundaries in only a few cases, including the border between Brodmann’s areas 4 and 3a, and the anterior and posterior borders of primary auditory cortex. Notably, Zilles et al. (1997) explicitly state that the V1 border does not share a consistent relationship with the folds.
The work presented here demonstrates that the location of V1 can be predicted with high accuracy from the cortical folds. The discrepancy between these results and the conclusions of Zilles et al. (1997) can be explained in part by the different methods used. Here, the entire mosaic of the primary folds was used to draw correspondence between the cortical geometry of distinct individuals and the alignment quality of V1 was then measured in a common space. However, Zilles et al. (1997) examined the correlation between individual sulci and gyri in individual brains. While the methods used were not described in detail, a spatially offset but consistent relationship between the location of primary folds and the boundary V1 or the inclusion of secondary or tertiary folds in the analysis could have obscured a consistent relationship between the folds and V1, and thus between the folds and other areas as well.
In addition, Zilles et al. (1997) concentrated on co-localization of folds and cortical area boundaries, while the work presented here examined alignment of the area as a whole. Since V1 is a large cortical area, good alignment may result even if there is a fairly large amount of spread in the location of the area boundary itself. While the results of the work presented here suggest that the location of cortical areas can be predicted from the primary folds, the achievable alignment quality for other cortical areas is an open question that will be addressed in future work.
4.3 Intersubject registration methods
The goal of intersubject registration is to normalize variability in the geometry of brain structures to align features in a common reference space. It is widely assumed that mapping brain geometry into a reference space is effective at superimposition of common functional areas among subjects. This assumption is the basis for the use of intersubject registration to increase the power of statistical tests used to identify regions of neural activation in functional imaging experiments. Despite widespread use of intersubject registration in functional imaging studies, very little validation of these methods has been performed (Crum et al., 2003).
Volume-based intersubject registration techniques match cerebral geometry among subjects in a three-dimensional Euclidean space, thus ignoring the intrinsic geometry of the cortical surface. Linear volume-based registration (Talairach and Tournoux, 1988) matches volumetric image intensities via 12-parameter affine transformations, while nonlinear volume-based registration methods (Evans et al., 1991; Miller et al., 1993; Roland et al., 1994; Friston et al., 1995; Davatzikos et al., 1996; Schormann and Zilles, 1998; Woods et al., 1998) use highly constrained nonlinear transformations to match tissue types across individuals.
4.3.1 Linear volume-based registration
Amunts et al. (2000) used linear volume-based registration to derive correspondence between the brains of ten individuals. Microanatomically identified V1 was aligned in stereotaxic coordinates, resulting in very low predictability, with a percent overlap of V1 for all ten subjects of 0.13%. The volume of the region of probability 1.0 of V1 inclusion was only 30 mm3. As stated by Amunts et al. (2000), this indicates that the location of V1 exhibits substantial variability in stereotaxic coordinates. A similar degree of variability was reported for area V2.
Alignment of functionally delineated cortical areas using linear volume-based registration has been compared directly to alignment using nonlinear surface-based registration by Fischl et al. (1999b). Standard fMRI-based visuotopic mapping techniques (Sereno et al., 1995) were used to determine the border of V1 in 11 subjects, then linear volume-based registration (Talairach and Tournoux, 1988) was performed to compare the alignment of V1 across subjects. The percent blurring for V1 calculated by Fischl et al. (1999b) was about 600% for Talairach registration, while the blurring after surface-based registration was about 80%, indicating that linear volume-based registration produces lower quality alignment than surface-based registration. There are two plausible explanations for the increased alignment quality they observed: that surface-based registration is superior to volume-based registration at superimposing cortical areas, or that the greater degrees of freedom in the nonlinear surface-based technique allowed better alignment of cortex. The contribution of these factors is examined below.
4.3.2 Nonlinear volume-based registration
Nonlinear volume-based registration methods (Christensen et al., 1994; Roland et al., 1994; Friston et al., 1995; Woods et al., 1998; Shen and Davatzikos, 2002) seek to improve the alignment quality of cortex in stereotaxic coordinates over that possible with linear volume-based methods by allowing local deformations of the brain volume. This is accomplished by applying a tightly constrained nonlinear transformation to the voxel intensities of an individual brain image such that the voxel intensities match those of a template volume. By allowing more degrees of freedom than linear methods, nonlinear registration methods seek to superimpose common tissue types with the goal of aligning common functional areas.
Alignment of cortical areas
Several studies have addressed the ability of nonlinear volume-based registration methods to align cortical areas. Roland et al. (1997) performed a similar study to that of Amunts et al. (2000) but compared the alignment of V1 and other cortical areas in five individuals using nonlinear volume-based registration (Roland et al., 1994) to that produced by linear volume-based registration. They found that substantially greater alignment accuracy could be achieved by using nonlinear volume-based registration, observing a 17.8% overlap for V1 of all subjects using the nonlinear method and only a 2.3% overlap using the linear method. Similar results were obtained for Brodmann’s Area 1/2, 3a, 3b, 4a, and 4p. While the findings of Roland et al. (1997) establish that nonlinear volume-based methods are superior to linear methods at aligning cortical areas, the substantially higher predictability observed in the present study under surface-based registration suggests that it outperforms nonlinear volume-based methods at aligning cortical areas.
