Abstract
Breast cancer metastasis suppressor 1 (BRMS1) inhibits formation of macroscopic lung metastases in breast, ovary, and melanoma xenograft models. Because it is unclear which step(s) of the metastatic cascade are affected by BRMS1, the major aim of this study was to determine when and how BRMS1 acts to suppress metastasis. We also examined whether BRMS1 expression globally blocks metastasis or selectively inhibits metastatic outgrowths in specific tissues. Metastatic human breast carcinoma cell lines MDA-MB-231 and -435 expressing enhanced green fluorescent protein (GFP; 231GFP and 435GFP) and cell lines transduced with the BRMS1 gene (231GFP-BRMS1 and 435GFP-BRMS1) were injected into the left cardiac ventricle to achieve the widest possible cellular distribution, by minimizing first-pass clearance in the lungs. Compared with parental cells, BRMS1-expressing clones formed significantly fewer metastases in all organs tested. When cells were injected directly into the vasculature, fewer of the BRMS1-expressing cells reached lungs or bone compared with parental cells, suggesting that restoration of BRMS1 expression increased cell death during transit. Susceptibility to anoikis was verified in vitro by demonstrating decreased survival on poly-hydroxyethyl methacrylate-coated dishes. Most of the BRMS1-expressing cells reaching secondary sites failed to proliferate, suggesting that BRMS1 also inhibits colonization. Coupled with previous reports showing modest effects of BRMS1 on adhesion and invasion, our results indicate that BRMS1 inhibits metastases in multiple organs by blocking several steps in the metastatic cascade.
The overwhelming majority of morbidity and mortality for patients with cancer is associated with metastatic disease. In breast cancer, metastases are relatively widely distributed, with the most common sites being bone, regional lymph nodes, lung, liver, and brain.1 Significant improvements in survival and quality of life have been realized over several decades due to earlier detection and more effective treatment of metastases. However, there is still much room for improvement.
A relatively new class of molecules, metastasis suppressors, hold promise for providing new avenues for therapeutic intervention. Metastasis suppressors are defined by the ability to prevent metastasis without blocking orthotopic tumor growth.2,3,4,5 Most have been discovered in the past decade, but their mechanisms of action remain largely unexplained.
Breast cancer metastasis suppressor 1 (BRMS1) was functionally defined by its ability to block lung and regional lymph node metastases in experimental breast, melanoma, and ovarian models.6,7,8,9,10 Decreased BRMS1 protein expression in human breast carcinomas has been correlated with reduced disease-free survival when stratified by loss of estrogen or progesterone receptor or HER2 overexpression.11 mRNA expression is inversely correlated with metastasis development in human cancers in most12,13,14 but not all15,16 studies. BRMS1 is a predominantly nuclear protein that is part of multiple SIN3:HDAC complexes.17,18 Based on its cellular location and interactions, BRMS1 is implicated in regulation of gene expression. BRMS1 in breast cancer cells results in decreases of nuclear factor-κB activity18,19,20,21; selective reduction in phosphatidylinositol-4,5-bisphosphate levels22; loss of osteopontin expression23,24; and alteration of gap junctional intercellular communication by changing connexin expression patterns.25
The functionality of metastasis suppressors has been defined mostly using assays that measure inhibition of lung and lymph node metastases. Few studies, however, have been undertaken to identify whether metastasis suppression is widespread (ie, whether metastasis to all organs is blocked) or selective (ie, metastasis is inhibited in some, but not all, organs). Part of the reason for this deficiency is the experimental model.26 First, with the exception of surgical orthotopic implantation,26,27 most orthotopic mouse breast or mammary cancer models infrequently metastasize to as many organs as in humans, presumably because they have been selected for rapid local tumor progression and growth.26 Expediency of primary tumor growth often means that total tumor burden is lethal before some metastases fully develop. Second, experimental models involving intravenous inoculation of cells are limited by first-pass clearance of tumor cells or emboli in the lungs, thereby reducing the seeding of extrapulmonary sites. Third, it is difficult and cost prohibitive to track tumor cells in every tissue using histology. Fourth, the mere presence of tumor cells in a tissue does not necessarily mean that metastases will develop.28,29,30,31To overcome these obstacles, we injected enhanced green fluorescent protein-expressing breast cancer cells into the left ventricle of the heart. The route of injection obviates the first-pass clearance in the lungs, and fluorescence allows ex vivo detection of single cells in most tissues.31,32,33 These modifications afforded us the opportunity to address whether BRMS1 blocks metastasis to all or selective organs and in vivo assessment of steps in the metastatic cascade impacted by restoration of BRMS1.
Materials and Methods
Cell Lines and Culture
Metastatic human breast carcinoma cell lines MDA-MB-231 and MDA-MB-435 are human estrogen- and progesterone receptor-negative breast carcinoma cell lines derived from metastatic infiltrating ductal breast carcinomas.34 The origin of MDA-MB-435 has been questioned in recent literature based on microarray results35,36; however, its expression of milk proteins37 and propensity to metastasize from mammary gland but not from subcutaneous sites38 are consistent with it being a breast carcinoma.
Both cell lines form metastases to multiple organs when injected intracardially.39,40 To facilitate detection,32,33 both cell lines were stably transduced with a HIV-1-based, lentiviral vector system constitutively expressing enhanced green fluorescent protein (GFP), as previously described.41,42 The GFP-tagged cells were further transduced with a lentiviral vector expressing BRMS1 with an N-terminal myc epitope tag. Single-cell clones expressing BRMS1 and GFP protein were obtained by limiting dilution.
Cell cultures were maintained in an equal mixture of Dulbecco’s-modified Eagle’s medium and Ham’s-F12 medium (Invitrogen, Carlsbad, CA) supplemented with 2 mmol/L l-glutamine, 1 mmol/L sodium pyruvate, 0.02 mmol/L nonessential amino acids, 5% fetal bovine serum (Atlanta Biologicals, Norcross, GA) plus puromycin (500 ng/ml; Fisher Scientific, Hampton, NH). All cultures were confirmed negative for Mycoplasma spp. infection using a PCR-based test (TaKaRa, Shiga, Japan).
To measure in vitro proliferation, cells were plated in triplicate in 24-well plates. Thereafter, at 24-hour intervals, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide was added to each well at a final concentration of 0.5 mg/ml and incubated for 3 hours. After incubation, media was gently removed and formazan crystals were dissolved with 1 ml of dimethyl sulfoxide, and absorbance was recorded at 490 nm. Growth curve experiments with triplicate samples were repeated independently.
