Summary
The extent to which the nuclear organisation of a gene impacts on its ability to be expressed, or whether nuclear organisation merely reflects gene expression states, remains an important but unresolved issue. A model system that has been instrumental in investigating this question is the murine Hox clusters. Nuclear reorganisation and chromatin decondensation, initiated towards the 3' end of the clusters, accompanies activation of Hox genes in both differentiation and development, and may be linked to mechanisms underlying colinearity. To investigate this, and to delineate the cis-acting elements involved, here we analyse the nuclear behaviour of a 3' Hoxb1 transgene transposed to the 5' end of the Hoxd cluster. We show that this transgene contains the cis-acting elements sufficient to initiate ectopic local nuclear reorganisation and chromatin decondensation, and to break Hoxd colinearity, in the primitive streak region of the early embryo. Significantly, in rhombomere 4 the transgene is able to induce attenuated nuclear reorganisation and decondensation of Hoxd even though there is no detectable expression of the transgene at this site. This shows that chromosome territory reorganisation and chromatin decondensation can be uncoupled from transcription itself, and suggests that they can therefore operate upstream of gene expression.
Keywords: Chromatin condensation, Chromosome territory, Colinearity, Hox, Nuclear Organisation
Introduction
Several facets of nuclear organisation have been correlated with gene expression at both constitutively active regions of the genome and at regions subject to co-ordinate regulation. Firstly, when active, these regions appear decondensed at a cytological level (Chambeyron and Bickmore, 2004; Christova et al., 2007; Goetze et al., 2007). Secondly, even though transcription and transcription factories can be seen within chromosome territories (CTs) (Abranches et al., 1998; Branco and Pombo, 2006; Verschure et al., 1999), many active genomic loci have been seen outside of their CTs (Brown et al., 2006; Chambeyron et al., 2005; Chambeyron and Bickmore, 2004; Mahy et al., 2002; Morey et al., 2007; Volpi et al., 2000; Williams et al., 2002). A key issue remains whether these forms of nuclear organisation are just a consequence of transcriptional activation or whether they have a causative role (Heard and Bickmore, 2007). The fact that the incidence of localisation outside of CTs decreases when transcription is blocked (Mahy et al., 2002) suggests that relocalisation is, at least in part, driven by transcription itself. On the other hand, looping out from the CT is not just a downstream consequence of a specific gene's activation. Along the primary (rostro-caudal) axis of the developing mouse embryo, activation of murine Hoxb and Hoxd loci is accompanied by both their decondensation and looping out from the CT (Chambeyron et al., 2005; Morey et al., 2007). However, in the limb bud, Hoxd activation and chromatin decondensation occur without relocalisation of this locus to the outside of its CT. We had suggested that this difference in nuclear behaviour of the same locus, when it is activated in different developmental contexts, maybe due to the different regulatory pathways and cis-acting sequences acting upon it (Morey et al., 2007).
This latter example illustrates how useful the study of the Hox gene clusters is in providing insights into the dynamic repositioning of a locus in the nucleus during spatial and temporal patterning of gene expression. Within mammalian Hox clusters there is a correspondence between the linear order of the genes and their sequence of activation in development - colinearity (Kmita and Duboule, 2003). Genes located at the 3' end of the clusters are expressed earlier, and more anteriorly, than genes located more 5'. It has been proposed that a transition from an inactive to an active chromatin state, propagated through Hox clusters from 3' to 5', might underlie colinear activation (Kondo and Duboule, 1999; Roelen et al., 2002; van der Hoeven et al., 1996). The relocalisation of Hox genes outside of their CTs does indeed initiate toward the 3' end of the clusters (Chambeyron et al., 2005; Chambeyron and Bickmore, 2004; Morey et al., 2007).
Transgenic experiments have revealed both local (e.g Marshall et al., 1994; Gould et al., 1997) and more distant (Gonzalez et al., 2007; Spitz et al., 2003; Spitz et al., 2005; Tarchini and Duboule, 2006; Zakany et al., 2004) cis-regulatory elements involved in the control of Hox gene expression. Manipulation of mouse Hox clusters, transferring genes from one position to another, has also indicated that part of this regulation depends on the position of a given gene in the cluster (Kmita et al., 2000; Kmita et al., 2002). To determine whether cis-acting elements responsible for initiating large-scale changes in chromatin structure are located at the 3' end of Hox clusters, here we have analysed the nuclear behaviour of the anterior (3') Hoxb1 gene transposed to the posterior (5') end of Hoxd. We used transgenic mouse embryos carrying a Hoxb1/LacZ reporter inserted by homologous recombination upstream of Hoxd13 (Kmita et al., 2000) (Fig. 1A). This transgene has the regulatory elements necessary for its autonomous expression when randomly integrated in the genome (Graaff et al., 2003), including 5' and 3' retinoic acid response elements (RAREs) and the rhombomere 4 (r4) autoregulatory enhancer (Marshall et al., 1994; Popperl et al., 1995; Studer et al., 1994). When transposed this transgene breaks the colinearity of Hoxd, inducing ectopic Hoxd13 expression in the primitive streak region (PS) of E7.5 embryos in a manner reminiscent of endogenous Hoxb1 expression (Kmita et al., 2000). However, ectopic activation of the Hoxb1/lacZ transgene occurs in the distal part of the E10.5 developing limb bud, where Hoxb1 is not normally expressed. Conversely expression of the transgene is absent in r4 of the E9.5 embryo, a site where both endogenous Hoxb1 and Hoxb1/lacZ transgenes inserted randomly into the genome, are expressed (Table 1).
Figure 1.
