Abstract
Rationale: Microarray technology is widely employed for studying the molecular mechanisms underlying complex diseases. However, analyses of individual diseases or models of diseases frequently yield extensive lists of differentially expressed genes with uncertain relationships to disease pathogenesis.
Objectives: To compare gene expression changes in a heterogeneous set of lung disease models in order to identify common gene expression changes seen in diverse forms of lung pathology, as well as relatively small subsets of genes likely to be involved in specific pathophysiological processes.
Methods: We profiled lung gene expression in 12 mouse models of infection, allergy, and lung injury. A linear model was used to estimate transcript expression changes for each model, and hierarchical clustering was used to compare expression patterns between models. Selected expression changes were verified by quantitative polymerase chain reaction.
Measurements and Main Results: A total of 24 transcripts, including many involved in inflammation and immune activation, were differentially expressed in a substantial majority (9 or more) of the models. Expression patterns distinguished three groups of models: (1) bacterial infection (n = 5), with changes in 89 transcripts, including many related to nuclear factor-κB signaling, cytokines, chemokines, and their receptors; (2) bleomycin-induced diseases (n = 2), with changes in 53 transcripts, including many related to matrix remodeling and Wnt signaling; and (3) T helper cell type 2 (allergic) inflammation (n = 5), with changes in 26 transcripts, including many encoding epithelial secreted molecules, ion channels, and transporters.
Conclusions: This multimodel dataset highlights novel genes likely involved in various pathophysiological processes and will be a valuable resource for the investigation of molecular mechanisms underlying lung disease pathogenesis.
Keywords: gene expression, infection, asthma, fibrosis
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
Gene expression profiling by microarray technology has become a widely used strategy for investigating the molecular mechanisms underlying many complex human diseases.
What This Study Adds to the Field
Gene expression profiling of a large set of diverse mouse lung disease models can be used to identify relatively small sets of genes that are associated with specific types of lung disease.
Gene expression profiling by microarray technology has become a widely used strategy for investigating the molecular mechanisms underlying many complex human diseases. Lung diseases that have been studied using microarrays include idiopathic pulmonary fibrosis (1, 2), hypersensitivity pneumonitis (2), nonspecific interstitial pneumonia (2), chronic obstructive pulmonary disease (3–5), primary pulmonary hypertension (6), asthma (7, 8), and others. These studies have resulted in many novel observations, and are a promising step toward development of individually targeted therapies (9). However, microarray studies of human disease have important limitations, including the heterogeneity of human populations and diseases, the inability to manipulate and examine specific pathways in humans, and the limited ability to safely acquire samples from affected tissues.
To overcome some limitations of human studies, many investigators have used microarrays to study animal models. Arrays have been used to study many models, including influenza infection (10), acute lung injury (11–13), bronchopulmonary dysplasia (14), idiopathic pulmonary fibrosis (15), bleomycin-induced lung injury (16), and asthma (17–19). These studies have their own limitations, as animal models typically exhibit some features of a disease but not others, and sometimes even features that are not components of the human disease being modeled. The search for relevant gene expression changes is further challenged by the identification of very large and unwieldy datasets of differentially expressed genes associated with these animal studies, which result, in part, from the use of genetically homogeneous populations and standardized approaches for inducing disease. These studies typically involve a single animal model, or, occasionally, a small number of models of the same disease. The use of genetically homogeneous populations and standardized approaches for inducing disease and collecting tissue for analysis facilitates the identification of large numbers of differentially expressed genes. In some cases, these approaches have identified individual genes (20) or small sets of genes (19) of special interest. More often, these studies have generated long lists of common gene expression changes (21, 22), making it difficult to determine which gene expression changes are important for key pathologic features of the disease being modeled, and also how various models relate to one another. In the present study, we hypothesized that relationships between lung gene expression and molecular mechanisms underlying pulmonary disease pathogenesis could be elucidated by comparing gene expression changes seen in a wide variety of models of lung pathology. To this end, we used a novel microarray-based approach using a heterogeneous set of models of various lung diseases, including bacterial infection, fibrosis, and allergic inflammation. By using a consistent approach to study each of these models, and by including a wide variety of models rather than one or two models of a single lung disease, we identified small sets of signature gene expression profiles associated with various forms of lung disease. Many of the genes identified here have not previously been associated with these lung diseases. Some of the results of these studies have been previously reported in the form of an abstract (23).
METHODS
Mouse Models of Lung Disease
Animal experiments were performed at the University of California, San Francisco, Vanderbilt University, and the University of Texas, Houston, and were approved by the institutional animal care and use committees at these institutions. Table 1 provides a summary of the 12 mouse models of lung disease that were used to assemble the dataset, with references to the published protocol descriptions and phenotypes of each. Microarray data from 2 of the 12 models (IL-13 and ovalbumin allergy in BALB/c mice) have been previously reported as part of a study focusing on the role of IL-13 in allergic airway disease (19), and were reanalyzed for inclusion in the large dataset reported here. Data from the other 10 of these 12 models have not been reported previously. In general, models were analyzed at a single time point, as specified in Table 1. These time points were chosen based upon previous studies and represent time points commonly used by investigators employing the models for analysis of pathophysiology. The one exception was that we studied mice at both 7 and 21 days after bleomycin injury, as earlier time points have been considered as models of acute lung injury, whereas later time points are used as models of the pulmonary fibrosis model. For each model, experimental groups were studied, along with control groups that were matched for genetic background, age, and sex.
TABLE 1.
MOUSE MODELS OF LUNG DISEASE
| Model | Description | Phenotype | Reference(s) |
|---|---|---|---|
| Aspergillus extract | C57BL/6 mice were sensitized via intranasal administration of Aspergillus fumigatus antigen mixture (n = 4) or saline alone (n = 3) 5 times at 4-d intervals and killed 4 h after the final challenge. | Th2 cytokine production, eosinophilic inflammation, airway hyperreactivity, goblet cell hyperplasia, elevated serum IgE, airway fibrosis | 52 |
| Bleomycin-induced acute lung injury | A single dose of bleomycin (1.1 U/kg body weight in 60 μl saline; n = 5) or saline alone (n = 4) was administered intratracheally to C57BL/6 mice. Mice were killed at 7 d. For bleomycin; n = 5 for both 7 and 21 d; for controls; n = 4 for 7 d and n = 2 for 21 d. | Inflammation (neutrophils, lymphocytes, macrophages), pulmonary edema | 29 |
| Bleomycin-induced pulmonary fibrosis | A single dose of bleomycin (1.1 U/kg body weight in 60 μl saline; n = 5) or saline alone (n = 2) was administered intratracheally to C57BL/6 mice. Mice were killed at 21 d. | Pulmonary fibrosis | 28 |
| IL-13 overexpression | Transgenic mice expressing IL-13 under the control of a Clara cell–specific promoter (n = 5) were studied at 6–8 wk of age. IL-13– overexpressing mice that lacked STAT6, a signaling molecule that is required for IL-13–induced pathology, were used as control animals (n = 4). | Eosinophilic inflammation, airway hyperreactivity, goblet cell hyperplasia, fibrosis, emphysema | 19, 49 |
| LPS aerosolized | E. coli LPS (7 ml of 1.0 mg/ml solution of LPS; n = 2) was delivered to C57BL/6 mice as a continuous aerosol (driving flow rate of 8 L/min) generated by a small-volume nebulizer over 30 min. Mice were killed 4 h after exposure. Controls were the LPS i.p. control mice, as described above. | Neutrophilic inflammation in lungs | (53) |
| LPS i.p. | E. coli LPS (5 μg/g BW; n = 3) or saline alone (n = 3) was administered to C57BL/6 mice via a single i.p. injection. Mice were killed 4 h after injection. | Systemic inflammation | 54 |
| M. pulmonis | C3H mice were inoculated intranasally with M. pulmonis (strain CT7; 105 cfu; n = 5) or PBS alone (n = 4). Mice were killed after 7 d. | Chronic inflammation, pulmonary edema, marked airway remodeling | 55 |
| M. tuberculosis | C57BL/6 mice were exposed to aerosols (5 ml suspension) containing M. tuberculosis (H37RV; 470 cfu; n = 3) or sterile DI water alone (n = 3) for 40 min and analyzed 21 d later. | Macrophage inflammation | 56 |
| N. brasiliensis | BALB/c mice were injected with 500 third-stage N. brasiliensis larvae (n = 3) or saline alone (n = 3) at the base of the tail and maintained on water supplemented with antibiotics (2 g/L/neomycin sulfate, 100 mg/L chloramphenicol) for 5 d before being killed. | Th2 cytokine production, eosinophil and basophil influx, goblet cell hyperplasia, airway fibrosis | 57, 58 |
| OVA allergy (C57BL/6 and BALB/c) | Mice were sensitized with OVA (50 μg in 10 mg alum, i.p.) or with alum alone (controls) on days 0, 7, and 14 and challenged with OVA (1 mg in 50 μl saline) or saline alone (controls) on days 21, 22, and 23. Mice were analyzed on Day 24. For C57BL/6 mice; n = 3 for OVA and 4 for controls; for BALB/c mice; n = 5 for OVA and controls. | Th2 cytokine production, eosinophilic inflammation, airway hyperreactivity, goblet cell hyperplasia, elevated serum IgE | 19, 49 |
| P. aeruginosa | P. aeruginosa (PA103: 5 × 107 cfu in 50 μl; n = 3) or lactated Ringer's solution alone (n = 2) was instilled intratracheally into BALB/c mice via the oropharynx. Mice were killed 2 h after treatment. | Severe alveolar epithelial cell injury, inflammation, edema formation | 59 |
Definition of abbreviations: i.p. = intraperitoneal; OVA = ovalbumin; PBS = phosphate-buffered saline.