Immunoblots
BRMS1 expression was determined by collecting total protein from 70 to 90% confluent cell cultures. After aspiration of medium, plates were rinsed three times with Ca+2- and Mg+2-free Dulbecco’s phosphate-buffered saline before addition of 0.5 ml of lysis buffer [25 mmol/L Tris-HCl, pH 7.4, 50 mmol/L β-glycerol phosphate, 0.5 mmol/L EDTA, 5% glycerol, 0.1% Triton X-100, 1 mmol/L sodium orthovanadate, 1 mmol/L benzamidine, and a proteinase inhibitor cocktail containing aprotinin, leupeptin, and phenylmethylsulfonyl fluoride (Roche, Indianapolis, IN)]. Lysates were centrifuged at 10,000 × g at 4°C for 10 minutes to remove insoluble material. Protein concentration was determined using the Bradford colorimetric assay (Pierce, Rockford, IL). Protein (25 to 80 μg/lane) was mixed with 5× loading buffer (50% glycerol and 1.5% bromophenol blue) and separated by 12% SDS-polyacrylamide gel electrophoresis. Protein was transferred to polyvinylidene difluoride membrane by wet transfer (0.36A, 75 minutes). The membrane was wetted in methanol, rinsed in 0.05% Tween 20, 20 mmol/L Tris, and 140 mmol/L NaCl, pH 7.6 (TTBS solution) and blocked in a TTBS solution containing 5% dry nonfat milk for 1 hour. BRMS1 was detected using 1:2500 dilution of mouse monoclonal antibody (3a1.2111) for 2 hours at room temperature under constant agitation. Membranes were then washed with TTBS and probed with 1:5000 dilution of sheep anti-mouse secondary antibody conjugated to horseradish peroxidase (Amersham-Pharmacia Biotech, Buckinghamshire, UK) in a solution of 5% nonfat dry milk/TTBS for 1 hour at room temperature before washing in TTBS. Bound secondary antibodies were detected using enhanced chemiluminescence (Amersham-Pharmacia Biotech).
Antibodies
Commercial antibodies were all purchased from Cell Signaling Technologies (Danvers, MA) [total Akt, Akt-phospho Ser473, total p42/p44 mitogen-activated protein kinase (MAPK), MAPK phospho Thr202/Tyr204, Bim, BclII, Bad, Bax, Bcl-xL, Bmf, poly(ADP-ribose) polymerase (PARP), and caspase-3] or Sigma-Aldrich (St. Louis, MO) (β-actin).
Animals and Metastasis Assays
Cells at 80 to 90% confluence were detached using a mixture of 0.5 mmol/L EDTA and 0.05% trypsin in Ca+2-, Mg+2-, and NaHC03-free Hanks’ balanced salt solution. Viable cells (three BRMS1-expressing clones plus parental and vector-only transfectants) were counted using a hemacytometer and resuspended at a final concentration of 1 × 106 cells/ml in ice-cold Hanks’ balanced salt solution. Female athymic mice aged between 4 and 6 weeks (Harlan Sprague-Dawley, Indianapolis, IN) were anesthetized by intramuscular administration of a mixture of 129 mg/kg Ketamine-HCl, and 4 mg/kg Xylazine. Cells (2 × 105 in 0.2 ml) were injected into the left ventricle of the heart between the third and fourth or fourth and fifth intercostal space. Presence of bright red, as opposed to burgundy, colored blood before and at the end of each inoculation confirmed injection of the entire volume into the arterial system. As observed previously, injection into the arterial circulation resulted in only rare lung metastases.40,43,44
To assay for metastases to lung, cells were injected in the lateral tail vein. Mice were necropsied at 5 weeks after inoculation after anesthesia with Ketamine:Xylazine and euthanasia by cervical dislocation. Major organs and all bones were removed and examined at low magnification (×2 to ×10) fluorescence stereomicroscopy. All studies were performed with 10 to 15 mice per group using three BRMS1-expressing clones derived from MDA-MB-435 and MDA-MB-231. Experiments were replicated independently at least once. Mice were maintained under the guidelines of the National Institutes of Health and the University of Alabama at Birmingham. All protocols were approved and monitored by the University of Alabama at Birmingham Institutional Animal Care and Use Committee.
Fluorescence Microscopy
To visualize cells or metastases, whole bones (dissected free of soft tissue using a no. 11 scalpel blade with gauze used to grip and remove tissue remnants) and organs were placed into Petri dishes containing ice-cold Ca+2- and Mg+2-free Dulbecco’s phosphate-buffered saline and examined by fluorescence microscopy using a Leica MZFLIII dissecting microscope with ×0.5 objective and GFP fluorescence filters (λexcitation, 480 ± 20 nm; λemission, 510-nm barrier; Leica, Deerfield, IL). Photomicrographs were collected using a MagnaFire digital camera (Optronics, Goleta, CA), and ImagePro Plus 5.1 software (Media Cybernetics, Silver Spring, MD). The total number of metastatic foci per organ was determined. The location and size of skeletal metastases were recorded on diagrams of murine bones and translated to a schematic overlay using a custom computer program.40 Fluorescence of tumor cells allowed visualization of single cells within brain, lung, bone, pancreas, kidneys, and adrenal glands without sectioning. Sectioning of random tissues with immunohistochemistry with anti-GFP antibodies confirmed that the fluorescent foci were tumor cells (Ref. 39; data not shown). Fluorescent focus size was determined using an ocular micrometer. Single cells and microscopic foci ranged in size from 20 to 35 μm.
Anoikis
Semiconfluent cells (70 to 90%) were washed twice using Ca+2- and Mg+2-free Dulbecco’s phosphate-buffered saline followed by detachment using a 2 mmol/L EDTA solution before resuspension in serum-free Dulbecco’s-modified Eagle’s medium and Ham’s-F12 medium at a concentration of 5 × 105 before plating onto 10-cm tissue culture plates coated with poly 2-hydroxyethyl methacrylate (poly-HEMA; 120 mg/ml in 95% ethanol; Sigma-Aldrich). At various times, apoptosis was evaluated using cell lysates (80 μg/lane) by immunoblotting (12% SDS-polyacrylamide gel electrophoresis) to detect caspase-3 and PARP cleavage or other apoptotic regulators, Bad, Bcl-xL, Bcl-2, Bim1, and Bmf. All antibodies were purchased from Cell Signaling Technologies.
Cells were suspended on poly-HEMA-coated plates for 30 minutes then seeded on tissue culture plates for 48 hours in serum-free media. Cell viability was determined using flow cytometry using propidium iodide (5 μg/ml) staining using a BD LSR II flow cytometer (BD Biosciences, San Jose, CA). Cells were also counted using a hemacytometer after Trypan blue staining to determine cell viability.
Statistics
Comparisons between groups were done by one-way analysis of variance with Holm-Sidak and Tukey’s post hoc tests using SigmaStat statistical analysis software for normally distributed data sets (SPSS Inc., Chicago, Il). Statistical significance was defined as a P ≤ 0.05.
Results and Discussion
The potential for metastasis suppressors to impact prevention and/or treatment of cancer metastasis is predicated on exploiting their mechanism of action.45 In previous studies, we found modest inhibition of adhesion and invasion by BRMS1-expressing cells9,10,46; but neither level of inhibition was adequate to account for the >90% suppression of lung and lymph node metastasis in vivo. Concurrent studies with another metastasis suppressor, KISS1, corroborated the inadequacies of in vitro models for predicting metastasis suppressor function,47 ie, in vitro assays showed minor differences when comparing parental and metastasis suppressor-expressing cells. Therefore, a more sensitive method to monitor cells in vivo was required. To monitor cell behavior in vivo more readily, we took advantage of the increased detection afforded by enhanced green fluorescent protein expression.33 We acknowledge that the studies presented here do not recapitulate the entire metastatic process because no orthotopic tumors were present. However, because prior studies showed that BRMS1-expressing cells grew in orthotopic sites and entered the vasculature at comparable rates,9,10 the studies reported here focus on steps of the metastatic cascade after intravasation.