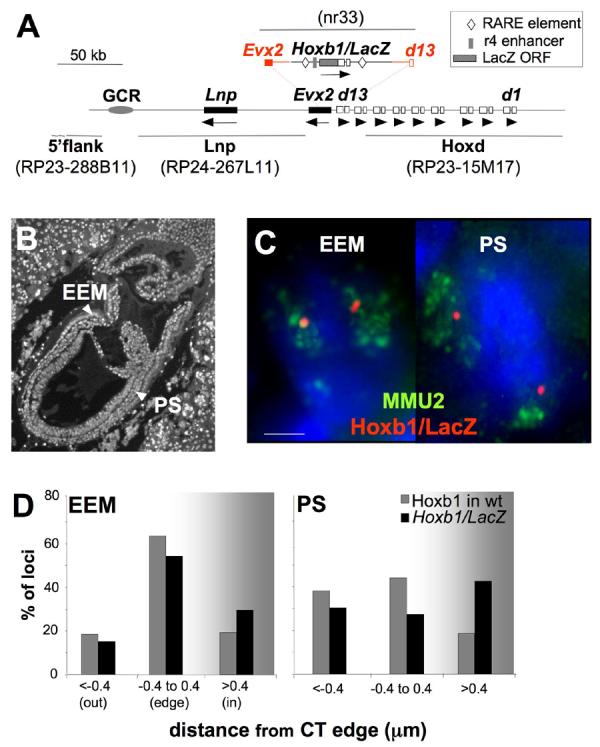
Looping out of Hoxb1/LacZ from the chromosome territory occurs in the PS of E7.5 transgenic embryos. (A) Map of the Hoxd locus on MMU2 showing the structure and the integration site of the Hoxb1/LacZ transgene. The extent of the homology arms used for recombination are shown in red and the Hoxb1 cis-regulatory retinoic acid response elements (RAREs) and the r4 enhancer included in the transgene are depicted (Kmita et al., 2000). Exons of Hoxd genes from Hoxd1 (d1) to Hoxd13 (d13) are shown as open boxes; other genes in the region are shown as black boxes. The orientation of transcription is indicated by arrows underneath the genes. The grey oval locates a region of non-coding sequence conservation that overlaps the Global Control Region (GCR) (Spitz et al., 2003). The location of the BACs and the Hoxb1/LacZ plasmid used as FISH probes are shown in grey. (B) DAPI staining of a 4μm E7.5 embryo section used for the FISH analysis showing the nuclei of the PS and EEM. (C) Maximal projection image after deconvolution of 3D FISH using the Hoxb1/LacZ probe (red) hybridised together with a MMU2 chromosome paint (green) on DAPI counterstained nuclei of the EEM or the PS of E7.5 Hoxb1/LacZ embryos. Scale bar: 2 μm. (D) Histograms showing the 3D position of hybridisation signals for the Hoxb1/LacZ transgene relative to the MMU2 CT edge (black bars), or for endogenous Hoxb1 relative to the edge of the MMU11 CT (grey bars) in nuclei from the EEM and PS of E7.5 transgenic or wild-type (wt) embryos respectively. Loci are defined as outside of the CT if the distance measured is > 0.4μm beyond the visible limits of the CT hybridisation signal.
Table 1.
Expression patterns of Hoxd genes, Hoxb1 and Hoxb1/LacZ transposed at Hoxd during embryogenesis.
| Embryonic stage |
Wild type embryos |
Hoxb1/LacZ embryos |
||||
|---|---|---|---|---|---|---|
| Tissue | 3′ Hoxd (d1-d10) | 5′ Hoxd (d11-d13) | Hoxb1 | LacZ | Hoxd13 | |
| E7.5 | extra-embryonic; anterior yolk sac mesoderm |
− | − | − | − | − |
| primitive streak and adjacent embryonic mesoderm |
− | − | + | + | + | |
| E9.5 | rhombomere 1 and 2 | − | − | − | − | − |
| rhombomere 4 | − | − | + | − | − | |
| tail bud | + | + | + | + (extends anteriorly) |
+ (reinforced in posterior embryo) |
|
| limb and genital buds | + | − | − | ND | − | |
| E10.5 | rhombomere 1 and 2 | − | − | − | − | − |
| rhombomere 4 | − | − | + | − | − | |
| tail bud | + | + | + | + (extends anteriorly) |
+ | |
| limb and genital buds | + | + | − | + (distal part of the buds) |
+ | |
−, not expressed; +, expressed; ND, not determined
Using fluorescence in situ hybridisation (FISH) on wild-type and transgenic embryos, we show that the transposed Hoxb1/LacZ recapitulates some of the behaviour of endogenous Hoxb1 in the PS of E7.5 embryos, suggesting that the transgene contains the minimal DNA elements necessary to initiate nuclear reorganisation and chromatin decondensation early in embryonic development, and that these elements can initiate nuclear reorganisation at an inappropriate genomic location (the 5' end of Hoxd) that is normally still silent at this early stage of embryogenesis. Moreover, the transgene can also initate these movements when ectopically activated in the limb bud later in development, where the rest of Hoxd does not loop out from the CT even though the Hoxd cluster is now active. In both of these cases (early embryo and limb bud) the nuclear organisation of the transposed Hoxb1/Lacz transgene completely correlates with its expression, so that transcription and nuclear organisation cannot be uncoupled from one another. However, in r4, the transgene is able to induce some attenuated, but still significant, nuclear reorganisation and chromatin decondensation of surrounding Hoxd regions, even though it is not itself detectably expressed. This suggests that chromosome territory reorganisation is not, in this case, simply a downstream consequence of transcription of the transgene.
Materials and methods
Mouse embryo sectioning and staging
Analysis on E7.5 and E9.5 embryos was as previously described (Chambeyron, 2005). We used embryos from crosses between Dct-LacZ homozygous transgenic CD1×CD1 mice as wild-type (wt) (Chambeyron et al., 2005; Morey et al., 2007). Hoxb1/LacZ homozygous embryos were collected from TgHb1×TgHb1 crosses (Kmita et al., 2000). The day on which the vaginal plug was detected was considered as 0.5 days of gestation (E0.5). Dct-LacZ and Hoxb1/LacZ E7.5 embryos in the deciduas and E9.5 embryos were fixed in 4% formaldehyde/PBS overnight at 4°C, dehydrated through a graded ethanol series, cleared in xylene and embedded in paraffin blocks. Adjacent serial sections were cut at 4μm and used for DNA-FISH and Haematoxylin-Eosin (HE) staining. HE stained sections from E7.5 embryos were used for embryo staging according to criteria previously established (Downs and Davies, 1993). A minimum of 3 independent embryos where used for the analyses.