Microarray Studies
Lung RNA from individual experimental or control mice was used to generate fluorophore-labeled cDNA, which was hybridized to 70-mer oligonucleotide microarrays, as previously described (19). Additional information about the array experiments is provided in the online supplement, and a complete description of the arrays, the experimental protocols, and the raw array data are available from the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo; accession number GSE4231). A total of 86 arrays (47 experimental samples and 39 control samples) passed quality control and were included in the main dataset.
Microarray Data Analysis
A linear model was used to estimate relative transcript expression (fold difference) for each experimental group compared with the corresponding control group, as described in the online supplement. Genes with missing values were excluded, and hierarchical clustering performed, as previously described (24).
PubMed Literature Database Mining
The PubMatrix literature mining tool (25) (http://pubmatrix.grc.nia.nih.gov/) was used to systematically determine whether genes identified in our microarray data analysis had been previously associated with lung diseases of interest. Details are provided in the online supplement.
Real-Time Polymerase Chain Reaction
Expression changes of selected genes were verified by quantitative real-time polymerase chain reaction (PCR), using either TaqMan probes or SYBR-Green for detection, as previously described (26). Gene expression differences were calculated from the mean values for experimental and control groups using the ΔΔCt (delta delta cycle threshold) method and three to four housekeeping genes. Additional details about PCR methods are provided in the online supplement.
RESULTS
Gene Transcript Expression Changes in 12 Different Lung Disease Models
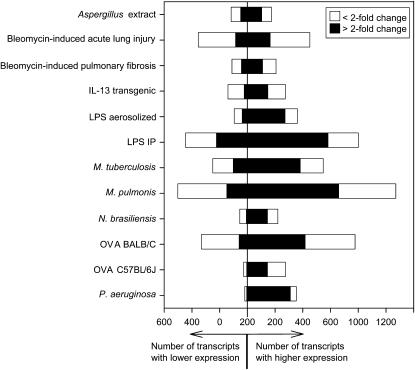
We obtained lung RNA from groups of mice representing 12 lung disease models and used DNA microarrays to analyze expression in these models. These models feature a wide range of disease-inducing stimuli, including infectious agents, toxins, allergens, and transgene overexpression that have varied effects on the lung (Table 1). RNA samples from individual mice were analyzed on separate DNA microarrays using a standardized protocol performed in a single laboratory. By using a standardized approach to sample processing, array hybridization, and data analysis, we were able to make extensive comparisons within the dataset. A final data set was assembled using results from 86 microarrays. The complete data set is publicly available from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE4231). Analyses of each of the 12 models in the dataset revealed numerous transcript expression changes in each model (Figure 1). The average number of statistically significant transcript expression changes within each of the 12 models (experimental group vs. the appropriate control group) was 663 (4% of the 16,463 transcripts represented on the arrays). On average, 460 transcripts had increased expression (including 289 that were increased more than twofold), and 203 transcripts had decreased expression (68 by more than twofold). The largest number of expression changes was seen in Mycoplasma pulmonis–infected lungs, which had 1,573 differentially expressed transcripts, including 812 changes that were twofold or greater. The smallest number of expression changes was observed in Nippostrongylus brasiliensis–infected mice (275 transcripts significantly different from control, including 157 changes that were twofold or greater).
Figure 1.
Transcript expression changes in 12 different lung disease models. Lung RNA samples from experimental and control mice representing all of the 12 models listed in Table 1 were analyzed using microarrays. A linear model was fit using results from all samples to estimate relative transcript expression (fold-difference) and determine statistical significance for each experimental group compared with its corresponding control group. The graph represents the number of significantly differentially expressed transcripts (adjusted P < 0.05) for each model. IP = intraperitoneal; OVA = ovalbumin.
Transcript Expression Changes Observed in Most or All Models
There was a small set of transcripts that had statistically significant changes in most of the 12 different models; 23 transcripts were increased in at least 9 of the 12 models examined (Table 2). Although the character of the inflammatory response seen in these 12 models is varied, many of the same transcripts associated with inflammation and immune response activation were induced in most of the models. These include chemokine ligands and receptors and proteins involved in leukocyte recruitment, homing, and phagocytosis, antigen recognition and processing, and in the acute inflammatory-phase response. Several of the genes among this set make proteins that mediate or are mediated by initial host immune responses, including early IFN-γ production (Ccl2, Ccr5, Cxcl9, Itgb2, Fcgr3, Ctss), oxidative burst (Ncf1), calcium mobilization (Ccl9, Lilrb4), and chemotaxis (Ccl2). Their up-regulation across most models suggests that these genes may play a role in the more common features of disease pathogenesis, and not the more distinct features associated with individual pathogens. Only one transcript, flavin, containing monooxygenase 3 (Fmo3), a xenobiotic-metabolizing enzyme that is normally expressed in the terminal bronchiolar epithelium of the postnatal lung (27), was decreased in all of the models. Thus, our results suggest that decreases in Fmo3 expression occur with many lung pathologies, which might lead to altered metabolism of xenobiotics, including drugs and nicotine in several lung diseases. Systematic analysis of the PubMed literature revealed that 19 of the 24 transcripts listed in Table 2 have been previously associated with lung disease(s), but 5 (21%) had no previously reported association with any of the types of lung disease represented by our models (Evi2b, Ms4a6d, 500001M20Rik, 2610307O08Rik, S100a4).
TABLE 2.