The nonlinear volume-based registration method of Roland et al. (1994) and Schormann and Zilles (1998) has been used to study the variability in stereotaxic coordinates for several other cortical areas. Low alignment quality was reported for inferior parietal cortical areas (Caspers et al., 2006), the middle temporal area (MT) (Wilms et al., 2005), motor cortical areas (Rademacher et al., 2001a), somatosensory cortical areas (Geyer et al., 1999; Grefkes et al., 2001; Eickhoff et al., 2007), the primary auditory cortex (Rademacher et al., 2001b; Morosan et al., 2001), and Broca’s region (Amunts et al., 1999). Less variability was observed in the location of amygdalar, hippocampal, and entorhinal allocortical regions (Amunts et al., 2005), although variability was still substantial. These studies make it clear that predictability of the location of cortical areas in stereotaxic coordinates is low when alignment via nonlinear volume-based methods is used.
Alignment of cortical folds
The results presented here indicate that there is a consistent relationship between the location of cortical area V1 and the location of cortical folds. Therefore, the ability of a registration method to match the folding pattern between individuals is a good indicator of whether cortical areas will be aligned, at least for V1. Grachev et al. (1999) compared alignment of expert-labeled anatomical landmarks under the linear volume-based method of Talairach and Tournoux (1988) and the nonlinear method of Woods et al. (1998), reporting higher alignment quality for cortical landmarks under nonlinear registration.
In a more complete study of the registration of cortical folds, Nieto-Castanon et al. (2003) parcellated cortex via hand tracing in nine subjects using the methods described by Caviness et al. (1996) and applied the nonlinear volume-based registration method provided in the SPM Toolbox (Friston et al., 1995). They found that for a single pair of subjects the average percent overlap for a given region was between 30% and 35%. As progressively more subjects were registered, the average percent overlap dropped to nearly zero percent for each brain region.
In a similar study, Hellier et al. (2003) compared the performance of several volume-based registration techniques using both global measures such as overlap of common tissue types (e.g., gray matter and white matter classification) as well as local measures such as the pattern of mean curvature and the position of individual gyri and sulci. They reported that nonlinear methods produce significantly more overlap of tissue types than linear methods, but that there were no significant differences between these methods on the local measures. In general, alignment quality of the local features of mean curvature and sulci and gyri was quite low under all methods.
Salmond et al. (2002) studied alignment quality of the medial temporal lobe using the nonlinear volume-based registration method of Friston et al. (1995). They studied localization in 20 normal children and examined the effect of various registration parameter values on the spatial co-localization of several manually delineated anatomical landmarks. Standard deviation from the mean location of the landmarks was used as a measure of alignment quality. For nonlinear registration using the optimal registration parameters, Salmond et al. (2002) report about a 1 mm standard deviation, indicating a 34% overlap for the 20 subjects, assuming an isotropic voxel size of 1 mm and a normally distributed registration error. This is a higher degree of overlap than has been observed in other studies of co-localization using nonlinear volume-based registration, however the allocortical anatomical landmarks used by Salmond et al. (2002) exhibit low spatial variability compared to neocortical landmarks (Bailey and von Bonin, 1951; Amunts et al., 2005). However, consistent with the results of the present study, Salmond et al. (2002) report that a nonlinear warping that results in less distortion, accomplished by using fewer basis functions, and thus fewer degrees of freedom, produced the best alignment.
4.3.3 Surface-based registration
Previously, surface-based registration has been shown to outperform linear volume-based registration methods at aligning functionally defined visual areas (Fischl et al., 1999b), as well as the location of some sulci and gyri (Van Essen, 2005). However, the work presented in this report represents the first direct demonstration of surface-based alignment of anatomically delineated cortical areas via the cortical folds.
The percent blurring of about 35% observed here for anatomically delineated V1 is much lower than that of about 80% observed previously for functionally defined V1 (Fischl et al., 1999b) using the same registration methods. The indirect nature of cortical area boundary measurement performed by Fischl et al. (1999b), and the increased distortions in fMRI, contribute to this discrepancy. Also, Fischl et al. (1999b) only measured the representation of the central visual field in V1. Differences both in the amount of visual field that was stimulated and in the amount of cortex devoted to its representation contribute to intersubject V1 size variation that is independent of the relationship of the entire cortical area to the cortical folds. Dougherty et al. (2003) performed a study of size variation among early visual areas within and across individuals that suffers from a similar problem because visual areas were delineated via fMRI without validation via anatomy.