3D DNA-FISH
Sagittal sections of E7.5 embryos that contained the PS, and sections from E9.5 embryos containing r1/r2, r4, forelimb buds and the tailbud were selected for FISH analysis. Sections laid on Superfrost slides (Menzel) were heated to 60°C for 20 minutes and washed four times in xylene for 10 minutes each before rehydration through an ethanol series. They were then microwaved for 20 minutes in 0.1 M citrate pH6 buffer, washed in water and rinsed once in 2×SSC before use. For FISH, slides were incubated in 2×SSC for 5 minutes at 75°C, denatured for 3 minutes at 75°C in 70% formamide/2×SSC, plunged into ice-cold 70% ethanol for 3 minutes, dehydrated through an ethanol series and air-dried. Hybridisation, washes and detection were as described previously (Chambeyron and Bickmore, 2004).
The BACs RP23-288B11 (5'flank), RP24-267L11 (Lnp), RP23-15M17 (Hoxd) and the MMU2 chromosome paint used as probes in this study were as previously described (Morey et al., 2007). The nr33 (Hoxb1/LacZ) probe consisted of the vector used for homologous recombination and therefore includes the genomic region extending from the 3' end of Evx2 up to the 5' end of Hoxd13 and the Hoxb1/LacZ reporter construct (Kmita et al., 2000)(Fig. 1A). Hoxb1 and Hoxb9 probes, were as previously described (Chambeyron et al., 2005; Chambeyron and Bickmore, 2004). Probes were prepared and labelled as described previously (Morey et al., 2007). Approximately 200 ng of biotin-paint and 100 ng of digoxigenin- or biotin-labelled BAC or plasmid probes were used per slide together with 10 μg mouse Cot1 DNA (Invitrogen) and 5 μg sonicated salmon sperm DNA.
Image capture and analysis
For 3D FISH, slides were examined using a Zeiss Axioskop fluorescence microscope with Plan-neofluar (numerical aperture = 1.3) or Plan apochromat ×100 objectives, a 50 W Hg source (Carl Zeiss, Welwyn Garden City, UK) and Chroma #83000 triple band pass filter set (Chroma Technology Corp., Rockingham, VT) with the excitation filters installed in a motorised filter wheel (Ludl Electronic Products, Hawthorne, NY). A piezoelectrically driven objective mount (PIFOCi model P-721, Physik Instrumente GmbH & Co, Karlsruhe) and a Princeton Instruments Micromax CCD camera with Kodak 1400e sensor (Universal Imaging, Maldon, UK), were used to control movement in the z dimension and collect image stacks with a 0.2 μm step. Hardware control, image capture and analysis were performed using in-house scripts written for IPLab Spectrum (Scanalytics Corp, Fairfax, VA). Images were deconvolved using a calculated PSF with the constrained iterative volume algorithm of Microtome (Scanalytics Corp, Fairfax, VA). 3D distance measurements were as described previously (Chambeyron et al., 2005). A minimum of 50 nuclei was analysed for each tissue. Chromatin condensation/decondensation was then assessed from the relationship between mean-squared interphase distance (d2 in μm2) between probes of known genomic separation (in Mb) (Chambeyron et al., 2005; Morey et al., 2007; Yokota et al., 1997).
Statistical analysis
The statistical relevance of data (Table 2) was assessed using the non-parametric Kolmogorov-Smirnov (KS) test to examine the null hypothesis that two sets of data show the same distribution. Data sets consisted of at least 50 nuclei (100 territories/loci) for each embryonic tissue, and for each combination of probes. A p-value < 0.05 was considered statistically significant.
Table 2.
Kolmogorov-Smirnov test applied to distances from CT edge and to interphase separations in E7.5 and E9.5 wt and Hoxb1/LacZ embryos.
|
p for comparison of distance from CT edge |
p for comparison of interphase separation |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Tissue |
Tissue |
||||||||
| Embryonic stage |
Embryo | Genomic region | PS vs EEM | Genomic region | PS vs EEM | ||||
| E7.5 | wt | Lnp | 0.895 | Hoxb | 0.011 | ||||
| Hoxb1 | 0.02 | Hoxd | 0.321 | ||||||
| Hoxd | 0.082 | ||||||||
| Hoxb1/LacZ | Lnp | 0.493 | 5′ of Hoxb1/LacZ | 0.005 | |||||
| Hoxb1/LacZ | 0.03 | 3′ of Hoxb1/LacZ | 0.001 | ||||||
| Hoxd | 0.999 | ||||||||
| E9.5 | r4 vs r1-r2 |
tail bud vs r1/r2 |
limb bud vs r1/r2 |
r4 vs r1/r2 |
tail bud vs r1/r2 |
limb bud vs r1/r2 |
|||
| wt | Lnp | 0.169 | 0.018 | 0.482 | Hoxb | <10−3 | 0.009 | ND | |
| Hoxb1 | <10−3 | 0.013 | ND | Hoxd | 0.852 | <10−3 | <10−3 | ||
| Hoxd | 0.313 | 0.021 | 0.28 | ||||||
| Hoxb1/LacZ | Lnp | 0.006 | <10−3 | 0.992 | 5′ of Hoxb1/LacZ | 0.254 | <10−3 | 0.02 | |
| Hoxb1/LacZ | 0.28 | <10−3 | <10−3 | 3′ of Hoxb1/LacZ | 0.036 | <10−3 | <10−3 | ||
| Hoxd | 0.004 | <10−3 | 0.149 | ||||||
Significant p-values are shown in bold. In E7.5 embryos, data distribution in the PS is compared to data distribution in the EEM. In E9.5 embryos, data distributions in the r4, tail bud and limb bud is compared to the distribution in r1/r2 control tissue.