GENES THAT WERE DIFFERENTIALLY EXPRESSED IN AT LEAST 9 OF THE 12 MODELS
| Description | Symbol | Aspergillus Extract | Bleomycin- induced Acute Lung Injury (7 d) | Bleomycin- induced Fibrosis (21 d) | IL-13, Transgenic | LPS, Aerosolized | LPS, i.p. | M. pulmonis | M. tuberculosis | N. brasiliensis | OVA BALB/C | OVA C57BL/6J | P. aeruginosa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chemokines/chemotaxis | |||||||||||||
| CC chemokine ligand 2 | Ccl2 | 1.5 | 4.8* | 3.6* | 3.7* | 7.4* | 14.7* | 15.0* | 10.8* | 2.5 | 5.5* | 4.4* | 13.4* |
| C-C chemokine ligand 8 | Ccl8 | 16.9* | 3.1* | 4.8* | 2.7* | 1.0 | 0.9 | 13.4* | 8.6* | 17.8* | 25.0* | 10.3* | 0.8 |
| C-C chemokine ligand 9 | Ccl9 | 4.6* | 2.3* | 3.0* | 4.9* | 3.1* | 1.8 | 2.4* | 1.5 | 3.3* | 3.6* | 2.9* | 1.5 |
| C-C chemokine receptor 5 | Ccr5 | 1.8 | 3.2* | 3.2* | 2.5* | 2.9* | 5.4* | 14.7* | 2.3* | 1.9 | 6.3* | 4.2* | 1.4 |
| CXC chemokine ligand 9 | Cxcl9 | 1.0 | 2.0* | 2.5* | 1.1 | 2.4* | 21.0* | 17.0* | 65.3* | 2.7* | 6.8* | 4.4* | 2.4 |
| Leukocyte glycoproteins | |||||||||||||
| Glycoprotein 49A | Gp49a | 1.9 | 3.2* | 4.1* | 4.8* | 4.3* | 9.3* | 10.3* | 3.3* | 2.5 | 4.3* | 1.7 | 4.7* |
| Lymphocyte antigen 86 | Ly86 | 2.2* | 2.7* | 2.7* | 2.3* | 0.9 | 3.0* | 3.2* | 3.2* | 2.6* | 5.0* | 3.6* | 1.0 |
| Leukocyte homing | |||||||||||||
| Integrin β2 | Itgb2 | 2.9* | 2.1* | 2.0* | 3.1* | 1.2 | 1.6 | 3.1* | 3.7* | 2.4* | 2.3* | 2.2* | 1.7 |
| Phagocytosis | |||||||||||||
| C-type lectin domain protein, family 7 member a | Clec7a | 3.4* | 2.4* | 3.4* | 4.9* | 1.6 | 0.8 | 2.8* | 5.5* | 2.4* | 5.2* | 3.9* | 3.0* |
| Fc receptor, IgG, low affinity III | Fcgr3 | 2.2* | 2.4* | 3.6* | 2.1* | 1.8 | 3.1* | 4.9* | 3.9* | 2.5* | 2.2* | 2.2* | 1.9 |
| Neutrophil cytosolic factor 1 | Ncf1 | 1.6* | 1.5* | 1.6* | 1.1 | 1.5 | 1.9* | 2.8* | 2.2* | 1.3 | 2.1* | 1.6* | 2.3* |
| Leukocyte Ig-like receptor, subfamily B, member 3 | Lilrb3 | 1.8* | 1.7* | 2.6* | 1.1 | 2.3* | 3.8* | 4.8* | 3.7* | 1.8 | 2.2* | 2.0* | 2.3* |
| Monooxygenases/electron transport | |||||||||||||
| Thromboxane A synthase 1, platelet | Tbxas1 | 1.8* | 1.6* | 1.9* | 2.3* | 1.0 | 1.1 | 2.1* | 1.8* | 2.4* | 4.5* | 2.1* | 0.9 |
| Flavin containing monooxygenase 3 | Fmo3 | −2.8* | −2.3* | −1.8 | −4.3* | −4.0* | −1.8 | −3.8* | −3.6* | −2.3* | −2.4* | −1.9* | −1.4 |
| Antigen recognition/processing | |||||||||||||
| Cathepsin S | Ctss | 3.4* | 3.8* | 4.2* | 3.2* | 1.0 | 1.4 | 4.0* | 8.9* | 2.8* | 4.0* | 3.2* | 1.2 |
| Leukocyte Ig-like receptor, subfamily B, member 4 | Lilrb4 | 2.6* | 3.1* | 3.8* | 4.4* | 5.9* | 10.0* | 11.7* | 5.2* | 2.7* | 3.1* | 3.0* | 5.8* |
| Acute-phase response | |||||||||||||
| Serine (or cysteine) peptidase inhibitor, clade A, member 3N | Serpina3n | 1.7 | 3.4* | 2.3 | 2.7* | 4.2* | 6.5* | 6.0* | 2.0 | 5.6* | 3.0* | 3.2* | 4.8* |
| Serum amyloid A3 | Saa3 | 2.6 | 4.6* | 6.6* | 9.6* | 46.7* | 31.9* | 55.8* | 13.8* | 3.8 | 8.7* | 14.0* | 10.6* |
| Others | |||||||||||||
| Ecotropic viral integration site 2b | Evi2b | 1.8* | 1.5* | 1.7* | 1.3 | 1.0 | 1.0 | 2.0* | 1.9* | 1.8* | 1.9* | 1.9* | 2.4* |
| Membrane-spanning 4 domains, subfamily A, member 6D† | Ms4a6d | 2.5* | 2.6* | 1.5 | 2.6* | 1.5 | 5.9* | 6.1* | 3.5* | 2.7* | 2.4* | 2.4* | 1.5 |
| Solute carrier family 26, member 4† | Slc26a4 | 15.8* | 3.2* | 3.3 | 15.6* | 14.1* | 4.5* | 4.0* | 4.1* | 13.8* | 8.7* | 4.8* | 3.5 |
| RIKEN cDNA 1500001M20 gene† | 1500001M20Rik | 1.7 | 3.7* | 2.5* | 2.3* | 4.6* | 6.5* | 6.9* | 2.6 | 4.9* | 2.5* | 2.7* | 4.2* |
| RIKEN cDNA 2610307O08 gene† | 2610307O08Rik | 1.2 | 2.1* | 2.0* | 1.8* | 1.8 | 2.8* | 3.5* | 2.9* | 1.9* | 2.2* | 1.9* | 2.2 |
| S100 Ca-binding protein A4 | S100a4 | 2.0* | 2.4* | 3.7* | 2.2* | 1.5 | 1.5 | 2.8* | 2.6* | 2.4* | 2.9* | 3.4* | 1.2 |
Definition of abbreviations: i.p. = intraperitoneal; OVA = ovalbumin; PBS = phosphate-buffered saline; P. aeruginosa = Pseudomonas aeruginosa; Th = T helper cell.
Values represent fold change compared with controls.
Significantly different from controls by t test.
This gene was not found to be associated with asthma, allergy, bacterial infection, bleomycin, or pulmonary fibrosis in the PubMed database.
Relationships between the 12 Lung Disease Models
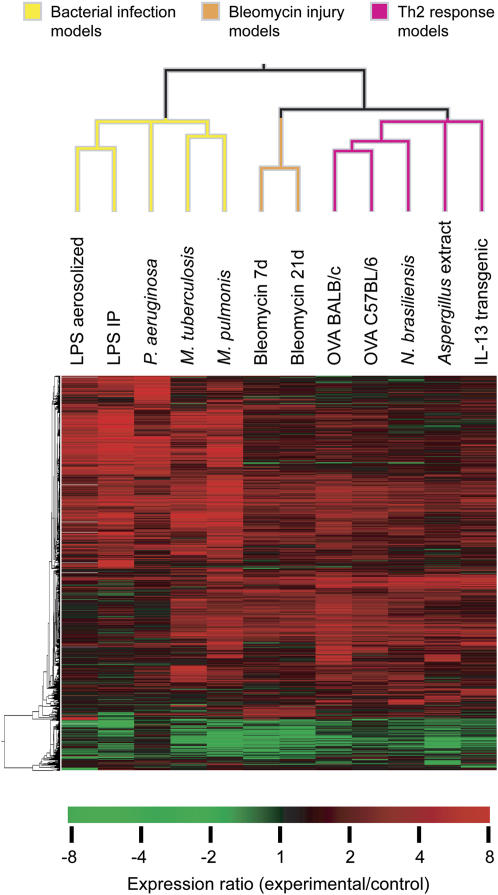
The models we studied were heterogeneous, and could be divided into groups in a number of ways. For example, groups could be defined based upon the nature of the stimulus (infectious organism, allergen, or other) or the duration of the stimulus (acute, subacute, or chronic). Rather than using a predetermined set of groupings based upon selected characteristics of the models, we instead chose to form groups based on similarities in gene expression patterns. Hierarchical clustering was used to examine the relationships between transcript expression changes in the 12 models (Figure 2). Clustering analysis of the 50 most highly differentially expressed transcripts from each model revealed 3 major clusters of expression patterns. A cluster of five models had expression patterns that were similar to one another, but quite distinct from the other seven models. The five models in that cluster were all models of bacterial infection, including three induced by infection with live bacteria (M. pulmonis, Mycobacterium tuberculosis, and Pseudomonas aeruginosa) and two triggered by administration of the bacterial toxin, LPS, either by aerosol inhalation or by intraperitoneal injection. The remaining seven models could be further subdivided into two clusters of two and five models each. The cluster with two models included models that represent different phases (7 and 21 d) of the response to intratracheal administration of the toxic chemotherapeutic agent, bleomycin. These are often used as models of acute lung injury and pulmonary fibrosis, respectively (28, 29). The final cluster of five models included all three of the allergic models studied (ovalbumin challenge in two different mouse strains and Aspergillus extract challenge), along with mice infected with the nematode parasite N. brasiliensis and mice with transgenic overexpression of the cytokine, IL-13. Increased production and activity of T helper type 2 (Th2) cytokines, including IL-13, are known to be central features of each of these five models. Although the clustering shown in Figure 2 resulted from analysis of the 50 most highly differentially expressed genes from each of the models, similar clusters were seen when fewer transcripts (25 transcripts) or more transcripts (100 or 200 transcripts) were used for clustering (data not shown), indicating that the relationships between models were reasonably robust. These clustering results suggest that there are patterns of transcript expression changes that are characteristic of the response to bacterial infection, bleomycin-induced lung injury and repair, and Th2 inflammation, and suggested that the data set could be used to identify those characteristic expression changes.
