Abstract
The distribution of selenium in mammals has been recently shown to be mediated primarily by selenoprotein P. Even in the absence of selenoprotein P, selenium is distributed from the liver into all organs and tissues when supplemented in the diet. The form of selenium that is actively taken up by mammalian cells at trace concentrations has yet to be determined. We used a human keratinocyte model to determine whether reduction of the oxyanion selenite (SeO32−) to the more reduced form of selenide (HSe−) would affect uptake. Indeed a reduced form of selenium, presumably selenide, was actively transported into keratinocytes and displayed saturation kinetics with an apparent Km of 279 nM. ATPase inhibitors blocked the uptake of selenide, as did the competing anions molybdate and chromate, but not sulfate. These results suggest that the small molecule form of selenium that is distributed in tissues is hydrogen selenide, despite its sensitivity to oxygen and reactivity to thiols.
Keywords: selenium, anion transport, selenite, selenide, keratinocyte
Introduction
The metalloid selenium is a required micronutrient in mammals needed for insertion into specific selenoproteins [1]. Two groups of selenoenzymes, isoenzymes of glutathione peroxidase and thioredoxin reductase, are the best studied of these selenoproteins[1]. Given the importance of these enzymes in antioxidant defense it is clear that optimal selenium nutrition has an impact on the balance of oxidants to antioxidants. The adsorption of selenium has been well studied in animals and humans, usually comparing selenate, selenite and selenomethionine for their ability to serve as nutritional sources of selenium[2,3]. Although the pathways for insertion of selenium into selenoproteins are becoming better understood in mammals [4,5], the transport of selenium at the molecular level into individual cells is poorly understood.
The metabolism of selenium upstream of the enzyme selenophosphate synthetase (SPS) [6,7] is poorly understood, yet it is clear that selenium must first be reduced to selenide since this substrate is needed for SPS enzymes from bacteria, archaebacteria and mammals [5,8]. This reduction has long been presumed to be mediated by thiols in a reaction known as the Painter reaction [9]. Painter described the reduction of selenite to selenide after reduction by cysteine, and subsequent work by Ganther revealed the reactivity with the abundant cellular thiol glutathione [10-12]. A selenotrisulfide intermediate in this reductive pathway has been characterized for a variety of thiols [10,13-15]. It has been speculated that this form of selenium (RSSeSR) might be abundant in the cell, although in vitro evidence suggests these compounds are quite sensitive to high concentrations of reduced thiols [10-13]. Nonetheless, through the Painter reaction, efficient reduction of selenium to selenide can occur if reduced thiols such as glutathione are abundant. Reduced glutathione is known to be present in millimolar quantities in the cytosol of prokaryotic or eukaryotic cells [16].
Studies that have focused on uptake of selenium have generally compared the transport of selenite or selenate to competing anions in model systems. Selenite was taken up by the same anion transporter allowing transport of selenate and sulfate in Salmonella typhimurium [17]. In both Clostridium pasteurianum and in Selenonomas ruminatium selenite uptake was reduced in the presence of uncoupling agents N,N'-dicyclohexylcarbodiimide (DCCD) or carbonyl cyanide m-chlorophenylhydrazone (CCCP) [18,19]. A yeast model revealed two apparent systems with affinities in the low micromolar range and the millimolar range [20], yet yeast do not make specific selenoproteins and as such the physiological relevance may not correlate to higher eukaryotes. Taken together, the results from several model systems suggest that many organisms have the ability to accumulate selenium with varying affinities and likely using several different uptake systems.
In mammals selenite uptake has been reported in red blood cells and shown to be inhibited by an anion transport inhibitor 4,4'-diisothiocyanatostilbene-2,2'-disulfonic (DIDS) [21]. It is unclear as to whether selenium oxyanions (selenite or selenate) would occur in vivo given the level of reduced thiols that can trigger the Painter reaction. Recent evidence in animal knockout models has clearly shown that the distribution of selenium from the liver to tissues and organs is mediated by selenoprotein P [22-24]. It is also well established that even in the absence of selenoprotein P, selenium can be distributed from the liver to all organs by an as yet unknown mechanism [22]. Recently the ApoE2 receptor has been shown to bind to selenoprotein P, and to mediate uptake [25,26]. However since selenoprotein P is not found in appreciable amounts in most tissues, it is clear distribution within the tissue (after degradation of selenoprotein P), must be a small molecule form that has yet to be described.
In this report, we investigated the uptake of selenium either as an oxyanion (selenite) or a more reduced form (selenite with excess glutathione) using a keratinocyte model (HaCat).