Results
The Hoxb1/LacZ transgene recapitulates endogenous Hoxb1 nuclear reorganisation in E7.5 embryos
The earliest events of nuclear reorganisation at Hoxb have been seen in the PS of E7.5 embryos, where Hoxb1 expression is accompanied by a looping out of the gene from its CT (Chambeyron et al., 2005). To determine if a similar nuclear movement also occurs upon activation of the Hoxb1/LacZ transgene relocated within Hoxd, we performed 3D DNA-FISH on sagittal sections of E7.5 transgenic embryos at late streak-early bud and neural plate stages when Hoxb1 is expressed (Fig. 1B). Nuclear distances between a Hoxb1/LacZ (nr33) probe and the edge of the mouse chromosome 2 (MMU2) CT, where Hoxd is located, were determined as previously described (Chambeyron et al., 2005; Chambeyron and Bickmore, 2004; Morey et al., 2007). In this analysis, a probe signal is considered to be outside the CT when the distance to the CT edge is negative. Distances > 0 correspond to probe signals inside the CT.
As a control, we analysed nuclei from anterior extra-embryonic yolk sac mesoderm (EEM) that does not express either Hoxb1 or any Hoxd genes (Table 1). In this tissue, hybridisation signals from the transgene were mainly located at the edge of the CT. In contrast, in nuclei from the PS, where endogenous and transgenic Hoxb1 are expressed, the transgene is more frequently relocated towards the outside of the CT (p=0.03 compared to EEM) as is endogenous Hoxb1 on mouse chromosome 11 (MMU11) in embryos wild-type (wt) for Hoxd (Chambeyron et al., 2005) (Figs. 1C, 1D and Table 2). However, the extent of localisation outside of the CT is less for the transgene than for endogenous Hoxb1 (Fig. 1D). This suggests that the Hoxb1/LacZ transgene contains the cis-regulatory elements necessary to initiate some of its autonomous nuclear reorganisation during gastrulation.
Looping out is restricted to the transgene itself
The looping out from CTs, initiated towards the 3' ends of endogenous Hoxb and Hoxd, spreads to adjacent genomic regions (Morey et al., 2007). To determine if this is also the case for the ectopic looping out induced by the Hoxb1lacZ transgene, we measured the nuclear position of FISH signals from probes Lnp (BAC RP24-267L11) to the 5' side of the transgene and Hoxd (BAC RP23-15M17) in the 3' direction (Fig. 1A). In E7.5 transgenic embryos the Lnp region localised well inside of the MMU2 CT, both in the EEM and the PS regions (Fig. 2A and Table 2). The 3' flanking Hoxd region was also located within, or at the edge of, the CT (Fig. 2B). No significant differences in the nuclear behaviour of Lnp or Hoxd could be detected between wt and transgenic embryos (p>0.05 for all four distributions). Therefore, Hoxb1/LacZ looping out from the CT is a localised event and does not propagate to flanking genomic regions.
Figure 2.
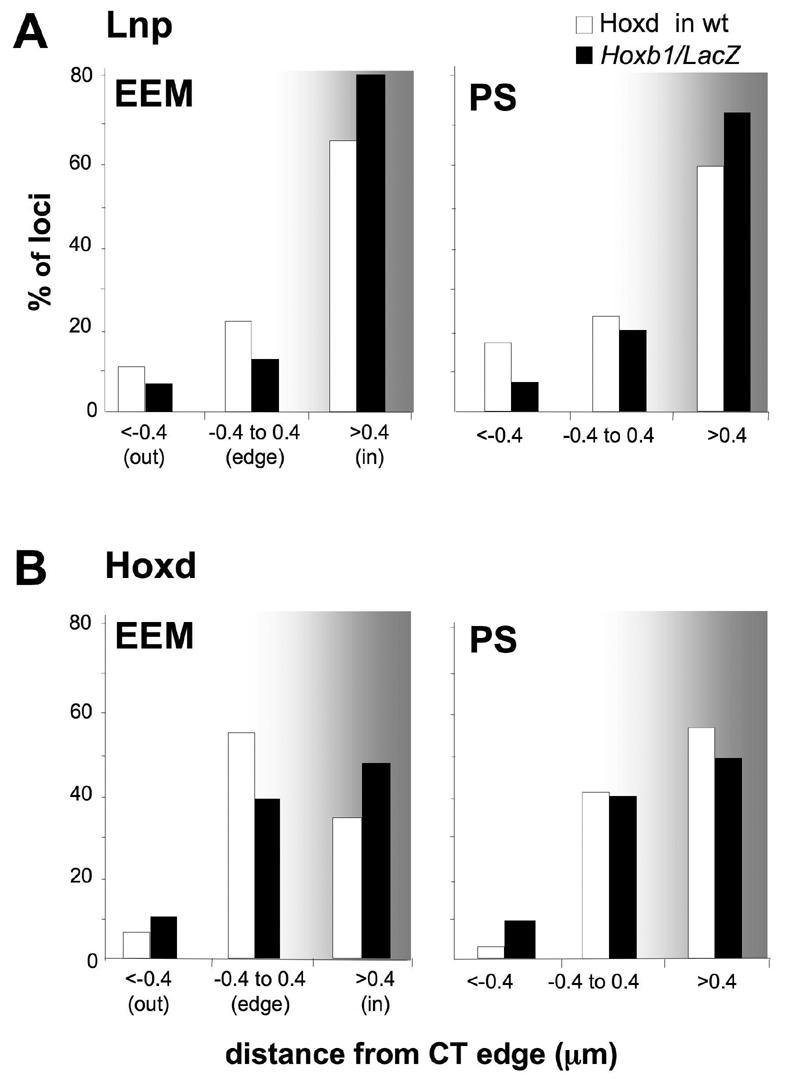
Hoxb1/LacZ nuclear reorganisation at E7.5 does not spread to flanking regions. Histograms showing the 3D positions of hybridisation signals for A) the Lnp and B) the Hoxd probe in nuclei from the EEM and the PS of wt (endog. Hoxd, white bars) or transgenic (black bars) E7.5 embryo.