Figure 2.
Transcript expression relationships among 12 lung disease models. Hierarchical clustering was performed using the most highly differentially expressed transcripts for each model. For each model, all transcripts with statistically significant expression changes (adjusted P < 0.05) were identified. The 50 differentially expressed transcripts with the largest fold change (either increases or decreases) were selected from each model, resulting in 510 total unique transcripts, and these were used for clustering. IP = intraperitoneal; OVA = ovalbumin.
Transcript Expression Changes in Bacterial Infection Models
We used information from all 12 models to identify transcript expression changes that were seen in most or all bacterial infection models, but not in other models. A total of 90 gene transcripts were significantly increased in at least four of the five bacterial infection models, but not in any of the other seven models (Table 3 and Table E1 in the online supplement). A substantial fraction of these (27 transcripts) have a functional relationship with the nuclear factor (NF)-κB pathway, which plays a central role in innate immunity and can be triggered both by LPS (via Toll-like receptors [TLRs]) and by host cytokines, including tumor necrosis factor (TNF) and IL-1. Transcripts that were selectively increased in bacterial infection models encoded IL-1α, IL-1β, TNF, an IL-1 receptor (IL-1R2), and the receptor, CD14, which is important for LPS recognition by TLR4. Transcripts encoding MyD88, which is critical for proximal TLR signaling, and several proteins that are related to the TNF receptor–associated factors were also increased selectively in these models, as were transcripts encoding NF-κB2 and Rel, the IκB family member, Nfkbiz, and two NF-κB/IκB regulators (Bcl10 and Map3k8). Several of the transcripts that were selectively increased in bacterial infection models are regulated by NF-κB in various systems (see Table 3). Transcripts related to other cytokines, especially IFNs, were also well represented in the set. Many of the genes that were selectively induced in bacterial infection models have known roles in leukocyte recruitment and activation in various systems. These include chemoattractants, other secreted molecules, cell adhesion molecules, and other surface receptors. For example, the transcript for the chemokine, Cxcl9 (monokine induced by IFN-γ), which is a chemoattractant for Th1 lymphocytes, was increased 17- and 65-fold in lungs infected by M. pulmonis and M. tuberculosis. Only two transcripts were decreased in at least four bacterial infection models, but not in any of the other models (Table E1). These are HOXA5, a transcription factor that suppresses angiogenesis (30), and D0H4S114, which encodes P311, a protein that may regulate alveolar regeneration (31). We found that 69 of the 89 transcripts (78%) were associated with bacterial infection in the published literature. However, our systematic review of PubMed found no previous reports of associations between the remaining 20 transcripts (indicated by a “†” symbol in Table 3 and “‡” in Table E1) and bacterial infection, LPS, or pneumonia.
TABLE 3.
GENE EXPRESSION CHANGES CHARACTERISTIC OF BACTERIAL INFECTION MODELS
| Description | Symbol | LPS, Aerosolized | LPS, i.p. | P. aeruginosa | M. tuberculosis | M. pulmonis |
|---|---|---|---|---|---|---|
| NF-κB pathway–related genes | ||||||
| Ligands | ||||||
| IL-1α | Il1a | 2.5* | 1.9 | 12.3* | 2.5* | 11.5* |
| IL-1β | Il1b | 7.2* | 15.3* | 18.1* | 3.5* | 20.7* |
| TNF | Tnf | 5.3* | 5.8* | 36.9* | 7.7* | 29.7* |
| Receptors | ||||||
| CD14 | Cd14 | 10.4* | 8.2* | 5.6* | 2.9* | 12.7* |
| IL-1 receptor type II | Il1r2 | 5.7* | 5.8* | 6.1* | 1.6 | 15.7* |
| Signaling from receptors to NF-κB | ||||||
| Myeloid differentiation primary response gene 88 | Myd88 | 2.9* | 3.4* | 2.4* | 1.7 | 3.0* |
| Sphingosine kinase 1† | Sphk1 | 2.9* | 4.1* | 3.0* | 1.1 | 2.1* |
| TRAF family member–associated NF-κB activator | Tank | 1.8* | 2.6* | 1.5 | 1.7* | 1.7* |
| TRAF type zinc finger domain containing 1 | Trafd1 | 2.2* | 3.2* | 1.4 | 2.5* | 3.1* |
| Baculoviral IAP repeat-containing 2 | Birc2 | 1.8* | 2.8* | 2.1* | 1.1 | 1.6* |
| Baculoviral IAP repeat-containing 3 | Birc3 | 2.3* | 2.0* | 3.9* | 1.1 | 2.0* |
| NF-κB/Rel/IkB family members | ||||||
| NF of κ light polypeptide gene enhancer in B-cells 2, p49/p100 | Nfkb2 | 4.0* | 4.3* | 3.4* | 1.6 | 2.8* |
| NF of κ light polypeptide gene enhancer in B-cell inhibitor, zeta | Nfkbiz | 3.9* | 8.7* | 17.6* | 2.5* | 6.5* |
| Avian reticuloendotheliosis viral (v-rel) oncogene related B | Relb | 3.8* | 3.5* | 3.5* | 1.7 | 2.4* |
| Regulators of NF-κB/IκB | ||||||
| B-cell leukemia/lymphoma 10 | Bcl10 | 1.8 | 2.1* | 2.3* | 1.8* | 1.9* |
| Mitogen-activated protein kinase kinase kinase 8 | Map3k8 | 2.2* | 4.5* | 3.0* | 1.4 | 3.7* |
| Transglutaminase 2, C polypeptide | Tgm2 | 2.4* | 2.4* | 1.3 | 2.2* | 2.5* |
| Genes induced by the NF-κB pathway | ||||||
| Growth arrest and DNA damage–inducible 45β | Gadd45b | 2.6* | 2.6* | 4.5* | 1.0 | 3.3* |
| MARCKS-like 1 | Marcksl1 | 4.0* | 10.2* | 3.2* | 1.4 | 4.5* |
| Pentaxin-related gene | Ptx3 | 4.1* | 6.2* | 4.2* | 1.4 | 3.8* |
| Prostaglandin-endoperoxide synthase 2 | Ptgs2 | 1.9 | 4.6* | 14.3* | 2.5* | 3.8* |
| Radical S-adenosyl methionine domain containing 2 | Rsad2 | 6.4* | 30.1* | 1.9 | 2.9* | 5.9* |
| Superoxide dismutase 2, mitochondrial | Sod2 | 3.3* | 4.6* | 2.6* | 2.0* | 3.5* |
| Thymidylate kinase family LPS-inducible member | Tyki | 5.6* | 19.6* | 1.3 | 3.7* | 3.9* |
| TNF-α–induced protein 2 | Tnfaip2 | 8.3* | 5.5* | 5.4* | 3.7* | 4.4* |
| TNF-α–induced protein 3 | Tnfaip3 | 3.3* | 9.3* | 14.6* | 2.1* | 7.4* |
| IFN-related genes | ||||||
| IFN-α and -β receptor 2 | Ifnar2 | 1.6 | 2.4* | 2.0* | 2.1* | 1.9* |
| IFN-dependent positive-acting transcription factor 3g | Isgf3g | 2.2* | 2.9* | 1.5 | 2.0* | 2.1* |
| IFN-γ–induced GTPase | Igtp | 2.8* | 4.6* | 1.7 | 2.6* | 2.9* |
| IFN regulatory factor 1 | Irf1 | 2.9* | 4.9* | 4.9* | 3.6* | 4.2* |
| IFN-induced protein 35† | Ifi35 | 1.9* | 3.1* | 1.2 | 2.0* | 1.8* |
| IFN-induced protein with tetratricopeptide repeats 3 | Ifit3 | 3.2* | 6.1* | 1.4 | 2.1* | 2.7* |
| IL-18 receptor accessory protein | Il18rap | 1.9* | 2.3* | 3.7* | 1.6 | 3.5* |
| Ubiquitin-specific protease 18 | Usp18 | 3.4* | 9.9* | 1.2 | 2.5* | 3.4* |
| Other cytokines and receptors | ||||||
| Colony-stimulating factor 1 (macrophage) | Csf1 | 3.0* | 3.3* | 4.4* | 2.2* | 1.5 |
| Colony-stimulating factor 3 (granulocyte) | Csf3 | 3.8* | 8.6* | 7.5* | 1.6 | 4.2* |
| NF, IL-3, regulated | Nfil3 | 2.5* | 4.5* | 4.4* | 1.2 | 2.0* |
| Pre–B-cell colony–enhancing factor 1 | Pbef1 | 2.1* | 4.1* | 1.8 | 2.3* | 3.3* |
| Leukocyte adhesion, migration, and activation | ||||||
| CD80 antigen | Cd80 | 2.3* | 2.5* | 2.7* | 1.2 | 1.7* |
| Chemokine (C-C motif) ligand 20 | Ccl20 | 4.8* | 2.6* | 17.1* | 1.7 | 7.8* |
| C-type lectin domain family 4, member e | Clec4e | 1.5 | 2.1* | 4.0* | 1.6* | 9.3* |
| Killer cell lectin-like receptor, subfamily A, member 2 | Klra2 | 1.0 | 2.2* | 1.9* | 3.9* | 3.2* |
| Lectin, galactose binding, soluble 9 | Lgals9 | 2.1* | 5.2* | 1.1 | 3.5* | 2.2* |
| Matrix metallopeptidase 8 | Mmp8 | 4.0* | 11.3* | 7.1* | 1.5 | 18.0* |
| Paired Ig-like type 2 receptor α | Pilra | 2.2* | 2.0* | 2.5* | 2.6* | 3.6* |