BRMS1- and GFP-co-expressing cells were generated using lentiviral infection. Three clones each from MDA-MB-435GFP and -231GFP were isolated and verified to express BRMS1 (Figure 1) using a recently developed monoclonal antibody (Supplemental Figure 1, see http://ajp.amjpathol.org). In vitro and in vivo tumor cell growth rates were not significantlyaffected (Supplemental Figures 2 and 3, see http://ajp.amjpathol.org).
Figure 1.
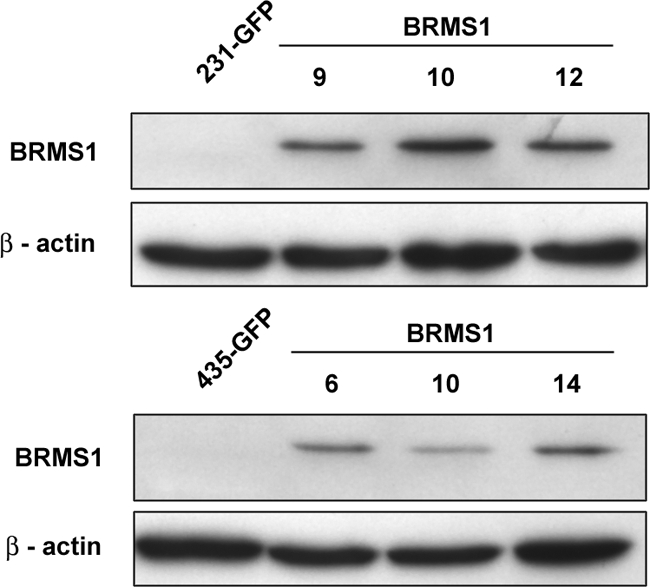
BRMS1 protein expression measured by immunoblot for three single cell clones obtained after transduction of 231GFP (top) and 435GFP (bottom) with a lentiviral vector.
MDA-MB-435GFP and -231GFP cells metastasized at comparable frequencies with untransfected cells when injected orthotopically or intravenously.39,40 However, expression of GFP allowed easier detection of microscopic metastases in various tissues, even permitting visualization of single cells in isolated organs.44,47 Because previous studies had shown significant reduction in lung metastases when BRMS1 expression was restored, the focus of the current study was on metastases to other sites. By injecting cells directly into the arterial circulation, wider distribution of cells was achieved. MDA-MB-231GFP and -435GFP cells most commonly metastasized to the bone, brain, kidney, adrenal gland, and pancreas. Less frequently, ovary and mesentery were involved.
BRMS1 Suppresses Metastases to Multiple Organs
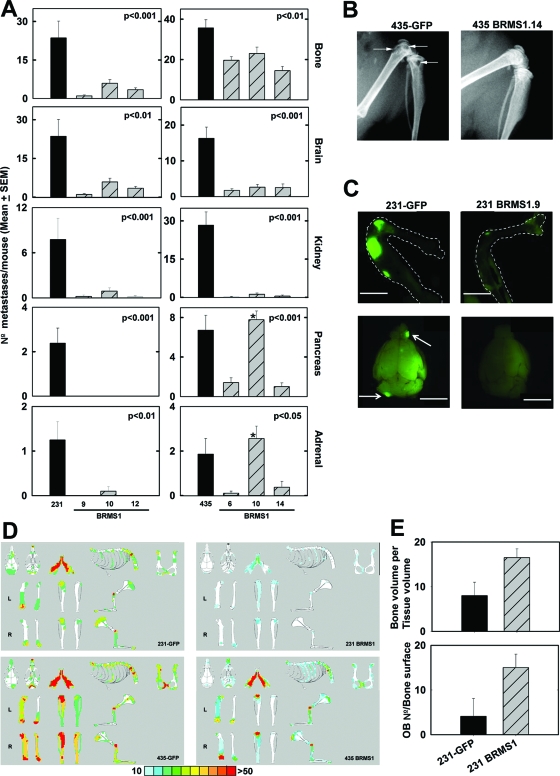
To test whether BRMS1 suppressed metastases to multiple organs, parental 231GFP and 435GFP cells and three cell clones expressing BRMS1 (231GFP-BRMS1 and 435GFP-BRMS1) were injected intracardially in female athymic mice. The number of metastases in each organ were counted and analyzed. 231GFP-BRMS1 and 435GFP-BRMS1 cells formed significantly fewer brain metastases (P ≤ 0.001) compared with parental cells (Figure 2, A and C). BRMS1 expression also resulted in fewer tumor cell foci within the kidneys (Figure 2A). Although most of renal metastases were found within the parenchyma, occasionally tumors were detected in the pelvi-calyceal system, which tended to be larger than those within the kidney tissue.
Figure 2.
BRMS1 suppresses breast cancer metastases to multiple organs after intracardiac injection. A: After injection, mice were necropsied 4 to 6 weeks later. All major organs were harvested, and the number of macroscopic metastatic foci in each organ was counted. Although not depicted, occasional metastases in the ovaries and mesentery were seen. *435GFP-BRMS1.10 was not significant in the pancreas and adrenal gland. B: Representative X-ray radiographical images showing osteolysis induced by 435GFP cells in the femur and tibia (arrows) without evidence of osteolysis in 435GFP-BRMS1.14. C: Representative fluorescence micrographs showing presence of 231GFP metastases in bone (right femur and tibia) and brain (arrows). Bar = 5 mm. D: Cumulative position and size of bone metastases from 231GFP, 435GFP, 231GFP-BRMS1, and 435GFP-BRMS1 cell-injected mice. The location and size of fluorescent cells or foci were entered onto schematic diagrams of murine bones. The maps were overlain to depict the frequency of tumor cells located at specific sites. The percentage of mice are depicted by color (red, >50% of mice). E: Histomorphometric analyses of osseous metastases in 231GFP and 231GFP-BRMS1 (a representative image for clone 9 is shown) cell-injected mice shows that re-expression of BRMS1 corresponds with more osteoblasts present and decreased osteolysis. The osteoblast number per bone surface area and mean bone volume-to-tissue volume ratio are shown. Similar histomorphometry findings were observed for MDA-MB-435.
Metastases to pancreas and adrenal glands were also significantly reduced (P ≤ 0.05), except 435GFP-BRMS1.10, which did not suppress metastasis to either organ (Figure 2A). Whether this observation reflects clonal heterogeneity or dose-dependent suppression (435GFP-BRMS1.10 had the lowest BRMS1 expression) is unclear. Although fewer metastases were found in ovary and mesentery in 231GFP-BRMS1 and 435GFP-BRMS1 groups compared with parent groups, the values did not reach statistical significance even if pooling data from replicate experiments.