The transgene induces chromatin decondensation in the primitive streak
Looping out of endogenous Hoxb1 in the PS is also accompanied by visible chromatin decondensation (Chambeyron et al., 2005). However, these two facets of nuclear organisation can be dissociated from one another, at least at Hoxd (Morey et al., 2007). Changes in chromatin condensation can be assessed by measuring the interprobe distance (d) between the signals from two FISH probes of known genomic separation (Yokota et al., 1997) and we have previously shown that this method can be used to detect chromatin decondensation at Hox loci in mouse embryos (Chambeyron et al., 2005; Morey et al., 2007). To analyse chromatin condensation around the transgene we measured the interphase distance in E7.5 transgenic embryos between FISH signals for Hoxb1/LacZ and for probes either 5' (Lnp) or 3' (Hoxd) of the transgene. The distribution of these distances was compared to those obtained at endogenous Hoxb (Chambeyron et al., 2005) and Hoxd clusters in wt E7.5 embryos (Fig. 3A). Interphase distances measured in this way follow a Rayleigh distribution, and there is generally considered to be a linear relationship between the mean-squared interphase distances (d2 in μm2) and genomic separation for probes that are <∼1Mb apart (Chambeyron and Bickmore, 2004; Yokota et al., 1995). Therefore, to normalise for differences in the genomic separations being compared between Hoxb and Hoxd, and between the wt and transgenic Hoxd loci, we divided the d2 by the genomic separation (in Mb) of the probe pairs being analysed (Fig. 3B). In EEM, the distribution of interphase distances, and so the level of chromatin condensation, 5' of Hoxb1/LacZ is similar to that at endogenous Hoxb and Hoxd (p=0.095 and p=0.173 respectively). The region 3' of Hoxb1/LacZ also showed a not dissimilar chromatin condensation level as Hoxd (p=0.052) and was significantly more condensed than endogenous Hoxb in this tissue (p=0.005) with an absence of alleles separated by large interphase distances (d2/Mb >2.25) (Fig. 3B). This suggests that the chromatin condensation in non-expressing tissue is determined by the genomic context of the transgene. In contrast, in PS cells there was significant shift in interphase separation to larger values, indicative of chromatin decondensation both 5' and 3' of Hoxb1/LacZ, as compared to the situation in the EEM or to the endogenous Hoxd region in wt PS (Fig. 3B and Table 2). However, decondensation did not reach the levels seen at endogenous Hoxb (p<10−3 for both 5' and 3' Hoxb1/LacZ). Therefore other elements from the Hoxb locus, absent from the transgene, may be necessary to recapitulate full levels of chromatin decondensation. Nevertheless, these results suggest that the Hoxb1/LacZ transgene contains the elements that can mediate some long-range decondensation at Hoxd.
Figure 3.
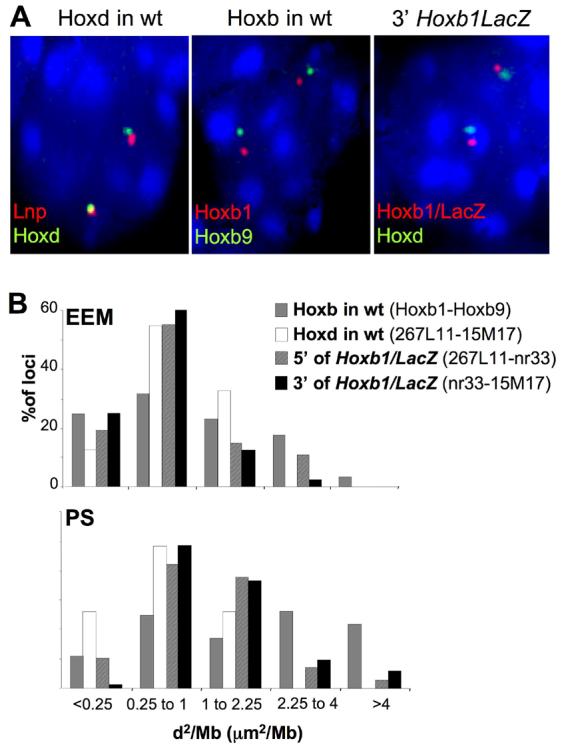
Chromatin decondensation in E7.5 transgenic embryos. (A) 3D DNA-FISH of, left panel; Lnp and Hoxd, middle panel; Hoxb1 and Hoxb9 probes on nuclei from the PS of E7.5 wild-type embryos, and of, right panel; Hoxb1/LacZ and Hoxd probes on nuclei from the PS of an E7.5 Hoxb1/LacZ embryo. Nuclei were counterstained with DAPI. Images are maximal projections of 3D stacks after deconvolution. (B) Distributions of mean-squared interphase distances (d2) in μm2 normalised to genomic separation (in Mb) measured; between the Hoxb1 and Hoxb9 probes (grey bars) at endogenous Hoxb and between Lnp and Hoxd probes (white bars) at endogenous Hoxd in EEM and PS nuclei from E7.5 wt embryos, or between the Lnp and Hoxb1/LacZ probes (hatched bars) and between the Hoxb1/LacZ and Hoxd probes (black bars) in EEM and PS nuclei from E7.5 Hoxb1/LacZ embryos.
Ectopic nuclear reorganisation of the transgene in the limb bud
Data in Figs. 1, 2 and 3 suggest that the Hoxb1/lacZ transgene has a dominant effect on chromosome organisation and decondensation at the integrated Hoxd locus in the E7.5 embryo. We have previously shown that changes in nuclear organisation at Hoxd later on in development (E9.5) are dependent on the developmental context (Morey et al., 2007). In particular, 3' Hoxd genes are first induced in the emerging limb bud of E9.0 embryos, Hoxd13 expression is detected in the limb after E10.0 (Tarchini and Duboule, 2006) and Hoxb1 is never expressed in the limb (Table 1). Activation of Hoxd genes in E9.5 forelimbs is associated with chromatin decondensation but not with significant looping out of Hoxd from its CT as compared to control non-expressing tissues of the embryo (Morey et al., 2007). Similar to the behaviour of endogenous Hoxd, in the limb bud of E9.5 transgenic embryos, we also saw significant decondensation of the regions both 5' and 3' of the transgene in the limb bud compared to control tissue from r1/r2 (Fig. 4B and Table 2). However, whereas the Lnp and Hoxd regions flanking the transgene remain well inside of the CT, as at endogenous Hoxd, the transgene significantly relocalises to the outside of the CT in the limb bud compared to its position in control cells from r1/r2 (Fig. 4A and Table 2).