Estimates of visual area size and location derived from fMRI exhibit additional variability due to the substantial artifacts associated with fMRI-based visual area maps derived by estimating the field-sign computed from data acquired using phase-encoded visual stimuli (Sereno et al., 1995). These artifacts result in both false positives and negatives for area location and thus estimates of visual area size and location based on fMRI are less accurate than direct observation of the anatomical features of these areas.
4.4 Intrinsic geometry
Recently, it has been demonstrated that the shape of V1 intrinsic to the cortical surface is stereotyped across individuals (Hinds et al., 2005). Because surface-based analysis methods use a mesh representation of the cortical surface, they are able to represent its “intrinsic geometry” in the form of distances along the cortical surface itself. Since they consider the natural two-dimensional structure of the cortex, surface-based methods for intersubject registration have the potential to greatly outperform volume-based methods at registering V1 among individuals. However, registration quality also depends on the relationship of V1 to cortical features used to register, and the ability of the registration method to preserve the intrinsic geometry during the non-isometric transformations used to effect it.
There are two parameters in the registration method of Fischl et al. (1999b) that control the amount of distortion introduced during the warping process: λA controls area distortion and λd controls distance distortion. The higher these stiffness parameters are, the less the surface vertices are allowed to deform in relation to their neighbors, and the closer to isometric the registration process becomes. In addition to establishing that the primary folds have a consistent relationship to the location of V1, the work presented here establishes that higher values of λd than are typically employed lead to better alignment of V1. This 24 supports the conclusion of Hinds et al. (2005) that the intrinsic shape of V1 is similar across individuals because allowing less distance distortion preserves intrinsic shape. However, much lower values of λA were found to lead to optimal V1 alignment, which indicates that allowing area distortion during the registration process improves V1 alignment. Such a difference between these two parameter values is surprising because the area and distance distortion are not independent. Preserving the distances between neighboring vertices should also preserve the area of the surrounding triangle faces.
The results of this study also suggest that the amount of distance distortion that should be allowed to produce good V1 alignment varies with iterations of the template generation process. For early iterations, better alignment is observed using low values for both λA and λd, but with subsequent iterations higher values of λd produce better V1 alignment. Based on the results presented here, the parameter value pair that should be employed to produce good V1 alignment is λA = 0.4 and λd = 5.0 in the third iteration of template generation.
Prohibitively long computation times dictated that V1 alignment quality was assumed to be independent between iterations. This amounts to assuming that the registration parameter values that produced the best V1 alignment at a given iteration will result in the best template to be used for the next iteration. While a reasonable assumption, this need not be the case in general, and as computational power increases a full investigation of the dependent model should be undertaken. Until then, the general trend of increasing λd over iterations as shown in Table 3 should be followed.
Additionally, the dependence of the V1 alignment quality on the choice of subject used as the initial template was not possible due to prohibitively long computation times. The effect of initial template subject should be investigated in future work.
4.5 Probabilistic atlases
Because observation of the histological features of cortex necessary to delineate cortical areas is not possible with current imaging technologies, probabilistic atlases have been developed to predict the location of cortical areas based on features of cortical geometry (Mazziotta et al., 1995; Fischl et al., 1999b; Amunts et al., 2000; Rademacher et al., 2001b; Van Essen, 2005). As discussed above in Section 4.3, previous atlasing work has produced atlases with a high associated prediction error of cortical area location.
Probabilistic atlases have great potential benefit in understanding the functional organization of the normal and diseased brain. Locations of activation in functional brain imaging studies are routinely reported in terms of Brodmann areas (Brodmann, 1909), however methods for determining the location of Brodmann areas from anatomical brain images are qualitative. High quality probabilistic atlases such as the one produced in this study provide a better method for reporting these activations, and therefore support the integration and comparison of results between studies.
Because areas are assumed to fill distinct functional roles, the ability to locate them accurately in living subjects enables experiments that investigate the relationship between area features (e.g., size and shape) and subject performance on tasks related to a hypothesized functional role (Duncan and Boynton, 2003). Similarly, probabilistic atlases can provide information on the functional effects of structural brain changes associated with disease or brain injury. The work presented here represents the first probabilistic atlas of a functional cortical area that has a sufficiently low associated prediction error to be useful in these contexts.
In addition to V1, high-resolution MRI has been used to delineate cortical area MT (Walters et al., 2003), entorhinal cortex (Augustinack et al., 2005), and other areas (Fatterpekar et al., 2002). As imaging technologies advance, delineation of further cortical areas will become possible, allowing atlases to be constructed toward the goal of reproducing cortical area maps such as that of Brodmann (1909), Vogt (1911), and von Economo and Koskinas (1925). However, this new map will have two main advantages over previous maps: it provides a quantitative method for projecting the map onto novel individuals and an explicit error analysis allowing the confidence of prediction to be reported.