Materials and Methods
Materials
Sodium selenite, β-mercaptoethanol and DCCD) were purchased from Acros Organics (Fair Lawn, NJ). Dithiothreitol (DTT) was obtained from Amresco (Solon, OH). Reduced nicotinamide adenine dinucleotide phosphate (NADPH) and sodium orthovanadate was purchased from Alexis Biochemicals (Lausen, Switzerland). Sodium molybdate was obtained from ICN Biomedicals (Aurora, OH). Thioredoxin was obtained from Promega (Madison, WI). Thioredoxin reductase, DIDS, potassium chromate and iodoacetic acid were purchased from Sigma-Aldrich (St. Louis, MO). 75Se radioisotope was acquired from the University of Missouri Research Reactor (MURR, Columbia, MO).
Cell culture
HaCat cell line, obtained from Dr. Norbert Fusenig (German Cancer Research Institute) was cultivated in as a monolayer in Dulbecco's modification of Eagle's medium (DMEM) supplemented with L-glutamine, sodium pyruvate, 4.5g/L glucose, 10% fetal bovine serum (FBS), 100 μg/ml streptomycin and 100 IU/ml penicillin (Mediatech, Herndon, VA). Cells were incubated in a humidified CO2 (5%) incubator at 37°C. Cells were cultured and harvested as previously described [27].
75Se uptake assay
HaCat cells were plated in 24 well dishes at approximately 50,000 cells per well and allowed to reach confluence. Cells were washed with Dulbecco's modified phosphate buffered saline (DPBS), followed by addition of unlabeled sodium selenite and radioisotope (2 μCi) to give18 nM in DPBS. For the analysis of uptake of reduced selenium, reduced glutathione (GSH, 10 mM, pH 7.5) was added to the unlabeled sodium selenite and radioisotope at a molar ratio (thiol to selenium) of 550 to 1. The final concentration of GSH and radioisotope once added to cells in DPBS was 10 μM (GSH) and 18 nM (Se). Cells were incubated for 1 hour at 37°C with a humidified CO2 (5%) atmosphere. Cells were subsequently washed with 1 ml of DPBS supplemented with 10 μM selenite (to allow for quick exchange of loosely bound selenium to the outside of the cell) and twice with 1 ml of DPBS. DPBS was removed and cells were lysed with 0.5 M NaOH and transferred to 12 × 75 mm glass culture tubes. 75Se was determined using a Model 1470 Gamma counter (Perkin Elmer, Wellesley, ME).
Inhibition of selenium uptake
To monitor inhibition of selenium uptake the conditions described above were utilized with the exception that the inhibitor was added to each well for 10 minutes at 37°C with a humidified 5% CO2 atmosphere in DPBS. After this brief pre-incubation step, uptake of selenium was monitored as described above.
Selenium uptake with thiols other than GSH or an enzymatic thiol-reducing system
Uptake was carried out as described above with the exception that thiols other than GSH were mixed with unlabeled selenite and radioisotope selenium for 15 minutes prior to addition to cells in DPBS. Alternatively thioredoxin reductase (1.5 μg/ml), thioredoxin (1 μM) and NADPH (500 μM) were incubated together in combinations with selenium isotope and selenite for 15 minutes. Due to the acid present in the radioisotope, this reaction was buffered to neutrality by the addition of Tris (pH 8.0).