Figure 4.
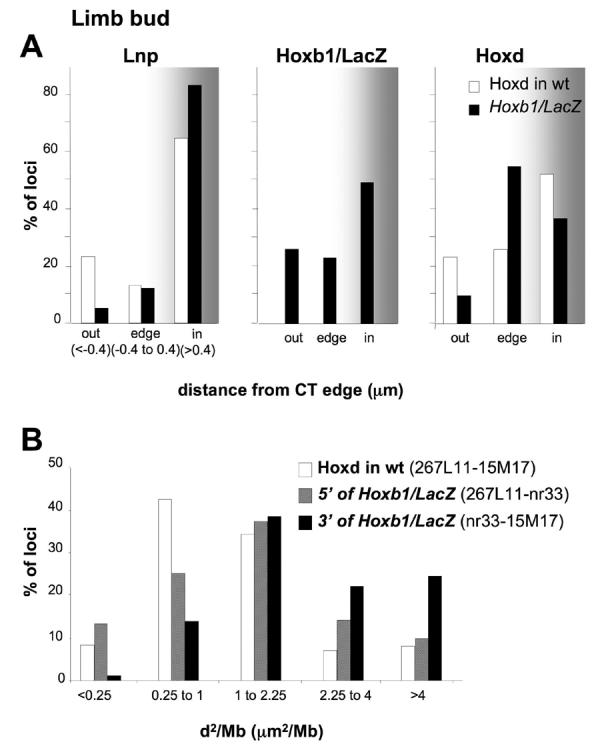
Nuclear reorganisation and chromatin decondensation in the limb bud of E9.5 Hoxb1/LacZ embryos. (A) Histograms showing the percentage of Lnp, Hoxb1/LacZ or Hoxd hybridisation signals inside, at the edge or outside of the MMU2 CT in the forelimb bud of E9.5 Hoxb1/LacZ embryos (black bars) compared to the endogenous Hoxd (white bars). The cut-offs used for the edge category are 0.4 μm and −0.4 μm from CT edge. (B) Distributions of squared interphase distances (d2) in μm2 standardised to genomic separation (in Mb) measured; between Lnp and Hoxd probes (white bars) at endogenous Hoxd in forelimb bud nuclei from E9.5 wt embryos, or between the Lnp and Hoxb1/LacZ probes (hatched bars) and between the Hoxb1/LacZ and Hoxd probes (black bars) in forelimb nuclei from E9.5 Hoxb1/LacZ embryos.
Hoxd nuclear organisation along the rostro-caudal axis in transgenic embryos
In the E7.5 embryo, and later on in the limb bud, ectopic nuclear reorganisation of the Hoxb1lacZ transgene correlates with its state of gene expression. In the first situation, transgene expression is occurring in the context of an otherwise normally silent Hoxd locus, in the second case ectopic transgene expression is induced in the activated Hoxd locus. The tailbud (E9.5) is a site in the embryo where both Hoxb and Hoxd are active and localise toward the edge/ outside of their respective CTs (Chambeyron et al., 2005; Morey et al., 2007). The transgene adopts a similar nuclear organisation (Fig. 5A and Table 2). Conversely, in rhombomeres 1 and 2 (r1/r2) of the hindbrain where neither endogenous Hoxb1, Hoxb1/LacZ, nor Hoxd genes, are expressed, the transgene was mainly located well inside (>0.4μm) of the CT, similar to the organisation of endogenous Hoxd (p=0.41), and significantly more internal in position than endogenous Hoxb1 (p<10−3). All regions were also condensed in r1/2 of both wt and transgenic embryos, and in comparison, were decondensed in the tailbud (Fig. 5C and Table 2). The levels of decondensation around the transgene were similar to those at the endogenous Hoxd in wt embryos (p=0.131 for the 5' side and p=0.076 to the 3'end).
Figure 5.
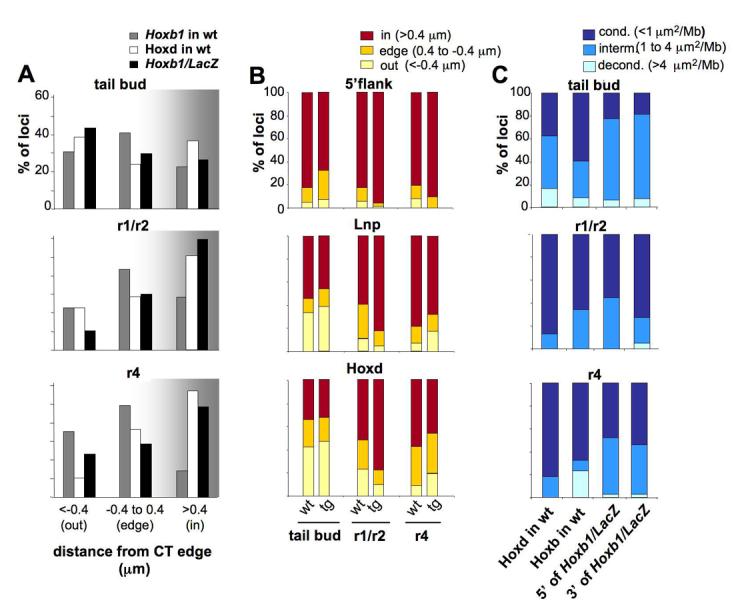
Nuclear reorganisation of the Hoxb1/LacZ transgene and surrounding regions along the antero-posterior axis of E9.5 embryos. (A) Histograms showing the percentage of Hoxb1/LacZ hybridisation signals inside, at the edge or outside of the MMU2 CT in the tailbud, r1/r2 or r4 or of E9.5 Hoxb1/LacZ embryos (black bars) compared to endogenous Hoxd (white bars) or endogenous Hoxb1 on MMU11 in wild-type E9.5 embryos (grey bars). (B) Histograms showing the percentage of signals located either inside (red bars), at the edge (orange bars) or outside (yellow bars) of the CT for the 5'flanking, the Lnp and the Hoxd probes in tailbud, r1/r2 or r4 of either wild-type (wt) or Hoxb1/LacZ transgenic (tg) embryos. (C) Percentages of nuclei showing condensed (dark blue), intermediate (blue) or decondensed (light blue) chromatin structures for the regions between Lnp and Hoxd probe signals (Hoxd wt) and between Hoxb1 and Hoxb9 probe signals (Hoxb wt) in tailbud, r1/r2 or r4 of wild-type embryos or between Lnp and Hoxb1/LacZ probe signals (5'Hoxb1/LacZ) and between Hoxb1/LacZ and Hoxd probe signals (3'Hoxb1/LacZ) in Hoxb1/LacZ embryos.