4.6 Functional imaging studies
Intersubject registration is routinely used in functional brain imaging studies as a tool to increase the power of statistical tests by enabling intersubject averaging. However, statistical power is only increased if the registration method is effective at producing alignment of common functional areas across subjects. Power can, in fact, decrease if the benefit from functional area overlap is offset by the increased experimental error introduced by intersubject registration.
As discussed in Section 4.3, previous studies have found that volume-based methods for intersubject registration produce little overlap of common functional areas. The results of the work presented here provide direct evidence that surface-based registrations via the cortical folds exhibit large regions of overlap, suggesting that they are effective at increasing the power of statistical tests in functional imaging experiments targeting cortical areas. It is important to note that these surface-based methods are only designed to register cortical areas, and are thus ill suited to register subcortical structures. Linear (Talairach and Tournoux, 1988) or nonlinear (Christensen et al., 1994; Friston et al., 1995; Shen and Davatzikos, 2002; Fischl et al., 2004) volume-based registration methods are more appropriate for this task.
4.7 Development of folds and arealization
The high predictability of the location of V1 from the primary folds observed here suggests that the development of the primary folds and the process of arealization are related, at least for the primary visual cortex. The calcarine sulcus is one of the first cortical folds to appear, presenting at about 16 weeks gestational age (Chi et al., 1977). V1 can be delineated cytoarchitecturally via differences in the development of the deep cortical layers after 20 or 21 weeks of gestation (Leuba and Kraftsik, 1994). In contrast, V1 does not begin the process of myelination until birth, and may take up to four years to become fully myelinated (Yakovlev and Lecours, 1967; Welker, 1990). Therefore, direct, noninvasive investigation of the development of V1 in relation to folding is not possible by imaging the stria of Gennari using MRI of myelin. More detailed analysis will require invasive methods or imaging of alternate features, such as cytoarchitecture, to investigate the relationship between the developmental mechanisms that give rise to areas and folds. It is interesting to note that the spatial relationship between the folds and V1 observed here is consistent with published theories of the effect of area location on the development of the folds (Van Essen, 1997; Hilgetag and Barbas, 2006).
5 Support and Acknowledgments
Support for this research was provided in part by the National Institute for Biomedical Imaging and Bioengineering (R01 EB001550), Mass. ADRC grant (5P50 AG05134), the National Center for Research Resources (P41 RR14075, R01 RR16594 and the NCRR BIRN Morphometric Project BIRN002, U24 RR021382), the National Institute for Neurological Disorders and Stroke (R01 NS052585) as well as the Mental Illness and Neuroscience Discovery (MIND) Institute, and is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. We thank the Massachusetts Alzheimer Disease Neuropathology Core (P50 AG005134) for tissue specimens used in this study.
The authors thank Megan L. Blackwell, Christopher J. Wiggins, André J.W. van der Kouwe, and Irina Sigalovsky for their technical assistance, and Mukund Balasubramanian, Jason W. Bohland, Leo Grady, and Jason Tourville for discussion and helpful insights.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amunts K, Kedo O, Kindler M, Pieperhoff P, Mohlberg H, Shah N, Habel U, Schneider F, Zilles K. Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps. Anatomy and Embryology. 2005;210(5):343–352. doi: 10.1007/s00429-005-0025-5. [DOI] [PubMed] [Google Scholar]
- Amunts K, Malikovic A, Mohlberg H, Schormann T, Zilles K. Brodmann’s areas 17 and 18 brought into stereotaxic space-where and how variable? Neuroimage. 2000;11(1):66–84. doi: 10.1006/nimg.1999.0516. [DOI] [PubMed] [Google Scholar]