Results
High affinity uptake of selenium after reduction by GSH
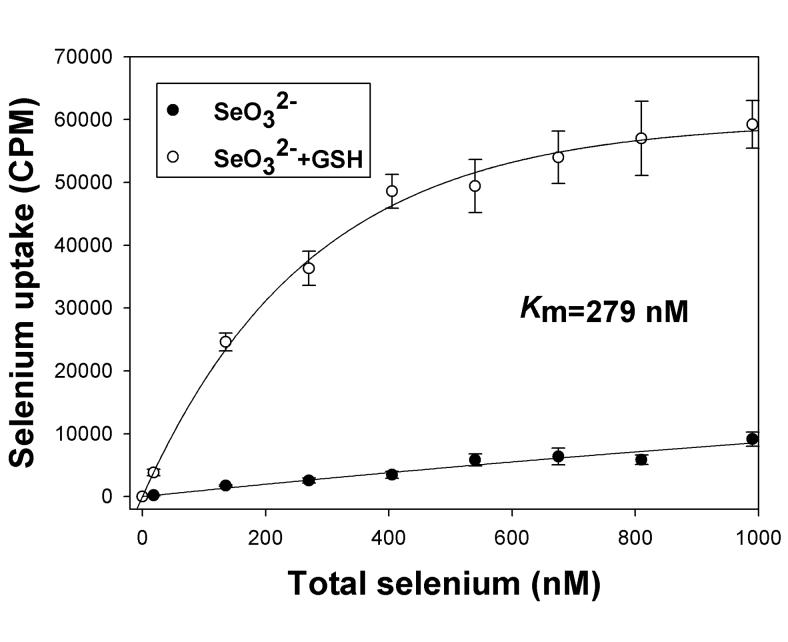
In our initial experiments, the uptake of radioisotope selenium (75Se) was followed in DPBS in adherent HaCat cells over a period of one hour. Uptake of selenite was essentially linear over the course of one hour (data not shown). Therefore we assessed the uptake of selenium at a fixed one hour time point in all subsequent experiments to determine the characteristics of selenium transport in the HaCat model. The uptake of selenite increased in a linear fashion when added to the culture in concentrations ranging from low nanomolar to 1 micromolar (Figure 1). Micromolar or higher concentrations should not be considered physiologically relevant for the trace element selenium. Thus it appears that the oxyanion selenite is not actively transported in HaCat cells. In contrast, addition of glutathione to the selenite just prior to addition to cells in excess led to efficient uptake of selenium at nanomolar concentrations (Figure 1). Moreover, the uptake of this reduced form of selenium displayed saturation kinetics with an apparent Km of 279 nM (Figure 1). These results strongly suggest that selenium, in a reduced form after reduction by the thiol glutathione, is actively transported by a high-affinity uptake system.
Figure 1. Glutathione reduction stimulates selenium uptake in HaCat cell model.
Uptake of selenite (SeO32−) and reduced selenium (SeO32− + GSH) was assessed after incubation of radiolabeled selenium with cells in DPBS for 60 minutes. The concentration of selenium was varied without changing the specific activity of the radioisotope as described in Methods. A representative experiment is shown, taken from two independent reproducible experiments (n=6). The data is shown as means with standard deviation plotted as error of three independent culture replicates.
Evidence supporting high affinity uptake of selenide
GSH is the most abundant reduced thiol present in mammals [16]. To determine whether selenide or a selenotrisulfide derivative of GSH (GSSeSG) is being taken up, we altered the thiol containing reductant in the pre-incubation with selenite. Table 1 summarizes these results. The total selenium retained by the cells was calculated and reported as compared to selenite, which is poorly taken up under these conditions (18 nM total selenium present, 0.36% of total selenium retained in the cells). Reduction of selenite with dithiothreitol (DTT) resulted in a significant stimulation of selenium uptake, as compared either to selenite or selenite with GSH (Table 1). Likewise, α-mercaptoethanol also significantly stimulated uptake, whereas oxidized thioredoxin had no effect (Table 1). When all the needed components of the TrxR/Trx system were present, selenium uptake was also stimulated (10 % of total available selenium, Table 1). It has been previously shown that TrxR/Trx is capable of reducing selenite to selenide in vitro [28,29]. Taken together, these results strongly suggest selenide is the form of selenium actively transported by the human HaCat cell model. These results do not support the hypothesis that selenotrisulfides are actively taken up, since the corresponding conjugates (RSSeSR') of DTT or thioredoxin would be significantly larger than the GSH derivative. We then further determined the properties of this high affinity transport using a series of common inhibitors.
Table 1.
Enhancement of selenium uptake by thiols in HaCat cells demonstrates evidence for high affinity uptake of selenide
| Rxn | Tested compoundsa | Selenium retained (picomoles) |
Selenium retainedb of total (%) |
|---|---|---|---|
| 1 | selenite (control) | 0.066 ± 0.011 | 0.36 ± 0.063 |
| 2 | selenite, 10μM glutathione | 0.50 ± 0.12 | 2.8 ± 0.66 |
| 3 | selenite, 10μM dithiothreitol | 5.3 ± 0.64 | 29 ±3.6 |
| 4 | selenite, 10μM β-mercatoethanol | 4.0 ± 0.46 | 22 ± 2.6 |
| 5 | selenite, 10μM Trx | 0.071 ±0.02 | 0.39 ± 0.11 |
| 6 | selenite, 15μM NADPH | 0.062 ± 0.015 | 0.34 ± 0.085 |
| 7 | selenite, 10μM Trx, 15μM NADPH | 0.076 ± 0.018 | 0.42 ± 0.099 |
| 8 | selenite, TrxR, Trx, NAPDH | 1.8 ± 0.28 | 10 ± 1.6 |
Thiols were pre-incubated with selenite for 10 minutes and then entire mixture was added to HaCat cells containing 1 mL of DPBS and incubated for 1 hour. Reactions 5-8 contained either 1 μM of thioredoxin, 1.5 μg of thioredoxin reductase or 500 μM of NADPH in a final volume of 30 μL, with only reaction 8 containing all the components to regenerate reduced thioredoxin catalytically.