Bone is the most common site of breast cancer metastasis.48 Presence of tumor cells in bones were visualized using GFP and X-ray (Figure 2, B and C). All bones were collected at necropsy 4 to 6 weeks after injection. Both 231GFP and 435GFP formed numerous osteolytic metastases as expected (Figure 2B), with an average of 24 ± 7 (mean ± SEM) metastases/mouse for 231GFP, and 36 ± 4 metastases/mouse by 435GFP. All 231GFP-BRMS1 clones formed significantly fewer skeletal metastases (P ≤ 0.001; Figure 2, A and D). A similar pattern of suppression was observed for 435GFP-BRMS1 clones (P ≤ 0.01; Figure 2, A and D).
To test whether BRMS1 affected the location and size of bone metastases, a custom software program40 was used. The most common sites for skeletal metastasis from 231GFP and 435GFP were proximal and distal femur, proximal tibia and humerus, lumbar and sacral vertebrae, scapulae, skull, and mandibles (Figure 2D), which are also frequent sites of metastases in women with breast cancer metastasis to bone. MDA-MB-231GFP-BRMS1 clones metastasized significantly less frequently to the vertebral column (P ≤ 0.001) or long bones like the femur and tibia (P ≤ 0.05). MDA-MB-435GFP-BRMS1 also showed a lower propensity to colonize these sites but the magnitude of inhibition was less (Figure 2A). Importantly, both 231GFP-BRMS1 and 435GFP-BRMS1 cells seeded bone, but the area of bone occupied by fluorescent cells at the time of necropsy was less than that of the parental cells (Figure 2C), suggesting that BRMS1 inhibits growth of tumor cells at the secondary site (ie, colonization). Additionally, BRMS1 also reduced the osteolysis as seen by X-ray (Figure 2B) and by histomorphometry. To illustrate, histomorphometric analysis showed that the ratio of bone volume to tissue volume, indicating bone loss, was lower in 231GFP cells (8.5 ± 2.1) compared with 231GFP-BRMS1 (16.2 ± 2.4; data not shown). Furthermore, mean osteoblast number/bone surface in 231GFP was 4.1 ± 3.7, far lower than 231GFP-BRMS1 (14.99 ± 2.4) (Figure 2E). The histomorphometry data support the concept that tumor cell-induced elimination of osteoblasts contributes to osteolysis.39,49 The distribution of skeletal metastases was not affected by BRMS1 (Figure 2D).
BRMS1 Decreases the Number of Cells Reaching the Secondary Site by Increasing Susceptibility to Anoikis
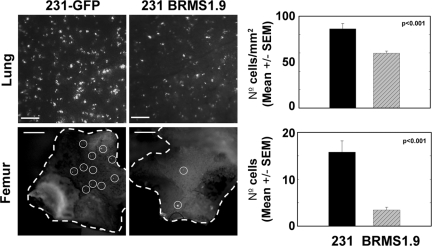
Whereas the bone metastasis data suggested that BRMS1 inhibits colonization, it was also possible that fewer cells reached the bone or other tissues after direct injection into the vasculature. To determine whether BRMS1 affected tumor cell seeding in bone, 231GFP and 231GFP-BRMS1.9 were injected intracardially, and both femurs were collected 1 hour after injection. Because 231GFP initially seed predominantly distal femur,39 cells were visualized and quantified at that site. Compared with 231GFP (16 ± 2), fewer 231GFP-BRMS1 cells (3 ± 0.6) were found (P ≤ 0.001; Figure 3). To test whether a similar reduction in cell seeding occurred in the lungs, GFP-labeled cells were injected into the lateral tail vein and counted in the lungs 1 hour after injection. The number of cells per mm2 were quantified. Similar to findings in the femur, fewer 231GFP-BRMS1 expressing cells (60 ± 2) were detected compared with 231GFP (86 ± 6) (P ≤ 0.001; Figure 3). Although persistence of cells without increasing size of green fluorescent foci (from the initial ∼25 μm) suggests that the seeded cells are in a dormant state, it is not possible with the present data to distinguish dormancy (lack of cell division) versus balanced growth and apoptosis.
Figure 3.
BRMS1 decreases seeding of tumor cells at secondary sites. 231GFP and 231GFP-BRMS1.9 cells were injected into the lateral tail vein to assess presence in the lungs or the left ventricle of the heart to evaluate seeding into femurs. Organs were collected 1 hour after injection and visualized using a fluorescent stereomicroscope. Representative fluoromicrographs from lung and femur are shown. Bars = 200 μm (lung) and 500 μm (femur). Because of autofluorescence in bone, single cells are highlighted by circles. The presence of single cells was quantified by direct counting and graphed. Significantly fewer cells reached both lungs (P < 0.001) and bone (P < 0.001). Cell seeding of other organs by 231GFP-BRMS1.9 was decreased compared with 231GFP, but the data were not quantified.
There are several potential explanations for decreased seeding by BRMS1-expressing cells. 1) BRMS1 could alter cell surfaces so that proclivity to adhere in specific organs is changed. This explanation is considered less likely than others for three reasons. In vitro adhesion to extracellular matrix components and endothelial cells was similar in BRMS1-expressing cells.9,10,46 In addition, there were no obvious changes in the distribution of metastases, which would be predicted if changes in adhesion molecules had occurred. A decrease in seeding was observed in the two most common and all other tissues (qualitative, but not quantified in this study), suggesting that the BRMS1-mediated seeding effects were not organ specific. Untested in the current study was whether BRMS1 alters strength of adhesion. 2) BRMS1 expression could affect how the cells respond to vascular dynamics (eg, sheer, pressure, and vessel size). Tumor cells often, because of their size, are physically trapped in lung capillaries.50,51 Similar entrapment in bone sinusoids is not as likely. The sluggish flow of blood in bone sinusoids52,53 would be predicted to cause less sheer than pulsatile flow in lung vessels. A combination of these considerations could account for the larger reduction in the number of BRMS1-expressing cells detected in bone compared with lung. 3) It is possible that cells are cleared more rapidly from trabecular bone than lung based on local microenvironment.54
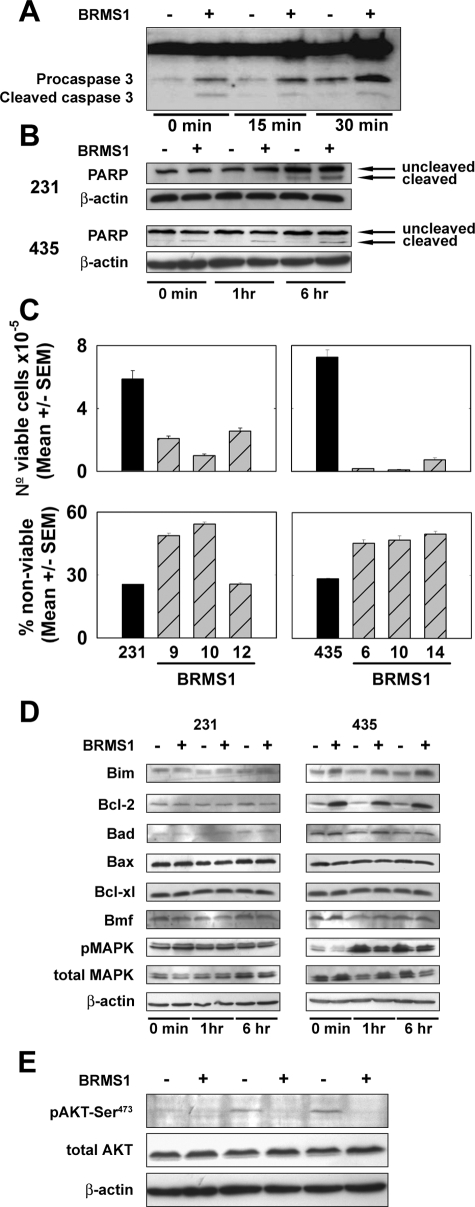
Another possible explanation is that BRMS1 expression results in increased cell death when cells are in suspension, detached from the surrounding matrix, or weakly adherent (ie, anoikis during transit). This explanation is consistent with prior observations that BRMS1-expressing cells are capable of invading and entering the vasculature.9,10 To test whether 231GFP-BRMS1 and 435GFP-BRMS1 cells were more susceptible to anoikis, 231GFP, 435GFP, and the respective BRMS1-expressing clones were placed, for varying periods of time, on poly-HEMA-coated plates and assessed for caspase-3 and PARP activation, commonly used indicators for the induction of apoptotic pathways. Activation of caspase-3 and PARP were detected in both 231GFP-BRMS1 and 435GFP-BRMS1 at higher levels than in 231GFP or 435GFP cells (Figure 4, A and B). Differential activation of caspase-3 or PARP was observed as early as 15 minutes after suspension (Figure 4B), but persisted for 1 and 6 hours.