- Amunts K, Schleicher A, Burgel U, Mohlberg H, Uylings HB, Zilles K. Broca’s region revisited: cytoarchitecture and intersubject variability. J Comp Neurol. 1999;412(2):319–341. doi: 10.1002/(sici)1096-9861(19990920)412:2<319::aid-cne10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Andrade A, Kherif F, Mangin J, Worsley K, Paradis A, Simon O, Dehaene S, Le Bihan D, Poline J. Detection of fMRI activation using cortical surface mapping. Hum Brain Mapp. 2001;12(2):79–93. doi: 10.1002/1097-0193(200102)12:2<79::AID-HBM1005>3.0.CO;2-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustinack JC, van der Kouwe AJ, Blackwell ML, Salat DH, Wiggins CJ, Frosch MP, Wiggins GC, Potthast A, Wald LL, Fischl BR. Detection of entorhinal layer II using 7 Tesla magnetic resonance imaging. Ann Neurol. 2005;57(4):489–494. doi: 10.1002/ana.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P, von Bonin G. The Isocortex of Man. University of Illinois; Urbana: 1951. [Google Scholar]
- Balasubramanian M, Polimeni JR, Schwartz EL. Quantitative evaluation and comparison of cortical flattening algorithms [Abstract] Society for Neuroscience Abstracts 2006 [Google Scholar]
- Barbier E, Marrett S, Danek A, Vortmeyer A, van Gelderen P, Duyn J, Bandettini P, Grafman J, Koretsky AP. Imaging cortical anatomy by high-resolution MR at 3.0T: detection of the stripe of Gennari in visual area 17. Magn Reson Med. 2002;48(4):735–738. doi: 10.1002/mrm.10255. [DOI] [PubMed] [Google Scholar]
- Boissonnat JD. Shape reconstruction from planar cross sections. Comput Vis Graph Image Process. 1988;44(1):1–29. [Google Scholar]
- Bolton JS. The exact histological localisation of the visual area of the human cerebral cortex. Philos Trans R Soc Lond B Biol Sci. 1900;193:165–222. [Google Scholar]
- Boyd J, Matsubara J. Repositioning the stria of Gennari [Abstract] Abstracts of the Society for Neuroscience 2005 [Google Scholar]
- Bridge H, Clare S, Jenkinson M, Jezzard P, Parker AJ, Matthews PM. Independent anatomical and functional measures of the V1/V2 boundary in human visual cortex. J Vis. 2005;5(2):93–102. doi: 10.1167/5.2.1. [DOI] [PubMed] [Google Scholar]
- Brodmann K. Vergleichende Lokalisationslehre der Grosshirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues. In: Laurence Garey., translator. Localisation in the Cerebral Cortex (1994) London: Smith-Gordon, new edition 1999, London: Imperial College Press; 1909. J. A. Barth, Leipzig. [Google Scholar]
- Campbell A. Histological studies on the localisation of cerebral function. Cambridge University Press; London: 1905. [Google Scholar]
- Caspers S, Geyer S, Schleicher A, Mohlberg H, Amunts K, Zilles K. The human inferior parietal cortex: Cytoarchitectonic parcellation and interindividual variability. NeuroImage. 2006;33:430–448. doi: 10.1016/j.neuroimage.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Caviness V, Meyer J, Makris N, Kennedy D. MRI-based topographic parcellation of human neocortex: an anatomically specified method with estimate of reliability. J Cog Neuro. 1996;8(6):566–587. doi: 10.1162/jocn.1996.8.6.566. [DOI] [PubMed] [Google Scholar]
- Chi JG, Dooling EC, Gilles FH. Gyral development of the human brain. Ann Neurol. 1977;1(1):86–93. doi: 10.1002/ana.410010109. [DOI] [PubMed] [Google Scholar]
- Christensen G, Rabbitt R, Miller M. Three-dimensional brain mapping using a deformable neuroanatomy. Physics in Medicine and Biology. 1994;39:609–618. doi: 10.1088/0031-9155/39/3/022. [DOI] [PubMed] [Google Scholar]
- Clark V, Courchesne E, Grafe M. In vivo myeloarchitectonic analysis of human striate and extrastriate cortex using magnetic resonance imaging. Cereb Cortex. 1992;2(5):417–424. doi: 10.1093/cercor/2.5.417. [DOI] [PubMed] [Google Scholar]
- Clark W. The laminar organization and cell content of the lateral geniculate body in the monkey. J Anat. 1941;75:419–433. [PMC free article] [PubMed] [Google Scholar]
- Crum WR, Griffin LD, Hill DLG, Hawkes DJ. Zen and the art of medical image registration: correspondence, homology, and quality. NeuroImage. 2003;20(3):1425–1437. doi: 10.1016/j.neuroimage.2003.07.014. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Dale AM, Sereno MI. Improved localization of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction:alinear approach. J Cogn Neurosci. 1993;5(2):162–176. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- Davatzikos C. Spatial transformation and registration of brain images using elastically deformable models. Comput Vis Image Underst. 1997;66(2):207–222. doi: 10.1006/cviu.1997.0605. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Bryan RN. Using a deformable surface model to obtain a shape representation of the cortex. IEEE Trans Med Imaging. 1996;15:785–795. doi: 10.1109/42.544496. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Prince J, Bryan R. Image registration based on boundary mapping. IEEE Trans Med Imaging. 1996;15(1):112–115. doi: 10.1109/42.481446. [DOI] [PubMed] [Google Scholar]