Total selenium retained was determined after several washes of the HaCat cell monolayer and subsequent lysis by NaOH as described in methods. The mean of two independent experiments (with multiple replicates) is shown with standard deviation as the error.
Uptake of reduced selenium is mediated by an anion transporter and blocked by acetylation
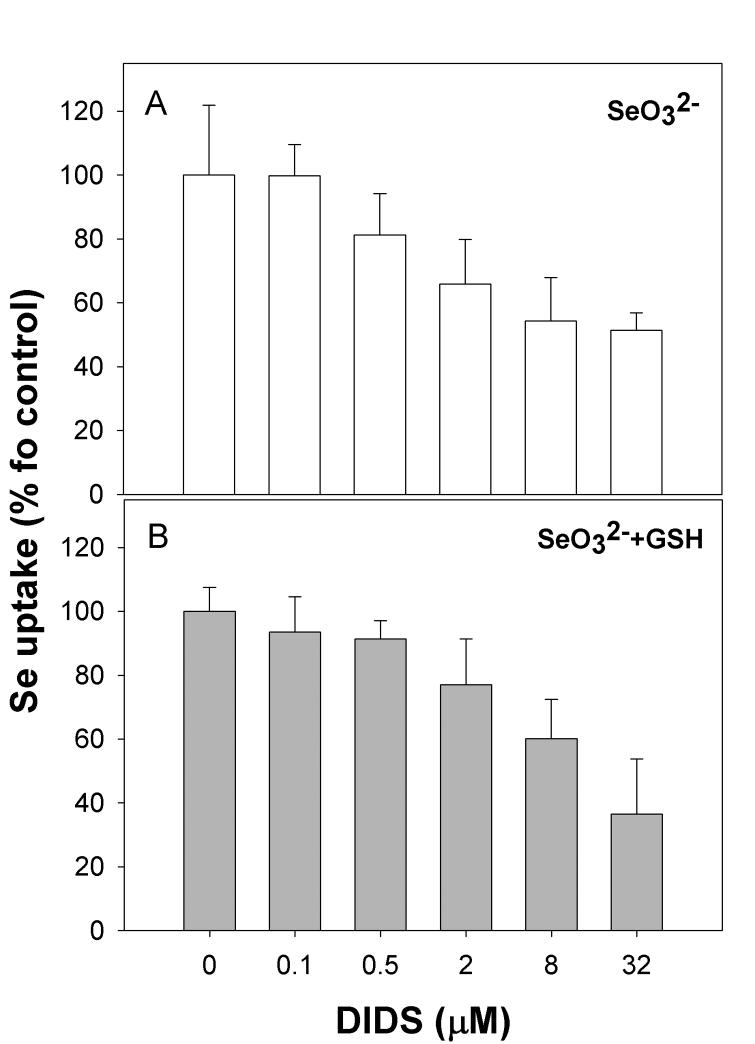
The reported pKa value for selenide (H2Se) is 3.8 [30], and as such this form of selenium carries a net negative charge at pH 7.4 (HSe−) in our experimental model. Given this potential anionic character, we evaluated whether DIDS, a specific inhibitor of mammalian anion transporters [21], would affect uptake of selenium. DIDS treatment of HaCat cells did indeed reduce the level of selenium taken up by cells, whether in the form of selenite or reduced selenium, in a concentration-dependent manner (Figure 2, A and B). These results suggest each form is allowed entry into the cell via an anion transporter. Pre-incubation of cells with micromolar levels of iodoacetic acid also inhibited uptake of reduced selenium, with no changes to selenite uptake (data not shown). This indicates that alkylation of selenide inhibits uptake, suggesting the high affinity system is specific for the selenide anion.
Figure 2. Selenite and selenide uptake is mediated by an anion transporter.
Cells were treated with DIDS at the concentrations indicated for 10 minutes prior to addition of selenium. (A) uptake of selenite, (B) uptake of selenite pre-incubated with excess GSH.