An interesting situation arises in r4. The Hoxb1/LacZ transgene, when randomly inserted in the genome, is able to recapitulate endogenous Hoxb1 expression in r4 (Marshall et al., 1994), but when transposed into 5'Hoxd, transgene expression in r4 is abolished (Kmita et al., 2000). We analysed the nuclear organisation, along the rostro-caudal axis of transgenic embryos, of the Lnp and Hoxd regions, and a genomic region extending further 5' of Lnp (5'flank; RP23-288B11, Fig. 1A) which did not show any nuclear movement in any of the tissues analysed so far in wt embryos (Morey et al., 2007). As expected, no CT looping out was observed with any of the regions in r1/ r2 (Fig. 5B), whereas there was a relocation of the Lnp and Hoxd regions, but not of the 5'flank region, to the outside of the CT in the tailbud (Fig. 5B and Table 2). However, in r4, where neither the Hoxd genes nor the transgene are expressed, a modest, but significant, relocalisation toward the edge of the CT of both Lnp and Hoxd regions was detected as compared to r1/r2 (Fig. 5B and Table 2). The transgene itself also appears to have a position less internal to the CT than it does in r1/2, however this does not reach statistical significance (p=0.28, Table 2). The nuclear position of the transgene in r4 is quite different from the behaviour of endogenous Hoxb1 (p<10−3), which extensively localises outside of its CT in this part of the hindbrain (Chambeyron et al., 2005) (Fig. 5A).
There was also some decondensation of the 3' Hoxb1/LacZ flanking region, but not of the 5' flanking region, in r4 compared to that in r1/r2 (Fig. 5C and Table 2). These results suggest that, even though not expressed, the Hoxb1/LacZ transgene exerts an influence on the nuclear organisation and chromatin decondensation of adjacent genomic regions of the Hoxd locus in r4, though this affect is much attenuated compared to the events seen at endogenous Hoxb1.
Discussion
In this paper, we have shown the contrasting behaviours of a Hoxb1/lacZ transgene, which has been transposed into the 5' end of the Hoxd locus, at different embryonic stages and in different tissues. In E7.5, the transgene is able to recapitulate some of the nuclear reorganisation and chromatin condensation seen at the endogenous Hoxb1 locus (Figs. 1 and 3). This indicates that the transgene contains the cis-acting elements needed to initiate these changes in higher-order chromatin structure during gastrulation, even though it is embedded in a Hoxd locus that is normally still silent at this stage. Whether the elements in the transgene responsible for chromatin decondensation are the same as those involved in CT reorganisation remains to be determined. The observation that looping out from the CT is restricted to the vicinity of the transgene (Fig. 2), but that transgene-induced chromatin decondensation spreads to adjacent genomic regions (Fig. 3), further serves to illustrate the fact that these two facets of nuclear reorganisation can be independent of one another (Morey et al., 2007). Interestingly, ectopic gene expression in the PS of the transgenic embryos is limited to the Hoxd13 gene immediately adjacent to the transgene (Kmita et al., 2000), suggesting that this may be a consequence of the nuclear repositioning of Hoxb1/LacZ.
We have previously shown that the normal Hoxd region, when activated in the E9.5 limb bud, decondenses but does not loop out from its CT (Morey et al., 2007). Therefore it is surprising here to find that the Hoxb1lacZ transgene does loop out from the CT in the limb bud. These results show that the transgene does not contain any DNA elements preventing its nuclear reorganisation and chromatin decondensation in tissues where Hoxb1 is normally silent, and that the absence of Hoxd looping out from the CT in the limb bud is not, as we had previously suggested (Morey et al., 2007) just due to the pathway activating Hoxd expression in this tissue, but that it is also dependent on the sequence of the Hoxd locus itself.
So far these experiments have shown that the nuclear behaviour of the transposed transgene cannot be dissociated from its pattern of gene expression. In the r4 hindbrain region of E9.5 embryos, although the extensive looping out and chromatin decondensation seen at endogenous Hoxb1 are attenuated at the transgene, coincident with the suppression of its expression here, we noticed a discrete effect of the transgene on adjacent Hoxd regions, which are relocated significantly more toward the outside of the CT, and are more decondensed, in r4 of transgenic embryos than are these regions in wild-type embryos (Fig. 5 and Table 2). Interestingly, this effect was more pronounced toward the 3' side of the transgene where the early mesoderm enhancer and the Hoxd cluster are located (Fig. 1A). This result suggests that the transgene, which does contain the regulatory elements necessary for its autonomous expression in r4 when randomly integrated in the genome, including the rhombomere 4 (r4) autoregulatory enhancer (Graaff et al., 2003; Marshall et al., 1994; Popperl et al., 1995; Studer et al., 1994), contains the elements that can initiate some degree of nuclear reorganisation at 5' Hoxd in r4 even in the absence of expression, and that it is likely that the chromatin environment of the rest of Hoxd in this tissue prevents subsequent events that are necessary for gene expression per se.
The nuclear re-organisation at the transposed Hoxd locus in r4 therefore suggests that chromatin decondensation, and looping out from the CT, can occur upstream of gene expression and so are unlikely to just be passive consequences of, for example, RNA polymerase II activity (though we cannot exclude that there may be non-coding transcription from the transgene). Similarly, a recent study has shown that decondensation and looping out from the CT of the human major histocompatibility complex, stimulated by interferon-γ via the JAK/STAT signalling pathway, also occurs upstream of transcriptional activity (Christova et al., 2007). Interestingly, given the presence of RAREs on the Hox1/LacZ transgene described here, this was correlated with the recruitment of the chromatin remodelling enzyme Brg1. Brg1 has been shown to interact with nuclear hormone receptors (Fryer and Archer, 1998). A better understanding of the molecular mechanisms driving nuclear reorganisation and chromatin decondensation of the Hoxb and d loci at different stages of development, are now required.