- De Leon MJ, George AE, Golomb J, Tarshish C, Convit A, Kluger A, De Santi S, McRae T, Ferris SH, Reisberg B, Ince C, Rusinek H, Bobinski M, Quinn B, Miller DC, Wisniewski HM. Frequency of hippocampal formation atrophy in normal aging and Alzheimer’s disease. Neurobiol Aging. 1997;18(1):1–11. doi: 10.1016/s0197-4580(96)00213-8. [DOI] [PubMed] [Google Scholar]
- Dijkstra E. A note on two problems in connexion with graphs. Numerische Mathematik. 1959;1(1):269–271. [Google Scholar]
- Dougherty RF, Koch VM, Brewer AA, Fischer B, Modersitzki J, Wandell BA. Visual field representations and locations of visual areas V1/2/3 in human visual cortex. Journal of Vision. 2003;3(10):586–598. doi: 10.1167/3.10.1. [DOI] [PubMed] [Google Scholar]
- Drury HA, Van Essen DC, Joshi SC, Miller MI. Analysis and comparison of areal partitioning schemes using two-dimensional fluid deformations. [Abstract] Neuroimage. 1996:S130. [Google Scholar]
- Duncan R, Boynton G. Cortical magnification within human primary visual cortex correlates with acuity thresholds. Neuron. 2003;38(4):659–671. doi: 10.1016/s0896-6273(03)00265-4. [DOI] [PubMed] [Google Scholar]
- Eickhoff S, Grefkes C, Zilles K, Fink G. The somatotopic organization of cytoarchitectonic areas on the human parietal operculum. Cereb Cortex. 2007;17(8):1800. doi: 10.1093/cercor/bhl090. [DOI] [PubMed] [Google Scholar]
- Evans A, Dai W, Collins L, Neelin P, Marrett S. Warping of a computerized 3-d atlas to match brain image volumes for quantitative neuroanatomical and functional analysis. SPIE Proc Imag Process. 1991;1445:236–246. [Google Scholar]
- Fatterpekar GM, Naidich TP, Delman BN, Aguinaldo JG, Gultekin SH, Sherwood CC, Hof PR, Drayer BP, Fayad ZA. Cytoarchitecture of the human cerebral cortex: MR microscopy of excised specimens at 9.4 Tesla. AJNR Am J Neuroradiol. 2002;23(8):1313–1321. [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat D, van der Kouwe A, Makris N, Segonne F, Quinn B, Dale A. Sequence-independent segmentation of magnetic resonance images. NeuroImage. 2004;23(Suppl 1):S69–84. doi: 10.1016/j.neuroimage.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999a;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 1999b;8(4):272–84. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flechsig P. Anatomie des Menschlichen Gehirns und Ruckenmarks auf Myelogenetischer. Thieme: Leipzig; 1920. [Google Scholar]
- Frisoni GB. Linear measures of atrophy in mild Alzheimer disease. Amer J Neurorad. 1996;17(5):913–923. [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Ashburner J, Frith CD, Poline J, Heather JD, Frackowiak RSJ. Spatial registration and normalization of images. Hum Brain Mapp. 1995;3(3):165–189. [Google Scholar]
- Geyer S, Schleicher A, Zilles K. Areas 3a, 3b, and 1 of human primary somatosensory cortex. NeuroImage. 1999;10(1):63–83. doi: 10.1006/nimg.1999.0440. [DOI] [PubMed] [Google Scholar]
- Goebel R, Esposito F, Formisano E. Analysis of FIAC data with BrainVoyager QX: from single-subject to cortically aligned group GLM analysis and self-organizing group ICA. Hum Brain Mapp. 2006;27(5):392–401. doi: 10.1002/hbm.20249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grachev I, Berdichevsky D, Rauch S, Heckers S, Kennedy D, Caviness V, Alpert N. A method for assessing the accuracy of intersubject registration of the human brain using anatomic landmarks. Neuroimage. 1999;9(2):250–268. doi: 10.1006/nimg.1998.0397. [DOI] [PubMed] [Google Scholar]
- Grefkes C, Geyer S, Schormann T, Roland P, Zilles K. Human somatosensory area 2: observer-independent cytoarchitectonic mapping, interindividual variability, and population map. NeuroImage. 2001;14(3):617–631. doi: 10.1006/nimg.2001.0858. [DOI] [PubMed] [Google Scholar]
- Halliday GM, McRitchie DA, Macdonald V, Double KL, Trent RJ, McCusker E. Regional specificity of brain atrophy in Huntington’s disease. Exp Neurol. 1998;154(2):663–672. doi: 10.1006/exnr.1998.6919. [DOI] [PubMed] [Google Scholar]
- Hasnain MK, Fox PT, Woldorff MG. Structure–function spatial covariance in the human visual cortex. Cereb Cortex. 2001;11(8):702–716. doi: 10.1093/cercor/11.8.702. [DOI] [PubMed] [Google Scholar]
- Hellier P, Barillot C, Corouge I, Gibaud B, Le Goualher G, Collins D, Evans A, Malandain G, Ayache N, Christensen G, et al. Retrospective evaluation of intersubject brain registration. IEEE Trans Med Imaging. 2003;22(9):1120–1130. doi: 10.1109/TMI.2003.816961. [DOI] [PubMed] [Google Scholar]