Evidence supporting ATP-dependent uptake of selenide
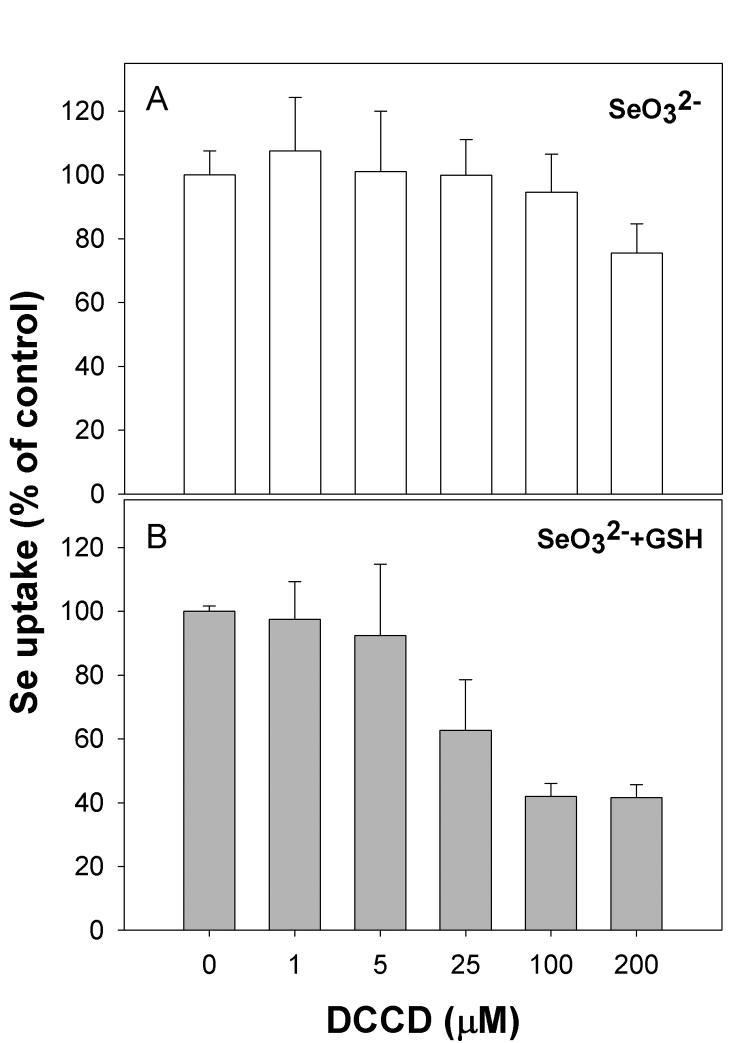
DCCD has been shown to act as a potent inhibitor of membrane-bound ATPase complexes [31]. Selenium uptake has also been shown to be sensitive to DCCD in both a bacterial and plant cell models [19,32]. In our keratinocyte model the uptake of selenide was inhibited in a dose-dependent manner by DCCD (Figure 3B). Inhibition of selenite uptake was only apparent when DCCD was present at 200 μM (Figure 3A). This suggests that uptake of selenide is mediated by a membrane-associated ATP-dependent transporter.
Figure 3. Sensitivity of selenium uptake to the ionophore DCCD.
Cells were treated with DCCD for 10 minutes prior to addition of selenium. (A) uptake of selenite, (B) uptake of selenite pre-incubated with excess GSH.
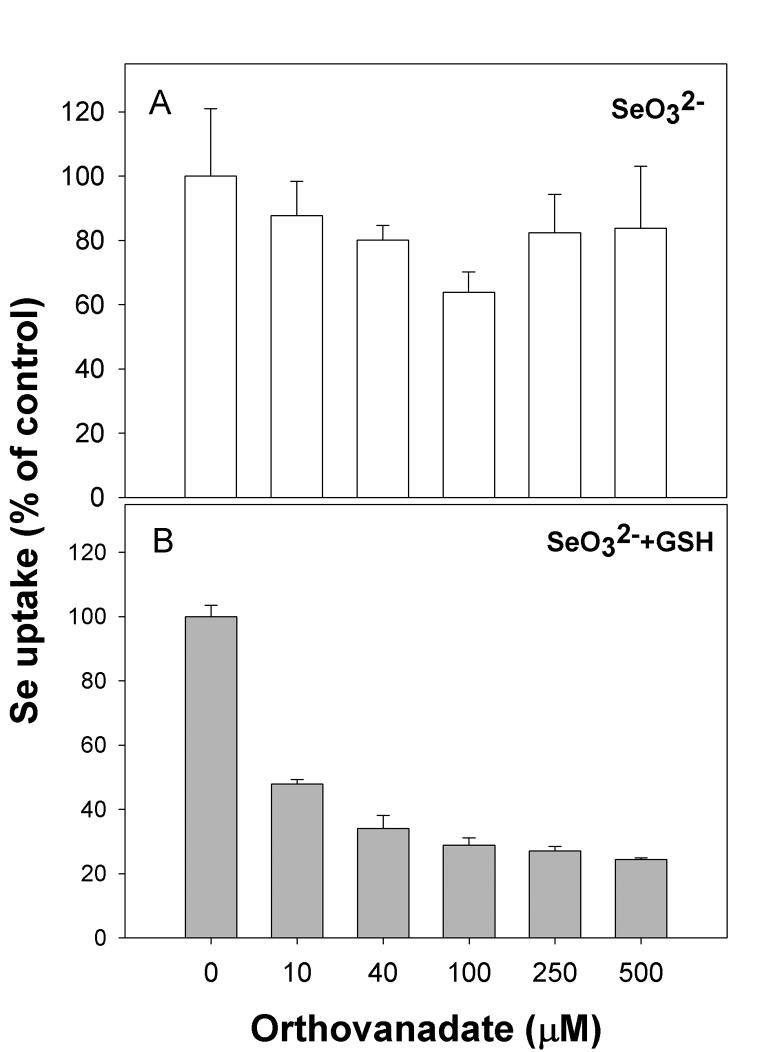
Another well-characterized inhibitor of ATPase enzymes, orthovanadate [33], also inhibited uptake of selenide (Figure 4A). Treatment of cells with orthovanadate did not alter the uptake of selenite (Figure 4B). Based on the results obtained in Figures 3 and 4, it appears that an ATP-dependent anion transporter is responsible for uptake of selenide, and this high affinity transporter does not play a significant role in the transport of selenite.