Figure 4.
BRMS1 expression in human breast cancer cells increases sensitivity to anoikis. 231GFP, 231GFP-BRMS1.9, 435GFP, and 435GFP-BRMS1.14 (as representative clones) were suspended on poly-HEMA-coated tissue culture plates for 0 to 6 hours and assessed for induction of apoptotic pathways, cell viability, and regulators of apoptosis. A: 231GFP and 231GFP-BRMS1.9 cells were lysed and analyzed for cleavage of caspase 3. B: 231GFP, 231GFP-BRMS1.9, 435GFP, and 435GFP-BRMS1.14 cells were lysed and analyzed for PARP cleavage by immunoblot. C: Re-expression of BRMS1 resulted in decreased numbers of viable cells. After 30-minute suspension on poly-HEMA-coated plates, cells were seeded on tissue culture plates in serum-free media for 48 hours, and viability was assessed by propidium iodide staining and flow cytometry. Nonviable (trypan blue positive) cells were also counted using a hemacytometer. Comparisons are made with vector-only transduced cells. Although a mixed population is used, the clones behave relatively homogeneously (see small SEM bars); therefore, only the mixed population result is presented for ease of comparison. D: Western blot analyses of multiple mediators of apoptosis after 30-minute suspension on poly-HEMA-coated plates. BclII and Bim1 expression were increased in 435GFP-BRMS1.14 cells, but similar changes were not observed in 231GFP-BRMS1.9 cells. Extracellular-regulated kinase 1/2 phosphorylation was decreased in 435GFP-BRMS1.14 cells but not 231GFP-BRMS1.9 cells. Results are representative of three independent experiments. E: Immunoblots reveal that phospho-Akt levels are reduced in 435GFP-BRMS1.14 suspended on poly-HEMA-coated plates for 1 or 6 hours. The blot was stripped and re-probed with a total-Akt antibody and β-actin as loading controls.
To evaluate whether the increase in caspase-3 and PARP cleavage in BRMS1-expressing cells translated into decreased cell viability, two different approaches were used. First, cells were suspended for 48 hours, and the percentage of nonviable cells stained by propidium iodide was analyzed by flow cytometry. Significantly higher numbers of nonviable cells were found in both 231GFP-BRMS1 (except clone 12) and 435GFP-BRMS1 compared with parent cells (Figure 4C). Second, cells were suspended for 30 minutes and placed onto 12-well tissue culture plates. After 48 hours, viable (trypan blue-negative) cells were counted, and the ratio of live to dead cells was determined. Fewer viable BRMS1-expressing cells were present (P ≤ 0.001; Figure 4C). This indicated that even a brief period of suspension can initiate the apoptotic cascade, resulting in cell death.
To begin testing which regulators of the apoptosis may be responsible for the anoikis, Western blot analyses were done for Bim, Bcl-2, Bad, Bax, Bcl-xL, and Bmf. The findings were inconsistent between the cell lines. Bim, a critical sensor of cell stress and a promoter of anoikis,55 and Bcl-2, an inhibitor of apoptosis, were significantly greater in 435GFP-BRMS1, but not 231GFP-BRMS1, compared with their respective metastatic counterparts (Figure 4D). For some, but not all, 435GFP-BRMS1 cell clones but none of the 231GFP-BRMS1 clones, Bad, a promoter of apoptosis, was increased. No increases in the pro-apoptotic protein Bax or the anti-apoptotic Bcl-xL were seen.
Although the biological findings are consistent, the molecular basis for BRMS1 increasing susceptibility to anoikis is paradoxical. Recently, HDAC inhibitors have been shown to increase Bim and Bmf protein expression.56,57 It was, therefore, somewhat surprising that Bim and Bmf expression were not consistently changed when BRMS1 was present because multiple SIN3:HDAC complexes include BRMS1. We speculate that BRMS1 presence in HDAC complexes is involved in gene-specific regulation rather than a more global affect seen after HDAC inhibitor treatment, although this hypothesis has yet to be tested. In breast cells, it is still not yet known whether BRMS1 increases, decreases, or even regulates HDAC activity. Also, the dependence of HDAC activity on transitory complexes, cell type, culture conditions, cell cycle, and other experimental parameters may account for some of the experimental differences observed.
It is curious that nuclear factor-κB stimulates Bim1 expression.58 But BRMS1 expression has been shown to significantly inhibit nuclear factor-κB expression23 and activity.19,21 In contrast, Bim is down-regulated via the MAPK signaling pathway in cells treated with epidermal growth factor.59 In 231GFP- and 231GFP-BRMS1, no differences in p42/44 MAPK phosphorylation are observed (Figure 4D) because MDA-MB-231 cells express a constitutively active Ki-ras mutant.60 In 435GFP-BRMS1 cells, levels of phosphorylated p42/44 MAPK were much lower after 1 and 6 hours suspension on poly-HEMA compared with 435GFP. These data collectively suggest that in the 435 cell line, BRMS1 may mediate increased susceptibility to anoikis through the differential expression of Bim downstream of extracellular-regulated kinase. However, Bim expression does not appear to be a universal mediator of BRMS1-enhanced susceptibility to anoikis.