| S100 calcium binding protein A8 (calgranulin A) | S100a8 | 8.2* | 5.7* | 5.1* | 1.7 | 18.7* |
| S100 calcium binding protein A9 (calgranulin B) | S100a9 | 15.8* | 42.0* | 8.9* | 2.3 | 19.3* |
| Selectin, lymphocyte | Sell | 3.0* | 9.0* | 3.9* | 1.2 | 4.2* |
| Sialic acid-binding Ig-like lectin E† | Siglece | 1.9* | 2.1* | 2.1* | 1.0 | 2.5* |
| Stress response signaling-related genes | ||||||
| Thioredoxin reductase 1† | Txnrd1 | 2.6* | 2.5* | 1.7* | 1.0 | 1.7* |
| Jnk signaling | ||||||
| Basic leucine zipper transcription factor, ATF-like | Batf | 2.4* | 4.8* | 2.4* | 1.8 | 6.3* |
| Dual specificity phosphatase 16 | Dusp16 | 1.9* | 3.5* | 2.4* | −1.3 | 2.2* |
| Zinc finger protein 36 | Zfp36 | 2.8* | 3.2* | 10.8* | 1.1 | 3.5* |
| p38 MAPK signaling | ||||||
| MAPK-activated protein kinase 2 | Mapkapk2 | 1.5 | 2.3* | 4.1* | 2.6* | 2.3* |
| p53 Signaling | ||||||
| Promyelocytic leukemia | Pml | 2.0* | 5.4* | 1.5 | 2.2* | 1.8* |
Definition of abbreviations: ATF = activating transcription factor; IAP = inhibitor of apoptosis protein; IκB = inhibitor of NF-κB; i.p. = intraperitoneal; MAPK = mitogen-activated protein kinase; MARCKS = myristoylated protein kinase C substrate; NF = nuclear factor; TNF = tumor necrosis factor; TRAF = TNF receptor–associated factor.
Data include selected genes that were significantly increased in at least four of the five bacterial infection models, but not in any of the other seven models. Values represent fold change compared with controls.
Significantly different from controls by t test.
This gene was not found to be associated with bacterial infection in the PubMed database.
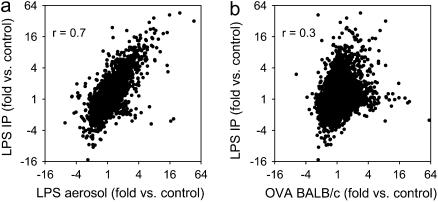
Within the set of five bacterial infection models, the two models with the most highly correlated expression pattern were the aerosolized and intraperitoneal LPS administration models (Figure 2). Comparison of expression of all transcripts represented on the arrays showed a high degree of correlation, but there were some transcripts that were highly increased in one of these models but not the other (Figure 3). Transcripts with statistically significant increases in the intraperitoneal LPS model, but not the aerosolized LPS model, included many likely to play a role in acute inflammatory responses, such as neutrophilic granule protein (23.9-fold for intraperitoneal vs. 1.2-fold for aerosolized), cathelicidin antimicrobial peptide (15.4 vs. 1.3-fold), myeloperoxidase (8.4- vs. −1.2-fold), inducible NO synthase (Nos2, 5.7- vs. 1.0-fold), E-selectin (7.9- vs. 1.9-fold), IL-6 (8.7- vs. 2.6-fold), and the chemokines Ccl3 (13.5- vs. 3.8-fold), Cxcl7 (4.4- vs. 1.2-fold), and Cxcl11 (9.0- vs. 3.3-fold). In contrast, transcripts with significant increases in the aerosolized LPS model, but not the intraperitoneal model, included several likely to be involved in the lung epithelial response to LPS, including keratin 4 (18.9-fold for aerosolized vs. −2.4-fold for intraperitoneal), keratin 13 (16.7- vs. −2.7-fold), small proline rich-like 1 (7.0- vs. −3.0-fold), and aquaporin 3 (2.1- vs. −1.2-fold).
Figure 3.
Similar pattern of gene expression changes in the intraperitoneal and aerosolized LPS models. Changes in gene expression in the intraperitoneal and aerosolized LPS models were highly correlated, although there were some genes that behaved differently in these two models (a). By comparison, expression changes in two models from different groups (intraperitoneal [IP] LPS and ovalbumin [OVA] challenge) were not highly correlated (b). Fold- difference measurements for all 16,463 transcripts represented on the arrays are shown in each plot, as are correlation coefficients (r).
Transcript Expression Changes in Bleomycin-induced Lung Disease
We used a similar approach to identify transcript expression changes that were characteristic of the bleomycin-induced acute lung injury (7 d after exposure) and bleomycin-induced pulmonary fibrosis (21 d after exposure) (32). Only 12 transcripts were significantly increased in these 2 models (7 and 21 d after bleomycin exposure), but were not significantly increased in any of the other 10 models. Because both allergic airway disease and bacterial infections can induce fibrosis, we reasoned that this approach was too restrictive. By including transcripts that were increased in both bleomycin models and in just 1 of the other 10 models, we identified 20 additional transcripts. The 32 transcripts that were increased in both bleomycin models and in at least 1 of the other 10 models are listed in Table 4. Although the transcript expression changes were all statistically significant, fold change values were mostly modest (∼1.5- to 3-fold). In fact, the list includes only 3 transcripts (elastin, pleckstrin homology-like domain family A member 3, and procollagen, type V, α2) that were among the 50 most highly up-regulated transcripts at 7 d after bleomycin, and 3 transcripts (elastin, laminin-α1, and latent transforming growth factor [TGF]-β binding protein) that were among the 50 most highly up-regulated transcripts at 21 days after bleomycin exposure. Nonetheless, previous studies make it clear that a substantial number of the 32 transcripts that were selectively increased after bleomycin exposure have functions relevant to fibrosis. These include seven transcripts that encode extracellular matrix proteins (Lamc2, Lama1, Col5a2, Col18a1, Eln, Emilin1, Fbn1), some of which have already been found to be up-regulated in pulmonary fibrosis (33, 34). Three additional transcripts are also involved in extracellular matrix formation and remodeling: Loxl2, a member of the lysyl oxidase family of matrix cross-linking enzymes that is already known to be dramatically increased after bleomycin exposure (1); Mmp2, a matrix metalloproteinase implicated in lung matrix remodeling (35); and stratifin, an inducer of the matrix metalloproteinases, Mmp1 and Mmp3. The profibrogenic cytokine TGF-β is believed to be the most important cytokine linked to the induction of pulmonary fibrosis (28). Our analysis identified one TGF-β superfamily member, growth differentiation factor 15, two TGF-β binding proteins (integrin-αv and latent TGF binding protein 2), and follistatin, an antagonist of the injury-repairing TGF-β superfamily member, activin, which blocks pulmonary fibrosis when administered to bleomycin-treated rats (36). Finally, we found that two transcripts encoding proteins in the Wnt (wingless-type MMTV integration site family) pathway (Wnt7b and Wnt1-inducible signaling pathway protein 1) were selectively up-regulated in the bleomycin models. The Wnt pathway, which regulates cell growth, differentiation, polarity, and adhesion, is key for numerous developmental processes, and has increasingly been implicated in the pathogenesis of diseases, including pulmonary fibrosis (37). The roles of other transcripts that were selectively increased in bleomycin lung injury, such as Klhdc8a, Nnat, and Psrc1, remain to be explored. We found PubMed reports of associations between pulmonary fibrosis or bleomycin and only eight (25%) of the transcripts that were selectively increased in the bleomycin models. The remaining 24 transcripts (indicated by a “†” symbol in Table 4) therefore represent novel candidates that may play roles in lung fibrosis.