Figure 4. Inhibition by orthovanadate further implicates an ATP-dependent process is transporting reduced selenium.
Cells were treated with orthovanadate at the concentrations indicated for 10 minutes prior to addition of selenium. (A) uptake of selenite, (B) uptake of selenite pre-incubated with excess GSH.
Differential inhibition of selenium uptake by competing anions
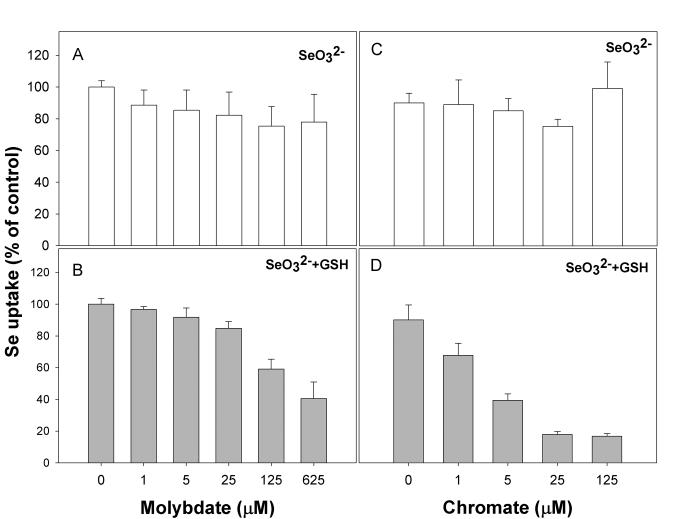
Previous studies in bacterial cell models have shown competing anions such as molybdate, chromate or sulfate can affect the uptake of selenite or selenate. In our model, the addition of sulfate did not inhibit uptake of either selenite or selenide (data not shown). This is surprising and somewhat contradicts the notion that selenium oxyanions can gain entry into cells via the sulfur transport mechanism. Nonetheless molybdate and chromate both inhibited uptake of selenide in a concentration-dependent manner (Figure 5B and 5D), yet had no significant effect on selenite transport (Figure 5A and 5C). These results suggest strongly that these anions also are taken up by the same transporter, although one can not rule out that they negatively affect the ATPase activity of a high-affinity selenide transporter.
Figure 5. Competing anions molybdate and chromate block uptake of reduced selenium, but not selenite, in a keratinocyte model.
Molybdate or chromate was added 10 minutes prior to addition of selenium at the concentrations indicated. (A) and (C), uptake of selenite, (B) and (D), uptake of selenite after addition of excess GSH.
Discussion
Recent animal models have solidified a role for selenoprotein P in the transport of selenium from the liver to other organs in mammals [22,23]. Nonetheless selenoprotein P deficient mice are quite capable of distributing selenium when it is supplemented in the diet [22]. A small molecule form of selenium is likely to be distributed into cells, yet little evidence exists in the literature to support this or to describe the form of selenium being distributed. Our findings suggest strongly hydrogen selenide is efficiently taken up by human cells with an apparent Km in the nanomolar range (Figure 1). Uptake of selenide was inhibited by DCCD and orthovanadate, suggesting the uptake is mediated by an ATP-dependent mechanism. A recent study using yeast as a model also revealed evidence selenide was effectively taken up, although this study was focused on selenium toxicity in a model (yeast) that does not make specific selenoproteins [34]. Differential inhibition by competing anions also indicates selenide is likely not transported by the same anion transporter responsible for selenite accumulation. Once taken up by the cell, selenide would be efficiently used for selenoprotein synthesis, as this is the form needed for SPS [5]. Future studies to identify the protein(s) responsible for this high affinity uptake of selenide will certainly aid in our understanding of selenium nutrition and distribution in mammals.