Acknowledgments
C.M. was an EMBO long-term fellow and a recipient of a Marie Curie Intra European Fellowship (MEIF-CT-2006-021308); W.A.B. is a Centennial fellow of the James S. McDonnell foundation. We thank Kirstie Lawson (MRC HGU) for her expert advice on mouse embryogenesis. The work was supported by the UK Medical Research Council and in part by the EU FP6 Network of Excellence Epigenome (LSH-CT-2004-503433).
Reference List
- Abranches R, Beven AF, ragon-Alcaide L, Shaw PJ. Transcription sites are not correlated with chromosome territories in wheat nuclei. J. Cell Biol. 1998;143:5–12. doi: 10.1083/jcb.143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco MR, Pombo A. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS. Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- Christova R, Jones T, Wu PJ, Bolzer A, Costa-Pereira AP, Watling D, Kerr IM, Sheer D. P-STAT1 mediates higher-order chromatin remodelling of the human MHC in response to IFN{gamma} J. Cell Sci. 2007;120:3262–3270. doi: 10.1242/jcs.012328. [DOI] [PubMed] [Google Scholar]
- Downs KM, Davies T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development. 1993;118:1255–1266. doi: 10.1242/dev.118.4.1255. [DOI] [PubMed] [Google Scholar]
- Fryer CJ, Archer TK. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- Goetze S, Mateos-Langerak J, Gierman HJ, de LW, Giromus O, Indemans MH, Koster J, Ondrej V, Versteeg R, van DR. The three-dimensional structure of human interphase chromosomes is related to the transcriptome map. Mol. Cell Biol. 2007;27:4475–4487. doi: 10.1128/MCB.00208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez F, Duboule D, Spitz F. Transgenic analysis of Hoxd gene regulation during digit development. Dev. Biol. 2007;306:847–859. doi: 10.1016/j.ydbio.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Gould A, Morrison A, Sproat G, White RA, Krumlauf R. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 1997;11:900–913. doi: 10.1101/gad.11.7.900. [DOI] [PubMed] [Google Scholar]
- Graaff W, Tomotsune D, Oosterveen T, Takihara Y, Koseki H, Deschamps J. Randomly inserted and targeted Hox/reporter fusions transcriptionally silenced in Polycomb mutants. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13362–13367. doi: 10.1073/pnas.2237046100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Bickmore W. The ins and outs of gene regulation and chromosome territory organisation. Curr. Opin. Cell Biol. 2007;19:311–316. doi: 10.1016/j.ceb.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301:331–333. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- Kmita M, Fraudeau N, Herault Y, Duboule D. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature. 2002;420:145–150. doi: 10.1038/nature01189. [DOI] [PubMed] [Google Scholar]
- Kmita M, van Der HF, Zakany J, Krumlauf R, Duboule D. Mechanisms of Hox gene colinearity: transposition of the anterior Hoxb1 gene into the posterior HoxD complex. Genes Dev. 2000;14:198–211. [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Duboule D. Breaking colinearity in the mouse HoxD complex. Cell. 1999;97:407–417. doi: 10.1016/s0092-8674(00)80749-7. [DOI] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Bickmore WA. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 2002;159:753–763. doi: 10.1083/jcb.200207115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall H, Studer M, Popperl H, Aparicio S, Kuroiwa A, Brenner S, Krumlauf R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994;370:567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- Roelen BA, de Graaff W, Forlani S, Deschamps J. Hox cluster polarity in early transcriptional availability: a high order regulatory level of clustered Hox genes in the mouse. Mech. Dev. 2002;119:81–90. doi: 10.1016/s0925-4773(02)00329-5. [DOI] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Spitz F, Herkenne C, Morris MA, Duboule D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat. Genet. 2005;37:889–893. doi: 10.1038/ng1597. [DOI] [PubMed] [Google Scholar]
- Studer M, Popperl H, Marshall H, Kuroiwa A, Krumlauf R. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science. 1994;265:1728–1732. doi: 10.1126/science.7916164. [DOI] [PubMed] [Google Scholar]
- Tarchini B, Duboule D. Control of Hoxd genes' collinearity during early limb development. Dev. Cell. 2006;10:93–103. doi: 10.1016/j.devcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- van Der HF, Zakany J, Duboule D. Gene transpositions in the HoxD complex reveal a hierarchy of regulatory controls. Cell. 1996;85:1025–1035. doi: 10.1016/s0092-8674(00)81303-3. [DOI] [PubMed] [Google Scholar]
- Verschure PJ, van DK,I, Manders EM, van DR. Spatial relationship between transcription sites and chromosome territories. J. Cell Biol. 1999;147:13–24. doi: 10.1083/jcb.147.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 2000;113(Pt 9):1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- Williams RR, Broad S, Sheer D, Ragoussis J. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 2002;272:163–175. doi: 10.1006/excr.2001.5400. [DOI] [PubMed] [Google Scholar]
- Yokota H, Singer MJ, van den Engh GJ, Trask BJ. Regional differences in the compaction of chromatin in human G0/G1 interphase nuclei. Chromosome. Res. 1997;5:157–166. doi: 10.1023/a:1018438729203. [DOI] [PubMed] [Google Scholar]
- Yokota H, van den EG, Hearst JE, Sachs RK, Trask BJ. Evidence for the organization of chromatin in megabase pair-sized loops arranged along a random walk path in the human G0/G1 interphase nucleus. J. Cell Biol. 1995;130:1239–1249. doi: 10.1083/jcb.130.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakany J, Kmita M, Duboule D. A dual role for Hox genes in limb anterior-posterior asymmetry. Science. 2004;304:1669–1672. doi: 10.1126/science.1096049. [DOI] [PubMed] [Google Scholar]