TABLE 4.
GENE EXPRESSION INCREASES CHARACTERISTIC OF BLEOMYCIN-INDUCED LUNG DISEASE
| Description | Symbol | Bleomycin-induced Acute Lung Injury (7 d) | Bleomycin-induced Fibrosis (21 d) | Other Model with Increased Expression |
|---|---|---|---|---|
| Extracellular matrix | ||||
| Elastin | Eln | 4.8* | 4.1* | Aspergillus extract 3.0* |
| Elastin microfibril interfacer 1† | Emilin1 | 1.7* | 1.7* | LPS aerosolized 2.1* |
| Fibrillin 1 | Fbn1 | 2.3* | 1.9* | M. pulmonis 1.6* |
| Laminin-α1 | Lama1 | 1.8* | 3.1* | — |
| Laminin-γ2 | Lamc2 | 2.0* | 2.0* | — |
| Procollagen, type V, α2† | Col5a2 | 2.9* | 2.2* | M. pulmonis 2.7* |
| Procollagen, type XVIII, α1 | Col18a1 | 1.5* | 1.7* | — |
| Regulation of extracellular matrix | ||||
| Lysyl oxidase–like 2† | Loxl2 | 1.9* | 2.0* | — |
| Matrix metallopeptidase 2 | Mmp2 | 2.3* | 2.3* | Aspergillus extract 2.1* |
| Stratifin† | Sfn | 2.6* | 2.3* | M. pulmonis 2.4* |
| TGF-β | ||||
| Growth differentiation factor 15 | Gdf15 | 2.4* | 2.0* | P. aeruginosa 2.4* |
| Integrin-αV | Itgav | 1.5* | 1.7* | LPS aerosolized 1.9* |
| Latent TGF-β binding protein 2† | Ltbp2 | 1.8* | 3.1* | Aspergillus extract 2.2* |
| Regulation of TGF-β | ||||
| Follistatin | Fst | 2.4* | 2.3* | — |
| Wnt signaling | ||||
| Wnt1-inducible signaling pathway protein 1† | Wisp1 | 1.9* | 1.9* | — |
| Wingless-related MMTV integration site 7B† | Wnt7b | 2.1* | 1.6* | — |
| Development/morphogenesis/cell cyle/differentiation | ||||
| Carboxypeptidase X1 (M14 family)† | Cpxm1 | 1.9* | 2.1* | OVA BALB/c 2.5* |
| Cyclin G1 | Ccng1 | 2.7* | 2.4* | — |
| Integral membrane protein 2A† | Itm2a | 2.2* | 2.5* | Aspergillus extract 1.9* |
| Keratin 18† | Krt18 | 1.8* | 2.1* | M. pulmonis 2.1* |
| MAP/microtubule affinity–regulating kinase 1† | Mark1 | 1.4* | 1.9* | — |
| Pleckstrin homology-like domain, family A, member 3† | Phlda3 | 3.4* | 2.2* | — |
| Transcription factor AP-2, α† | Tcfap2a | 1.4* | 1.7* | — |
| Other targeting/signaling | ||||
| Aldo-keto reductase family 1, member B8† | Akr1b8 | 1.7* | 2.4* | LPS aerosolized 2.2* |
| ATPase inhibitory factor 1† | Atpif1 | 1.6* | 1.8* | OVA BALB/c 1.5* |
| Chondrolectin† | Chodl | 1.8* | 2.0* | IL-13 overexpression 1.5* |
| Lactamase, β† | Lactb | 1.9* | 1.5* | — |
| S100 Ca-binding protein A14† | S100a14 | 2.6* | 2.1* | M. pulmonis 1.8* |
| S100 Ca-binding protein A6 (calcyclin) | S100a6 | 1.7* | 2.0* | M. pulmonis 1.9* |
| Others | ||||
| Kelch domain containing 8A† | Klhdc8a | 2.3* | 2.4* | M. pulmonis 1.6* |
| Neuronatin† | Nnat | 2.3* | 1.9* | Aspergillus extract 1.9* |
| Proline/serine-rich coiled-coil 1† | Psrc1 | 2.4* | 2.4* | Aspergillus extract 1.9* |
Definition of abbreviations: AP = activator protein; MAP = microtubule-associated protein; MMTV = mouse mammary tumor virus; OVA = ovalbumin; TGF = transforming growth factor; Wnt = wingless-type MMTV integration site family.
Data include selected genes that were significantly increased in both bleomycin models and only 1 or none of the other 10 models. Values represent fold change compared with controls.
Significantly different from controls by t test.
Genes not found to be associated with bleomycin or pulmonary fibrosis in the PubMed database.
Using a similar approach to search for transcripts with decreased expression, we identified 21 transcripts that were significantly decreased in both bleomycin models and in 1 or none of the other 10 models (Table E2). The large majority (19/21, or 90%) of these transcripts were not associated with pulmonary fibrosis or bleomycin in our systematic analysis of PubMed.
The two bleomycin-induced models had surprisingly similar overall patterns of gene expression even though they analyzed gene expression at much different time points after bleomycin administration (7 d for the acute lung injury model and 21 d for the fibrosis model). Studies of additional lung injury models will be required to determine which of these expression changes are selective for bleomycin-induced injury and which are seen in other models of acute lung injury and/or pulmonary fibrosis.
Transcript Expression Changes in Th2 Inflammation Models
The third group included five models with Th2 inflammation. Within this group, the two ovalbumin allergy models (in BALB/c and C57BL/6 mice) were the most closely related (Figure 2). However, it is remarkable that other models provoked by very different stimuli—infection with the parasite N. brasiliensis, overexpression of a single Th2 cytokine (IL-13), or a more complex antigen mixture from a fungus (Aspergillus)—also had similar effects on gene expression. We identified 26 transcripts that were characteristic of the Th2 inflammation models (Table 5). These include seven transcripts encoding secreted proteins produced by epithelial cells, including the mucin glycoproteins Muc5ac and Muc5b. All but one of these seven transcripts were up-regulated in Th2 models and also in M. pneumoniae infection, which all are characterized by prominent mucus overproduction. Five molecules involved in ion transport were also increased in Th2 models. Some of the 26 transcripts have previously been implicated in the pathogenesis of allergic airway disease and asthma or in additional forms of Th2 inflammation, including acidic chitinase (Chia) (38–41), the secreted mucins (19, 42, 43), the calcium-activated chloride channel, Clca3 (44), 12,15-lipoxygenase (Alox15) (7, 45), and arginase (Arg1) (46–48). Most of the transcripts in Table 5 that encode proteins that are secreted by epithelial cells or are involved in ion transport have previously shown to be induced in response to the direct effects of IL-13 on epithelial cells (19), which is believed to be central to the pathogenesis of allergen-induced mucus production (19, 49). There were no transcripts that were decreased in at least four of the five Th2 models and in one or none of the other models. Half (13/26) of the transcripts identified in this analysis were not found to be associated with allergy or asthma in the PubMed database (Table 5).
TABLE 5.