We previously showed that BRMS1 selectively decreases cell levels of phosphatidylinositol-4,5-bisphosphate22 and have extended those findings to show that 435GFP-BRMS1 cells have significantly reduced expression of epidermal growth factor receptor and a corresponding abrogation of downstream signaling through the phosphatidylinositol 3-kinase/Akt, but not the MAPK pathway (K.S. Vaidya and D.R. Welch, unpublished data). Reductions in phosphoinositide signaling may explain the loss of a potentially compensatory Akt survival response observed when BRMS1-expressing cells are plated onto poly-HEMA-coated plates (Figure 4E).
By using the intracardiac injection model coupled with GFP-labeled tumor cells, we have been able to evaluate the impact of BRMS1 on multiple steps of the metastatic cascade in vivo. The data clearly show that BRMS1 inhibits metastasis to multiple organs, in addition to lung and regional lymph nodes. The new data, along with previously published findings, further demonstrate that BRMS1 inhibits, to varying degrees in two human breast cancer cell lines, multiple steps of the metastatic cascade: adhesion, invasion, anoikis, and colonization. Although the precise biochemical and molecular mechanisms for inhibition at each step have not been fully defined, the implications for clinical control of cancer metastasis are significant. First, a global metastasis suppressor would be of greater generic use than a cocktail of organ-selective metastasis inhibitors. Second, inhibition at early, middle, and late steps of the metastatic cascade means that mimicry of BRMS1 metastasis suppressor mechanisms may have use for prevention of metastases as well as treatment of microscopic lesions that have yet to colonize ectopic sites.
Acknowledgments
We thank Dr. Janet Price (University of Texas M.D. Anderson Cancer Center) for generously providing the MDA-MB-435 and MDA-MB-231 cells, Drs. John Kappes and Yujiang Jia for generating the lentiviruses for infection, and Carrie Elzie and Pritish Pawar for helpful advice and suggestions. We are also grateful for the superb assistance provided by University of Alabama–Birmingham Comprehensive Cancer Center Epitope Recognition Immunoreagent Shared Resource (Dr. Mary Ann Accavitti-Loper) and the University of Alabama–Birmingham Center for Metabolic Bone Diseases Histomorphometry Core (Dr. Jay McDonald).
Footnotes
Address reprint requests to Danny R. Welch, Department of Pathology, VH-G019, University of Alabama at Birmingham, Birmingham, AL 35294-0019. E-mail: danwelch@uab.edu.
Supported by grants from the United States Public Health Service (CA087728 to D.R.W. and CA113037 National Research Service Award to D.R.H.), the US Army Medical Research and Materiel Command (DAMD17-02-1-0541 to D.R.W.), Susan G. Komen for the Cure (PDF1122006 to K.S.V.), and the National Foundation for Cancer Research.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This work is submitted in partial fulfillment of the requirements for the University of Alabama–Birmingham Graduate Program in Molecular and Cellular Pathology (P.A.P. and K.T.N.).
A guest editor acted as editor-in-chief for this manuscript. No person at the University of Alabama at Birmingham was involved in the peer review process or final disposition for this article.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- Rinker-Schaeffer CW, O'Keefe JP, Welch DR, Theodorescu D. Metastasis suppressor proteins: discovery, molecular mechanisms, and clinical application. Clin Cancer Res. 2006;12:3382–3389. doi: 10.1158/1078-0432.CCR-06-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevde LA, Welch DR. Metastasis suppressor pathways: an evolving paradigm. Cancer Lett. 2003;198:1–20. doi: 10.1016/s0304-3835(03)00304-5. [DOI] [PubMed] [Google Scholar]
- Steeg PS. Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer. 2003;3:55–63. doi: 10.1038/nrc967. [DOI] [PubMed] [Google Scholar]
- Vaidya KS, Welch DR. Metastasis suppressors and their roles in breast carcinoma. J Mammary Gland Biol Neoplasia. 2007;12:175–190. doi: 10.1007/s10911-007-9049-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samant RS, Debies MT, Hurst DR, Moore BP, Shevde LA, Welch DR. Suppression of murine mammary carcinoma metastasis by the murine ortholog of breast cancer metastasis suppressor 1 (Brms1). Cancer Lett. 2006;235:260–265. doi: 10.1016/j.canlet.2005.04.032. [DOI] [PubMed] [Google Scholar]
- Zhang S, Lin QD, Di W. Suppression of human ovarian carcinoma metastasis by the metastasis-suppressor gene, BRMS1. Int J Gynecol Cancer. 2006;16:522–531. doi: 10.1111/j.1525-1438.2006.00547.x. [DOI] [PubMed] [Google Scholar]
- Samant RS, Debies MT, Shevde LA, Verderame MF, Welch DR. Identification and characterization of murine ortholog (Brms1) of breast cancer metastasis suppressor 1 (BRMS1). Int J Cancer. 2002;97:15–20. doi: 10.1002/ijc.1569. [DOI] [PubMed] [Google Scholar]
- Shevde LA, Samant RS, Goldberg SF, Sikaneta T, Alessandrini A, Donahue HJ, Mauger DT, Welch DR. Suppression of human melanoma metastasis by the metastasis suppressor gene BRMS1. Exp Cell Res. 2002;273:229–239. doi: 10.1006/excr.2001.5452. [DOI] [PubMed] [Google Scholar]
- Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer Res. 2000;60:2764–2769. [PubMed] [Google Scholar]
- Hicks DG, Yoder BJ, Short S, Tarr S, Prescott N, Crowe JP, Dawson AE, Budd GT, Sizemore S, Cicek M, Choueiri T, Tubbs RR, Gaile D, Nowak N, Accavitti-Loper MA, Frost AR, Welch DR, Casey G. Loss of BRMS1 protein expression predicts reduced disease-free survival in hormone receptor negative and HER2 positive subsets of breast cancer. Clin Cancer Res. 2006;12:6702–6708. doi: 10.1158/1078-0432.CCR-06-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yamashita H, Toyama T, Yamamoto Y, Kawasoe T, Iwase H. Reduced expression of the breast cancer metastasis suppressor 1 mRNA is correlated with poor progress in breast cancer. Clin Cancer Res. 2006;12:6410–6414. doi: 10.1158/1078-0432.CCR-06-1347. [DOI] [PubMed] [Google Scholar]
- Ohta S, Lai EW, Pang ALY, Brouwers FM, Chan WY, Eisenhofer G, deKrijger R, Ksinantova L, Breza J, Blazicek P, Kvetnansky R, Wesley RA, Pacak K. Downregulation of metastasis suppressor genes in malignant pheochromocytoma. Int J Cancer. 2005;114:139–143. doi: 10.1002/ijc.20670. [DOI] [PubMed] [Google Scholar]