Acknowledgements
This research was supported by grants to WTS from the National Institutes of Health (ES014354) and the Florida Department of Health (05-NIR-10).
Abbreviations
- DIDS
4,4'-diisothiocyanatostilbene-2,2'-disulfonate
- DCCD
N,N'-dicyclohexylcarbodiimide
- RSSeSR
selenotrisulfide
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stadtman TC. Selenium biochemistry. Mammalian selenoenzymes. Ann N Y Acad Sci. 2000;899:399–402. doi: 10.1111/j.1749-6632.2000.tb06203.x. [DOI] [PubMed] [Google Scholar]
- 2.Raghib MH, Chan WY, Rennert OM. Comparative studies of selenium-75 (selenite and selenomethionine) absorption from various milk diets in suckling rats. J Nutr. 1986;116:1456–63. doi: 10.1093/jn/116.8.1456. [DOI] [PubMed] [Google Scholar]
- 3.Robinson MF, Thomson CD, Jenkinson CP, Luzhen G, Whanger PD. Long-term supplementation with selenate and selenomethionine: urinary excretion by New Zealand women. Br J Nutr. 1997;77:551–63. doi: 10.1079/bjn19970056. [DOI] [PubMed] [Google Scholar]
- 4.Small-Howard A, et al. Supramolecular complexes mediate selenocysteine incorporation in vivo. Mol Cell Biol. 2006;26:2337–46. doi: 10.1128/MCB.26.6.2337-2346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu XM, et al. Biosynthesis of Selenocysteine on Its tRNA in Eukaryotes. PLoS Biol. 2006;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim IY, Guimaraes MJ, Zlotnik A, Bazan JF, Stadtman TC. Fetal mouse selenophosphate synthetase 2 (SPS2): characterization of the cysteine mutant form overproduced in a baculovirus-insect cell system. Proc Natl Acad Sci U S A. 1997;94:418–21. doi: 10.1073/pnas.94.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogasawara Y, Lacourciere GM, Ishii K, Stadtman TC. Characterization of potential selenium-binding proteins in the selenophosphate synthetase system. Proc Natl Acad Sci U S A. 2005;102:1012–6. doi: 10.1073/pnas.0409042102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson BA, Xu XM, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc Natl Acad Sci U S A. 2004;101:12848–53. doi: 10.1073/pnas.0402636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Painter HE. The Chemistry and Toxicity of Selenium Compounds, with Special Reference to the Selenium Problem. Chem. Rev. 1941;28:179–213. [Google Scholar]
- 10.Ganther HE. Selenotrisulfides. Formation by the reaction of thiols with selenious acid. Biochemistry. 1968;7:2898–905. doi: 10.1021/bi00848a029. [DOI] [PubMed] [Google Scholar]
- 11.Ganther HE. Reduction of the selenotrisulfide derivative of glutathione to a persulfide analog by glutathione reductase. Biochemistry. 1971;10:4089–98. doi: 10.1021/bi00798a013. [DOI] [PubMed] [Google Scholar]
- 12.Ganther HE. Selenium metabolism, selenoproteins and mechanisms of cancer prevention: complexities with thioredoxin reductase. Carcinogenesis. 1999;20:1657–66. doi: 10.1093/carcin/20.9.1657. [DOI] [PubMed] [Google Scholar]
- 13.Self WT, Tsai L, Stadtman TC. Synthesis and characterization of selenotrisulfide-derivatives of lipoic acid and lipoamide. Proc Natl Acad Sci U S A. 2000;97:12481–6. doi: 10.1073/pnas.220426897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakagawa T, Hasegawa Y, Yamaguchi Y, Tanaka H, Chikuma M, Sakurai H, Nakayama M. Isolation, characterization and thiol exchange reaction of penicillamine selenotrisulfides. Biochem Biophys Res Commun. 1986;135:183–8. doi: 10.1016/0006-291x(86)90960-5. [DOI] [PubMed] [Google Scholar]
- 15.Haratake M, Fujimoto K, Ono M, Nakayama M. Selenium binding to human hemoglobin via selenotrisulfide. Biochim Biophys Acta. 2005;1723:215–20. doi: 10.1016/j.bbagen.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Kehrer JP, Lund LG. Cellular reducing equivalents and oxidative stress. Free Radic Biol Med. 1994;17:65–75. doi: 10.1016/0891-5849(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 17.Brown TA, Shrift A. Assimilation of selenate and selenite by Salmonella typhimurium. Can J Microbiol. 1980;26:671–5. doi: 10.1139/m80-117. [DOI] [PubMed] [Google Scholar]