GENE EXPRESSION CHANGES CHARACTERISTIC OF TYPE 2 T HELPER CELL MODELS
| Description | Symbol | OVA BALB/C | OVA C57BL/6J | N. brasiliensis | Aspergillus | IL-13 Transgenic | Other Model with Increased Expression |
|---|---|---|---|---|---|---|---|
| Secreted molecules | |||||||
| Anterior gradient 2 (Xenopus laevis)† | Agr2 | 3.9* | 6.1* | 2.2* | 2.5* | 4.4* | M. pulmonis 3.1* |
| Chitinase, acidic | Chia | 2.6* | 1.7 | 7.6* | 6.2* | 8.5* | — |
| Mucin 5, subtypes A and C, tracheobronchial/ gastric | Muc5ac | 13.0* | 10.9* | 5.4* | 3.8* | 4.2* | M. pulmonis 3.2* |
| Mucin 5, subtype B, tracheobronchial | Muc5b | 3.3* | 3.0* | 3.1* | 2.5* | 4.6* | M. pulmonis 11.1* |
| Regenerating islet–derived 3γ† | Reg3g | 5.5* | 3.3* | 1.7 | 2.7* | 3.7* | M. pulmonis 14.5* |
| Small proline-rich protein 2A | Sprr2a | 8.5* | 2.9 | 10.1* | 6.8* | 5.8* | M. pulmonis 5.4* |
| Trefoil factor 2 (spasmolytic protein 1) | Tff2 | 4.3* | 3.7* | 2.6* | 1.7 | 5.0* | M. pulmonis 6.4* |
| Channels/transporters | |||||||
| ATPase, Na+/K+ transporting, α3 polypeptide† | Atp1a3 | 2.7* | 1.6 | 2.9* | 3.5* | 2.2* | P. aeruginosa 2.9* |
| ATPase, Ca++ transporting, cardiac muscle, fast twitch 1† | Atp2a1 | 11.2* | 2 | 3.6* | 6.0* | 6.7* | — |
| Chloride channel, calcium activated 3 | Clca3 | 59.9* | 54.2* | 54.5* | 62.9* | 34.8* | — |
| FXYD domain-containing ion transport regulator 4† | Fxyd4 | 2.7* | 2.5 | 8.7* | 12.5* | 4.1* | — |
| Solute carrier family 5 (sodium/glucose cotransporter), member 1† | Slc5a1 | 1.6* | 1.2 | 2.5* | 2.1* | 1.9* | LPS i.p. 1.7* |
| Other genes | |||||||
| RIKEN cDNA 9130211I03 gene† | 9130211I03Rik | 2.5* | 2.1* | 1.8* | 1.8* | 1 | — |
| Arachidonate 15-lipoxygenase | Alox15 | 4.6* | 2.3* | 6.4* | 3.8* | 1 | — |
| Arginase 1, liver | Arg1 | 9.0* | 5.3* | 7.2* | 8.9* | 4.6* | M. pulmonis 9.4* |
| Eosinophil-associated, RNase A family, member 11 | Ear11 | 20.5* | 5.3* | 10.2* | 6.7* | 11.3* | — |
| Guanylate cyclase activator 1a (retina)† | Guca1a | 10.2* | 7.0* | 1.3 | 3.5* | 2.4* | M. pulmonis 3.9* |
| Heme-binding protein 2† | Hebp2 | 1.8* | 1.3 | 1.8* | 1.7* | 1.7* | LPS aerosolized 2.2* |
| Integrin-αX | Itgax | 2.2* | 2.5* | 3.3* | 2.6* | 3.7* | Bleomycin 21 d 3.4* |
| Megakaryocyte-associated tyrosine kinase† | Matk | 2.8* | 2.9* | 2.5* | 2.8* | 1.6 | — |
| Meteorin, glial cell differentiation regulator-like† | Metrnl | 2.5* | 1.8* | 1.7 | 2.1* | 1.9* | M. pulmonis 3.0* |
| Macrophage galactose N-acetyl-galactosamine specific lectin 1 | Mgl1 | 4.6* | 2.2* | 2.2* | 1.3 | 2.1* | Bleomycin 7 d 1.6* |
| NAD synthetase 1† | Nadsyn1 | 15.5* | 5.0* | 6.5* | 10.3* | 9.4* | Bleomycin 7 d 4.3* |
| Programmed cell death 1 ligand 2 | Pdcd1lg2 | 2.8* | 2.5* | 3.6* | 2.0* | 1.6 | M. pulmonis 2.2* |
| Scinderin | Scin | 1.5* | 2.2* | 1.8* | 1.9* | 2.4* | — |
| Zinc finger, CCCH-type with G patch domain† | Zgpat | 2.1* | 2.7* | 4.1* | 2.8* | 3.6* | — |
Definition of abbreviations: CCCH = cysteine-cysteine-cysteine-histidine; OVA = ovalbumin.
Genes differentially expressed in at least four of five Th2 models and one or none of the other models.
Significantly different from controls (t test).
Genes not found to be associated with asthma or allergy in the PubMed database.
Validation of Array-based Transcript Expression Measurements
To determine whether transcript expression changes measured using microarrays could be verified using a different method, we used quantitative real-time PCR to measure the expression of selected transcripts. We had sufficient material available to study three of the bacterial infection models, each of the two bleomycin models, and three of the Th2 models. In the 83 cases in which the arrays showed at least 1.5-fold higher expression of a transcript in a disease model, 75 of the PCR reactions (90%) also showed at least 1.5-fold higher expression (Table E3). Six of the eight PCR reactions that failed to show at least 1.5-fold higher expression were assays for Zgpat or Atp2a1 (each primer pair was used for the three allergic models), indicating a clear discrepancy between array and PCR results for these two transcripts. Array and PCR estimates of the magnitude of the fold difference often differed. The most dramatic differences were usually seen in cases where PCR measurements showed very large differences and arrays showed smaller differences. PCR typically has greater sensitivity and dynamic range than arrays (50, 51), and we used a conservative array analysis method (no background subtraction) that tends to underestimate fold-difference. We conclude that array findings of differential expression were very frequently confirmed by quantitative PCR, although the magnitude of the change was often underestimated by the arrays.
Conclusions
The results of the microarray analyses presented here demonstrate abundant transcript expression changes in the context of lung disease. The dataset of transcript expression profiles generated here is publicly available, and will provide a useful resource for investigators studying pulmonary disease pathogenesis. By using multiple models of varying lung disease, we identified transcript expression changes that were common to many forms of lung disease and transcripts whose expression was changed only among specific lung disease models. We described a series of characteristic transcript expression changes for the various categories of lung disease, including bacterial infection, bleomycin-induced lung disease, and Th2 (allergic) inflammation.
We chose to study a wide variety of experimental systems rather than studying a single system, an approach that has some different strengths and weaknesses than other array-based approaches. One important limitation of our approach is that we did not study most model systems at multiple time points, as it would not have been practical for us to do so with so many models. Instead, we generally chose a single time point when there is known to be established pathology, based on previous work by investigators who developed the models and use them to investigate mechanism. Therefore our approach will not detect transient gene expression changes that occur before the time point that we analyzed and may contribute to the development of pathology. A second limitation is that different types of mice were used in different models. Because models included age-, sex-, and strain-matched control animals, we were able to identify gene expression changes due to disease as opposed to those which were simply due to baseline differences between strains. Nonetheless, it is likely that genetic and environmental differences between the cohorts of mice used for the various models complicate the process of trying to identify common features between models. A third limitation is that our set of models included only two “lung injury” models, and in both cases the injury was initiated by bleomycin administration. Additional studies will be required to determine which of the bleomycin lung injury–associated gene expression changes seen in our studies are also found in other lung injury models. Like studies of lung gene expression in just one model, our approach is also limited because arrays are not sufficiently sensitive to detect some gene expression changes, and because changes in gene expression that affect relatively small populations of cells within the lung may not be detectable.
The major advantage of the multimodel approach that we used is that it provides a way to generate small lists of genes associated with related groups of disease models rather than a much longer list of genes that are differentially expressed in a single model and may or may not have an association with specific pathological features of that model. It is likely that the relatively short lists generated using our approach are highly enriched for genes that are involved in pathogenesis, based on data from previous studies involving other experimental approaches. Other genes in these lists have not previously linked to lung disease pathogenesis. Based on their consistent and relatively specific associations with specific forms of lung disease, many of these signature genes are good candidates for further study. Our systematic review of the PubMed database of published literature indicated that many of these genes have not previously been associated with the diseases that were represented in the multiple model dataset. Although our analysis approach focused largely on genes that were differentially expressed across groups of models, this kind of multiple model dataset could also be used to identify gene expression changes that are unique for specific disease models. For example, we were able to identify gene expression changes that were different between the intraperitoneal and aerosolized LPS models and give insights into how the route of exposure affects the response to this bacterial component. Some approaches to analyzing array data have relied on selection of the most highly differentially expressed transcripts for further hypothesis generation and testing, but those approaches do not necessarily lead to the identification of disease-specific gene expression changes that are likely to be good targets for therapeutic intervention. In conclusion, the present study shows that the combined analysis of multiple disease models is a powerful method for identifying transcript expression changes that are characteristic of specific pathophysiological processes, even when the degree of differential expression is relatively modest.
Supplementary Material
Acknowledgments
The authors thank Drs. Teiji Sawa and Jeanine Wiener-Kronish for generously providing samples from the Pseudomonas aeruginosa infection model. They also thank Dean Sheppard, Thiennu Vu, and Andrea Barczak for their advice, and Agnes Paquet, Michael Salazar, and the staffs of the Sandler Center Functional Genomics Core, the Sandler Animal Physiology and Morphology Core, and the University of California, San Francisco/National Heart, Lung, and Blood Institute Shared Microarray Facility for technical assistance.