- Henschen S. Klinische und anatomische Beiträge zur Pathologie des Gehirns. Nordiska Bokhandeln; Stockholm: 1920. [Google Scholar]
- Hilgetag C, Barbas H. Role of mechanical factors in the morphology of the primate cerebral cortex. PLoS Comput Biol. 2006;2(3):e22. doi: 10.1371/journal.pcbi.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds OP, Polimeni JR, Blackwell ML, Wiggins CJ, Wiggins GC, van der Kouwe A, Wald LL, Schwartz EL, Fischl B. Reconstruction and analysis of human V1 by imaging the stria of Gennari using MRI at 7T [Abstract] Society for Neuroscience Abstracts 2005 [Google Scholar]
- Jaccard P. Nouvelles reserches sur la distribution florale. Bull Soc Vaud Sci Nat. 1908;44:223–270. [Google Scholar]
- Jack CR, Petersen RC, Xu YC, Waring SC, O’Brien PC, Tangalos EG, Smith GE, Ivnik RJ, Kokmen E. Medial temporal atrophy on MRI in normal aging and very mild Alzheimer’s disease. Neurology. 1997;49(3):786–794. doi: 10.1212/wnl.49.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuba G, Kraftsik R. Changes in volume, surface estimate, three-dimensional shape and total number of neurons of the human primary visual cortex from midgestation until old age. Anatomy and Embryology. 1994;190(4):351–366. doi: 10.1007/BF00187293. [DOI] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: theory and rationale for its development. Neuroimage. 1995;2(2):89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- Miller M, Christensen G, Amit Y, Grenander U. Mathematical textbook of deformable neuroanatomies. Proc Natl Acad Sci U S A. 1993;90(24):11944–11948. doi: 10.1073/pnas.90.24.11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morosan P, Rademacher J, Schleicher A, Amunts K, Schormann T, Zilles K. Human primary auditory cortex: cytoarchitectonic subdivisions and mapping into a spatial reference system. NeuroImage. 2001;13(4):684–701. doi: 10.1006/nimg.2000.0715. [DOI] [PubMed] [Google Scholar]
- Nieto-Castanon A, Ghosh SS, Tourville JA, Guenther FH. Region of interest based analysis of functional imaging data. Neuroimage. 2003;19(4):1303–1316. doi: 10.1016/s1053-8119(03)00188-5. [DOI] [PubMed] [Google Scholar]
- Polyak S. A contribution to the cerebral representation of the retina. J Comp Neurol. 1933;57(3):541–617. [Google Scholar]
- Putnam T. Studies on the central visual system. IV: The details of the organization of the geniculostriate system in man. Arch Neurol Psychiatry. 1926;16:683–707. [Google Scholar]
- Rademacher J, Bürgel U, Geyer S, Schormann T, Schleicher A, Freund H, Zilles K. Variability and asymmetry in the human precentral motor system. Brain. 2001a;124(11):2232–2258. doi: 10.1093/brain/124.11.2232. [DOI] [PubMed] [Google Scholar]
- Rademacher J, Caviness VS, Steinmetz H, Galaburda AM. Topographical variation of the human primary cortices: implications for neuroimaging, brain mapping, and neurobiology. Cereb Cortex. 1993;3(4):313–329. doi: 10.1093/cercor/3.4.313. [DOI] [PubMed] [Google Scholar]
- Rademacher J, Morosan P, Schormann T, Schleicher A, Werner C, Freund HJ, Zilles K. Probabilistic mapping and volume measurement of human primary auditory cortex. Neuroimage. 2001b;13(4):669–683. doi: 10.1006/nimg.2000.0714. [DOI] [PubMed] [Google Scholar]
- Roland P, Geyer S, Amunts K, Schormann T, Schleicher A, Malikovic A, Zilles K. Cytoarchitectural maps of the human brain in standard anatomical space. Hum Brain Mapp. 1997;5(4):222–227. doi: 10.1002/(SICI)1097-0193(1997)5:4<222::AID-HBM3>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Roland PE, Graufelds CJ, Wacaronhlin J, Ingelman L, Andersson M, Ledberg A, Pedersen J, Akerman S, Dabringhaus A, Zilles K. Human brain atlas: For high-resolution functional and anatomical mapping. Hum Brain Mapp. 1994;1(3):173–184. doi: 10.1002/hbm.460010303. [DOI] [PubMed] [Google Scholar]
- Salmond C, Ashburner J, Vargha-Khadem F, Connelly A, Gadian D, Friston K. The precision of anatomical normalization in the medial temporal lobe using spatial basis functions. NeuroImage. 2002;17(1):507–512. doi: 10.1006/nimg.2002.1191. [DOI] [PubMed] [Google Scholar]
- Sanides F. Functional architecture of motor and sensory cortices in primates in the light of a new concept of neocortex evolution. In: Noback CRMW, editor. The Primate Brain: Advances in Primatology. Appleton-Century-Crofts; New York: 1970. [Google Scholar]
- Schormann T, Zilles K. Three-dimensional linear and nonlinear transformations: an integration of light microscopical and MRI data. Hum Brain Mapp. 1998;6(5–6):339–47. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<339::AID-HBM3>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz EL, Merker B. Computer-aided neuroanatomy: Differential geometry of cortical surfaces and an optimal flattening algorithm. IEEE Computer Graphics and Applications. 1986;6(3):36–44. [Google Scholar]