- 18.Bryant RD, Laishley EJ. Evidence for proton motive force dependent transport of selenite by Clostridium pasteurianum. Can J Microbiol. 1989;35:481–6. doi: 10.1139/m89-074. [DOI] [PubMed] [Google Scholar]
- 19.Hudman JF, Glenn AR. Selenite uptake and incorporation by Selenomonas ruminantium. Arch Microbiol. 1984;140:252–6. doi: 10.1007/BF00454937. [DOI] [PubMed] [Google Scholar]
- 20.Gharieb MM, Gadd GM. The kinetics of 75[Se]-selenite uptake by Saccharomyces cerevisiae and the vacuolization response to high concentrations. Mycol Res. 2004;108:1415–22. doi: 10.1017/s0953756204001418. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki KT, Shiobara Y, Itoh M, Ohmichi M. Selective uptake of selenite by red blood cells. Analyst. 1998;123:63–7. doi: 10.1039/a706230c. [DOI] [PubMed] [Google Scholar]
- 22.Burk RF, Hill KE. SELENOPROTEIN P: An Extracellular Protein with Unique Physical Characteristics and a Role in Selenium Homeostasis. Annu Rev Nutr. 2005;25:215–35. doi: 10.1146/annurev.nutr.24.012003.132120. [DOI] [PubMed] [Google Scholar]
- 23.Schomburg L, Schweizer U, Holtmann B, Flohe L, Sendtner M, Kohrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burk RF, Hill KE. Orphan selenoproteins. Bioessays. 1999;21:231–7. doi: 10.1002/(SICI)1521-1878(199903)21:3<231::AID-BIES7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 25.Burk RF, Hill KE, Olson GE, Weeber EJ, Motley AK, Winfrey VP, Austin LM. Deletion of apolipoprotein E receptor-2 in mice lowers brain selenium and causes severe neurological dysfunction and death when a low-selenium diet is fed. J Neurosci. 2007;27:6207–11. doi: 10.1523/JNEUROSCI.1153-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson GE, Winfrey VP, Nagdas SK, Hill KE, Burk RF. Apolipoprotein E receptor-2 (ApoER2) mediates selenium uptake from selenoprotein P by the mouse testis. J Biol Chem. 2007;282:12290–7. doi: 10.1074/jbc.M611403200. [DOI] [PubMed] [Google Scholar]
- 27.Ganyc D, Talbot S, Konate F, Jackson S, Schanen B, Cullen W, Self WT. Impact of trivalent arsenicals on selenoprotein synthesis. Environ Health Perspect. 2007;115:346–53. doi: 10.1289/ehp.9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bjornstedt M, Kumar S, Bjorkhem L, Spyrou G, Holmgren A. Selenium and the thioredoxin and glutaredoxin systems. Biomed Environ Sci. 1997;10:271–9. [PubMed] [Google Scholar]
- 29.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–9. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 30.Selenium In Nutrition. National Academies Press; 1983. [PubMed] [Google Scholar]
- 31.Azzi A, Casey RP, Nalecz MJ. The effect of N,N'-dicyclohexylcarbodiimide on enzymes of bioenergetic relevance. Biochim Biophys Acta. 1984;768:209–26. doi: 10.1016/0304-4173(84)90017-x. [DOI] [PubMed] [Google Scholar]
- 32.Smith IK. Characterization of Sulfate Transport in Cultured Tobacco Cells. Plant Physiol. 1976;58:358–362. doi: 10.1104/pp.58.3.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cantley LC, Jr., Josephson L, Warner R, Yanagisawa M, Lechene C, Guidotti G. Vanadate is a potent (Na,K)-ATPase inhibitor found in ATP derived from muscle. J Biol Chem. 1977;252:7421–3. [PubMed] [Google Scholar]
- 34.Tarze A, Dauplais M, Grigoras I, Lazard M, Ha-Duong NT, Barbier F, Blanquet S, Plateau P. Extracellular production of hydrogen selenide accounts for thiol-assisted toxicity of selenite against Saccharomyces cerevisiae. J Biol Chem. 2007;282:8759–67. doi: 10.1074/jbc.M610078200. [DOI] [PubMed] [Google Scholar]