- Stark AM, Tongers K, Maass N, Mehdorn HM, Held-Feindt J. Reduced metastasis-suppressor gene mRNA-expression in breast cancer brain metastases. J Cancer Res Clin Oncol. 2005;131:191–198. doi: 10.1007/s00432-004-0629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi G, Di Cristofano C, Capodanno A, Iorio MC, Aretini P, Isola P, Tancredi M, Collecchi P, Naccarato AG, Porta RP, Bevilacqua G, Caligo MA. High level of messenger RNA for BRMS1 in primary breast carcinomas is associated with poor prognosis. Int J Cancer. 2007;120:1169–1178. doi: 10.1002/ijc.22379. [DOI] [PubMed] [Google Scholar]
- Kelly LM, Buggy Y, Hill A, O'Donovan N, Duggan C, McDermott EW, O'Higgins NJ, Young L, Duffy MJ. Expression of the breast cancer metastasis suppressor gene, BRMS1, in human breast carcinoma: lack of correlation with metastasis to axillary lymph nodes. Tumour Biol. 2005;26:213–216. doi: 10.1159/000086955. [DOI] [PubMed] [Google Scholar]
- Hurst DR, Mehta A, Moore BP, Phadke PA, Meehan WJ, Accavitti MA, Shevde LA, Hopper JE, Xie Y, Welch DR, Samant RS. Breast cancer metastasis suppressor 1 (BRMS1) is stabilized by the Hsp90 chaperone. Biochem Biophys Res Comm. 2006;348:1429–1435. doi: 10.1016/j.bbrc.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan WJ, Samant RS, Hopper JE, Carrozza MJ, Shevde LA, Workman JL, Eckert KA, Verderame MF, Welch DR. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. J Biol Chem. 2004;279:1562–1569. doi: 10.1074/jbc.M307969200. [DOI] [PubMed] [Google Scholar]
- Liu Y, Smith PW, Mayo MW, Jones DR. Breast cancer metastasis suppressor 1 modulates NF-kappa B-dependent transcription through deacetylation of RelA/p65 in non-small cell lung cancer cells. Proc Am Assoc Cancer Res. 2006;47:4310. [Google Scholar]
- Liu Y, Smith PW, Jones DR. Breast cancer metastasis suppressor 1 functions as a corepressor by enhancing histone deacetylase 1-mediated deacetylation of RelA/p65 and promoting apoptosis. Mol Cell Biol. 2006;26:8683–8696. doi: 10.1128/MCB.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicek M, Fukuyama R, Welch DR, Sizemore N, Casey G. Breast cancer metastasis suppressor 1 inhibits gene expression by targeting nuclear factor-κB activity. Cancer Res. 2005;65:3586–3595. doi: 10.1158/0008-5472.CAN-04-3139. [DOI] [PubMed] [Google Scholar]
- DeWald DB, Torabinejad J, Samant RS, Johnston D, Erin N, Shope JC, Xie Y, Welch DR. Metastasis suppression by breast cancer metastasis suppressor 1 involves reduction of phosphoinositide signaling in MDA-MB-435 breast carcinoma cells. Cancer Res. 2005;65:713–717. [PubMed] [Google Scholar]
- Samant RS, Clark DW, Fillmore RA, Cicek M, Metge BJ, Chandramouli KH, Chambers AF, Casey G, Welch DR, Shevde LA. Breast cancer metastasis suppressor 1 (BRMS1) inhibits osteopontin transcription by abrogating NF-kappaB activation. Mol Cancer. 2007;6:6. doi: 10.1186/1476-4598-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevde LA, Samant RS, Paik JC, Metge BJ, Chambers AF, Casey G, Frost AR, Welch DR. Osteopontin knockdown suppresses tumorigenicity of human metastatic breast carcinoma. Clin Exp Metastasis. 2006;23:123–133. doi: 10.1007/s10585-006-9013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders MM, Seraj MJ, Li ZY, Zhou ZY, Winter CR, Welch DR, Donahue HJ. Breast cancer metastatic potential correlates with a breakdown in homospecific and heterospecific gap junctional intercellular communication. Cancer Res. 2001;61:1765–1767. [PubMed] [Google Scholar]
- Talmadge JE, Singh RK, Fidler IJ, Raz A. Murine models to evaluate novel and conventional therapeutic strategies for cancer. Am J Pathol. 2007;170:793–804. doi: 10.2353/ajpath.2007.060929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman RM. Orthotopic metastatic mouse models for anticancer drug discovery and evaluation: a bridge to the clinic. Invest New Drugs. 1999;17:343–359. doi: 10.1023/a:1006326203858. [DOI] [PubMed] [Google Scholar]
- Welch DR. Do we need to redefine a cancer metastasis and staging definitions? Breast Disease. 2007;26:3–12. doi: 10.3233/bd-2007-26102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch DR. Defining a cancer metastasis. Philadelphia: American Association for Cancer Research,; AACR Education Book 2006. 2006:pp 111–115. [Google Scholar]
- Goodison S, Kawai K, Hihara J, Jiang P, Yang M, Urquidi V, Hoffman RM, Tarin D. Prolonged dormancy and site-specific growth potential of cancer cells spontaneously disseminated from nonmetastatic breast tumors as revealed by labeling with green fluorescent protein. Clin Cancer Res. 2003;9:3808–3814. [PubMed] [Google Scholar]
- Suzuki M, Mose ES, Montel V, Tarin D. Dormant cancer cells retrieved from metastasis-free organs regain tumorigenic and metastatic potency. Am J Pathol. 2006;169:673–681. doi: 10.2353/ajpath.2006.060053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman RM. The multiple uses of fluorescent proteins to visualize cancer in vivo. Nat Rev Cancer. 2005;5:796–806. doi: 10.1038/nrc1717. [DOI] [PubMed] [Google Scholar]
- Hoffman RM. Imaging tumor angiogenesis with fluorescent proteins. APMIS. 2004;112:441–449. doi: 10.1111/j.1600-0463.2004.apm11207-0806.x. [DOI] [PubMed] [Google Scholar]
- Price JE, Polyzos A, Zhang RD, Daniels LM. Tumorigenicity and metastasis of human breast carcinoma cell lines in nude mice. Cancer Res. 1990;50:717–721. [PubMed] [Google Scholar]
- Rae JM, Creighton CJ, Meck JM, Haddad BR, Johnson MD. MDA-MB-435 cells are derived from M14 Melanoma cells: a loss for breast cancer, but a boon for melanoma research. Breast Cancer Res Treat. 2007;104:13–19. doi: 10.1007/s10549-006-9392-8. [DOI] [PubMed] [Google Scholar]
- Ellison G, Klinowska TCM, Westwood RF, Docter E, French T, Fox JC. Further evidence to support the melanocytic origin of MDA-MB-435. Mol Pathol. 2002;55:294–299. doi: 10.1136/mp.55.5.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellappan S, Grijalva R, Zhou XY, Yang WT, BarEli M, Mills GB, Yu DH. Lineage infidelity of MDA-MB-435 cells: expression of melanocyte proteins in a breast cancer cell line. Cancer Res. 2004;64:3479–3485. doi: 10.1158/0008-5472.CAN-3299-2. [DOI] [PubMed] [Google Scholar]