- Sereno M, Dale A, Liu A, Tootell R. A surface-based coordinate system for a canonical cortex. [Abstract] Neuroimage 1996 [Google Scholar]
- Sereno MI, Dale AM, Reppas JB, Kwong KK, Belliveau JW, Brady TJ, Rosen BR, Tootell RB. Borders of multiple visual areas in humans revealed by functional magnetic resonance imaging. Science. 1995;268(5212):889–93. doi: 10.1126/science.7754376. [DOI] [PubMed] [Google Scholar]
- Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Transactions on Medical Imaging. 2002;21(11):1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- Smith G. The morphology of the occipital region of the cerebral hemisphere in man and the apes. Anatomischer Anzeiger. 1904;24:436–451. [Google Scholar]
- Stensaas SS, Eddington DK, Dobelle WH. The topography and variability of the primary visual cortex in man. J Neurosurg. 1974;40(6):747–755. doi: 10.3171/jns.1974.40.6.0747. [DOI] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar Stereotaxic Atlas of the Human Brain. Thieme; New York: 1988. [Google Scholar]
- Thompson P, Toga A. A surface-based technique for warping three-dimensional images of the brain. IEEE Trans Med Imaging. 1996;15(4):402–417. doi: 10.1109/42.511745. [DOI] [PubMed] [Google Scholar]
- Tovi M, Ericsson A. Measurements of T1 and T2 over time in formalin-fixed human whole-brain specimens. Acta Radiol. 1992;33(5):400–404. [PubMed] [Google Scholar]
- Van Essen DC. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997;385(6614):313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- Van Essen DC. A Population-Average, Landmark- and Surface-based (PALS) atlas of human cerebral cortex. Neuroimage. 2005;28(3):635–662. doi: 10.1016/j.neuroimage.2005.06.058. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Drury HA. Structural and functional analyses of human cerebral cortex using a surface-based atlas. J Neurosci. 1997;17(18):7079–7102. doi: 10.1523/JNEUROSCI.17-18-07079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt O. Die myeloarchitektonik des isocortex parietalis. J Psychol Neurol. 1911;18:107–118. [Google Scholar]
- von Economo C, Koskinas G. Die Cytoarchitektonik der Hirnrinde des erwachsenen Menschen. Springer; Vienna/Berlin: 1925. [Google Scholar]
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57(5):369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Wald LL, Wiggins GC, Potthast A, Wiggins CJ, Triantafyllou C. Design considerations and coil comparisons for 7 T brain imaging. Applied Magnetic Resonance. 2005;29(1):19–37. [Google Scholar]
- Walters NB, Egan GF, Kril JJ, Kean M, Waley P, Jenkinson M, Watson JD. In vivo identification of human cortical areas using high-resolution MRI: an approach to cerebral structure-function correlation. Proc Natl Acad Sci U S A. 2003;100(5):2981–2986. doi: 10.1073/pnas.0437896100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker W. Why does cerebral cortex fissure and fold? A review of determinants of gyri and sulci. Cerebral cortex. 1990;8:3–136. [Google Scholar]
- Wilms M, Eickhoff S, Specht K, Amunts K, Shah N, Malikovic A, Fink G. Human V5/MT+: comparison of functional and cytoarchitectonic data. Anatomy and Embryology. 2005;210(5):485–495. doi: 10.1007/s00429-005-0064-y. [DOI] [PubMed] [Google Scholar]
- Woods R, Grafton S, Holmes C, Cherry S, Mazziotta J. Automated image registration: I. general methods and intrasubject, intramodality validation. J Comput Assist Tomogr. 1998;22(1):139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- Yakovlev P, Lecours A. The myelogenetic cycles of regional maturation of the brain. In: Minkowski A, editor. Regional development of the brain in early life. F.A. Davis Co; Philadelphia: 1967. [Google Scholar]
- Zeki S. Interhemispheric connections of prestriate cortex in monkey. Brain Research. 1970;19(1):63–75. doi: 10.1016/0006-8993(70)90237-4. [DOI] [PubMed] [Google Scholar]
- Zilles K, Schleicher A, Langemann C, Amunts K, Morosan P, Palomero-Gallagher N, Schormann T, Mohlberg H, Buergel U, Steinmetz H. Quantitative analysis of sulci in the human cerebral cortex: Development, regional heterogeneity, gender difference, asymmetry, intersubject variability and cortical architecture. Human Brain Mapping. 1997;5(4):218–221. doi: 10.1002/(SICI)1097-0193(1997)5:4<218::AID-HBM2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.