- Welch DR. Technical considerations for studying cancer metastasis in vivo. Clin Exp Metastasis. 1997;15:272–306. doi: 10.1023/a:1018477516367. [DOI] [PubMed] [Google Scholar]
- Phadke PA, Mercer RR, Harms JF, Jia Y, Frost AR, Jewell J, Bussard KM, Nelson S, Moore C, Kappes JC, Gay CV, Mastro AM, Welch DR. Kinetics of metastatic breast cancer cell trafficking in bone. Clin Cancer Res. 2006;12:1431–1440. doi: 10.1158/1078-0432.CCR-05-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms JF, Welch DR. MDA-MB-435 human breast carcinoma metastasis to bone. Clin Exp Metastasis. 2003;20:327–334. doi: 10.1023/a:1024062911144. [DOI] [PubMed] [Google Scholar]
- Van Tine BA, Kappes JC, Banerjee NS, Knops J, Lai L, Steenbergen RD, Meijer CL, Snijders PJ, Chatis P, Broker TR, Moen PT, Jr, Chow LT. Clonal selection for transcriptionally active viral oncogenes during progression to cancer. J Virol. 2004;78:11172–11186. doi: 10.1128/JVI.78.20.11172-11186.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Wu X, Levasseur DN, Liu H, Lai L, Kappes JC, Townes TM. Lentiviral vector transduction of hematopoietic stem cells that mediate long-term reconstitution of lethally irradiated mice. Stem Cells. 2000;18:352–359. doi: 10.1634/stemcells.18-5-352. [DOI] [PubMed] [Google Scholar]
- Harms JF, Welch DR, Samant RS, Shevde LA, Miele ME, Babu GR, Goldberg SF, Gilman VR, Sosnowski DM, Campo DA, Gay CV, Budgeon LR, Mercer R, Jewell J, Mastro AM, Donahue HJ, Erin N, Debies MT, Meehan WJ, Jones AL, Mbalaviele G, Nickols A, Christensen ND, Melly R, Beck LN, Kent J, Rader RK, Kotyk JJ, Pagel MD, Westlin WF, Griggs DW. A small molecule antagonist of the alpha-v, beta-3 integrin suppresses MDA-MB-435 skeletal metastasis. Clin Exp Metastasis. 2004;21:119–128. doi: 10.1023/b:clin.0000024763.69809.64. [DOI] [PubMed] [Google Scholar]
- Goldberg SF, Harms JF, Quon K, Welch DR. Metastasis-suppressed C8161 melanoma cells arrest in lung but fail to proliferate. Clin Exp Metastasis. 1999;17:601–607. doi: 10.1023/a:1006718800891. [DOI] [PubMed] [Google Scholar]
- Eccles SA, Welch DR. Metastasis: recent discoveries and novel treatment strategies. Lancet. 2007;369:1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samant RS, Seraj MJ, Saunders MM, Sakamaki T, Shevde LA, Harms JF, Leonard TO, Goldberg SF, Budgeon LR, Meehan WJ, Winter CR, Christensen ND, Verderame MF, Donahue HJ, Welch DR. Analysis of mechanisms underlying BRMS1 suppression of metastasis. Clin Exp Metastasis. 2000;18:683–693. doi: 10.1023/a:1013124725690. [DOI] [PubMed] [Google Scholar]
- Nash KT, Phadke PA, Navenot J-M, Hurst DR, Accavitti-Loper MA, Sztul E, Vaidya KS, Frost AR, Kappes JC, Peiper SC, Welch DR. KISS1 metastasis suppressor secretion, multiple organ metastasis suppression, and maintenance of tumor dormancy. J Natl Cancer Inst. 2007;99:309–321. doi: 10.1093/jnci/djk053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roodman GD. Mechanisms of disease: mechanisms of bone metastasis. N Engl J Med. 2004;350:1655–1664. doi: 10.1056/NEJMra030831. [DOI] [PubMed] [Google Scholar]
- Mastro AM, Gay CV, Welch DR, Donahue HJ, Jewell J, Mercer R, DiGirolamo D, Chislock EM, Guttridge K. Breast cancer cells induce osteoblast apoptosis: a possible contributor to bone degradation. J Cell Biochem. 2004;91:265–276. doi: 10.1002/jcb.10746. [DOI] [PubMed] [Google Scholar]
- Weiss L, Orr FW, Honn KW. Interactions between cancer cells and the microvasculature: a rate regulator for metastasis. Clin Exp Metastasis. 1989;7:127–167. doi: 10.1007/BF01787020. [DOI] [PubMed] [Google Scholar]
- Weiss L, Schmid-Schonbein GW. Biomechanical interactions of cancer cells with the microvasculature during metastasis. Cell Biophys. 1989;14:187–215. doi: 10.1007/BF02797133. [DOI] [PubMed] [Google Scholar]
- Mastro AM, Gay CV, Welch DR. The skeleton as a unique environment for breast cancer cells. Clin Exp Metastasis. 2003;20:275–284. doi: 10.1023/a:1022995403081. [DOI] [PubMed] [Google Scholar]
- Welch DR, Harms JF, Mastro AM, Gay CV. Breast cancer metastasis to bone: evolving models and research challenges. J Musculoskelet Neuronal Interact. 2003;3:30–38. [PubMed] [Google Scholar]
- Langley RR, Fidler IJ. Tumor cell-organ microenvironment interactions in the pathogenesis of cancer metastasis. Endocr Rev. 2007;28:297–321. doi: 10.1210/er.2006-0027. [DOI] [PubMed] [Google Scholar]
- Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J, Muthuswamy SK, Brugge JS. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat Cell Biol. 2003;5:733–740. doi: 10.1038/ncb1026. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Adachi M, Kawamura R, Imai K. Bmf is a possible mediator in histone deacetylase inhibitors FK228 and CBHA-induced apoptosis. Cell Death Differ. 2006;13:129–140. doi: 10.1038/sj.cdd.4401686. [DOI] [PubMed] [Google Scholar]
- Zantl N, Weirich G, Zall H, Seiffert BM, Fischer SF, Kirschnek S, Hartmann C, Fritsch RM, Gillissen B, Daniel PT, Hacker G. Frequent loss of expression of the pro-apoptotic protein Bim in renal cell carcinoma: evidence for contribution to apoptosis resistance. Oncogene. 2007;26:7038–7048. doi: 10.1038/sj.onc.1210510. [DOI] [PubMed] [Google Scholar]
- Inta I, Paxian S, Maegele I, Zhang W, Pizzi M, Spano P, Sarnico I, Muhammad S, Herrmann O, Inta D, Baumann B, Liou HC, Schmid RM, Schwaninger M. Bim and Noxa are candidates to mediate the deleterious effect of the NF-kappa B subunit RelA in cerebral ischemia. J Neurosci. 2006;26:12896–12903. doi: 10.1523/JNEUROSCI.3670-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadros MR, Connelly S, Kari C, Abrams MT, Wickstrom E, Rodeck U. EGFR-dependent downregulation of Bim in epithelial cells requires MAPK and PKC-delta activities. Cancer Biol Ther. 2006;5:498–504. doi: 10.4161/cbt.5.5.2567. [DOI] [PubMed] [Google Scholar]
- Fukazawa H, Noguchi K, Murakami Y, Uehara Y. Mitogen-activated protein/extracellular signal-regulated kinase kinase (MEK) inhibitors restore anoikis sensitivity in human breast cancer cell lines with a constitutively activated extracellular-regulated kinase (ERK) pathway. Mol Cancer Ther. 2002;1:303–309. [PubMed] [Google Scholar]