Supported by National Institutes of Health grants HL56835, HL72301, and HL85089 (to D.J.E.), HL24136 and HL59157 (to D.M.), a Junior Investigator Award from the Sandler Program for Asthma Research (to M.R.B.), and the UCSF Sandler Asthma Basic Research (SABRE) Center.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200702-333OC on November 20, 2007
Conflict of Interest Statement: C.C.L. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.Y.H.Y. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. X.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.K.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.R.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. P.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.M.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. T.S.B. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. V.N. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. W.P. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.V. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.J.E. received $2,500 in 2007 for serving on an advisory board for Johnson & Johnson.
References
- 1.Kaminski N, Allard JD, Pittet JF, Zuo F, Griffiths MJ, Morris D, Huang X, Sheppard D, Heller RA. Global analysis of gene expression in pulmonary fibrosis reveals distinct programs regulating inflammation and fibrosis. Proc Natl Acad Sci USA 2000;97:1778–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selman M, Pardo A, Barrera L, Estrada A, Watson SR, Wilson K, Aziz N, Kaminski N, Zlotnik A. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. Am J Respir Crit Care Med 2006;173:188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ning W, Li CJ, Kaminski N, Feghali-Bostwick CA, Alber SM, Di YP, Otterbein SL, Song R, Hayashi S, Zhou Z, et al. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci USA 2004;101:14895–14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, Palma J, Brody JS. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA 2004;101:10143–10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Golpon HA, Coldren CD, Zamora MR, Cosgrove GP, Moore MD, Tuder RM, Geraci MW, Voelkel NF. Emphysema lung tissue gene expression profiling. Am J Respir Cell Mol Biol 2004;31:595–600. [DOI] [PubMed] [Google Scholar]
- 6.Geraci MW, Moore M, Gesell T, Yeager ME, Alger L, Golpon H, Gao B, Loyd JE, Tuder RM, Voelkel NF. Gene expression patterns in the lungs of patients with primary pulmonary hypertension: a gene microarray analysis. Circ Res 2001;88:555–562. [DOI] [PubMed] [Google Scholar]
- 7.Laprise C, Sladek R, Ponton A, Bernier MC, Hudson TJ, Laviolette M. Functional classes of bronchial mucosa genes that are differentially expressed in asthma. BMC Genomics 2004;5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lilly CM, Tateno H, Oguma T, Israel E, Sonna LA. Effects of allergen challenge on airway epithelial cell gene expression. Am J Respir Crit Care Med 2005;171:579–586. [DOI] [PubMed] [Google Scholar]
- 9.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006;439:353–357. [DOI] [PubMed] [Google Scholar]
- 10.Baskin CR, Garcia-Sastre A, Tumpey TM, Bielefeldt-Ohmann H, Carter VS, Nistal-Villan E, Katze MG. Integration of clinical data, pathology, and cDNA microarrays in influenza virus–infected pigtailed macaques (Macaca nemestrina). J Virol 2004;78:10420–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leikauf GD, McDowell SA, Bachurski CJ, Aronow BJ, Gammon K, Wesselkamper SC, Hardie W, Wiest JS, Leikauf JE, Korfhagen TR, et al. Functional genomics of oxidant-induced lung injury. Adv Exp Med Biol 2001;500:479–487. [DOI] [PubMed] [Google Scholar]
- 12.Jeyaseelan S, Chu HW, Young SK, Worthen GS. Transcriptional profiling of lipopolysaccharide-induced acute lung injury. Infect Immun 2004;72:7247–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wesselkamper SC, Case LM, Henning LN, Borchers MT, Tichelaar JW, Mason JM, Dragin N, Medvedovic M, Sartor MA, Tomlinson CR, et al. Gene expression changes during the development of acute lung injury: role of transforming growth factor β. Am J Respir Crit Care Med 2005;172:1399–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagenaar GT, ter Horst SA, van Gastelen MA, Leijser LM, Mauad T, van der Velden PA, de Heer E, Hiemstra PS, Poorthuis BJ, Walther FJ. Gene expression profile and histopathology of experimental bronchopulmonary dysplasia induced by prolonged oxidative stress. Free Radic Biol Med 2004;36:782–801. [DOI] [PubMed] [Google Scholar]
- 15.Kaminski N, Zuo F, Cojocaro G, Yakhini Z, Ben-Dor A, Morris D, Sheppard D, Pardo A, Selman M, Heller RA. Use of oligonucleotide microarrays to analyze gene expression patterns in pulmonary fibrosis reveals distinct patterns of gene expression in mice and humans. Chest 2002;121(Suppl 3):31S–32S. [PubMed] [Google Scholar]
- 16.Katsuma S, Nishi K, Tanigawara K, Ikawa H, Shiojima S, Takagaki K, Kaminishi Y, Suzuki Y, Hirasawa A, Ohgi T, et al. Molecular monitoring of bleomycin-induced pulmonary fibrosis by cDNA microarray–based gene expression profiling. Biochem Biophys Res Commun 2001;288:747–751. [DOI] [PubMed] [Google Scholar]
- 17.Zou J, Young S, Zhu F, Gheyas F, Skeans S, Wan Y, Wang L, Ding W, Billah M, McClanahan T, et al. Microarray profile of differentially expressed genes in a monkey model of allergic asthma. Genome Biology [Internet]. 2002;3:research0020.1–0020.13 [Epub: April 11, 2002]. Available from: http://genomebiology.com/2002/3/5/research/0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmermann N, King NE, Laporte J, Yang M, Mishra A, Pope SM, Muntel EE, Witte DP, Pegg AA, Foster PS, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest 2003;111:1863–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuperman DA, Lewis CC, Woodruff PG, Rodriguez MW, Yang YH, Dolganov GM, Fahy JV, Erle DJ. Dissecting asthma using focused transgenic modeling and functional genomics. J Allergy Clin Immunol 2005;116:305–311. [DOI] [PubMed] [Google Scholar]
- 20.Karp CL, Grupe A, Schadt E, Ewart SL, Keane-Moore M, Cuomo PJ, Kohl J, Wahl L, Kuperman D, Germer S, et al. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immunol 2000;1:221–226. [DOI] [PubMed] [Google Scholar]
- 21.Nikolaidis NM, Zimmermann N, King NE, Mishra A, Pope SM, Finkelman FD, Rothenberg ME. Trefoil factor-2 is an allergen-induced gene regulated by Th2 cytokines and STAT6 in the lung. Am J Respir Cell Mol Biol 2003;29:458–464. [DOI] [PubMed] [Google Scholar]
- 22.Woodruff PG, Koth LL, Yang YH, Rodriguez MW, Favoreto S, Dolganov GM, Paquet AC, Erle DJ. A distinctive alveolar macrophage activation state induced by cigarette smoking. Am J Respir Crit Care Med 2005;172:1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis CC, Hang YH, Kuperman D, Huang Y, Sheppard D, Erle DJ. Gene expression profiling in multiple mouse models of lung disease reveals signature profiles for asthma, lung infection, and fibrosis [abstract]. Am J Respir Crit Care Med 2004;169:A53.
- 24.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 1998;95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker KG, Hosack DA, Dennis G Jr, Lempicki RA, Bright TJ, Cheadle C, Engel J. PubMatrix: a tool for multiplex literature mining. BMC Bioinformatics 2003;4:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponchel F, Toomes C, Bransfield K, Leong PT, Douglas SH, Field SL, Bell SM, Combaret V, Puisieux A, Mighell AJ, et al. Real-time PCR based on SYBR-Green I fluorescence: an alternative to the Taqman assay for a relative quantification of gene rearrangements, gene amplifications, and microgene deletions. BMC Biotechnol 2003;3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janmohamed A, Hernandez D, Phillips IR, Shephard EA. Cell-, tissue-, sex- and developmental stage–specific expression of mouse flavin-containing monooxygenases (Fmos). Biochem Pharmacol 2004;68:73–83. [DOI] [PubMed] [Google Scholar]
- 28.Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J, Pittet JF, Kaminski N, Garat C, Matthay MA, et al. The integrin αvβ6 binds and activates latent TGF β1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell 1999;96:319–328. [DOI] [PubMed] [Google Scholar]
- 29.Pittet JF, Griffiths MJ, Geiser T, Kaminski N, Dalton SL, Huang X, Brown LA, Gotwals PJ, Koteliansky VE, Matthay MA, et al. TGF-β is a critical mediator of acute lung injury. J Clin Invest 2001;107:1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhoads K, Arderiu G, Charboneau A, Hansen SL, Hoffman W, Boudreau N. A role for Hox A5 in regulating angiogenesis and vascular patterning. Lymphat Res Biol 2005;3:240–252. [DOI] [PubMed] [Google Scholar]
- 31.Zhao L, Leung JK, Yamamoto H, Goswami S, Kheradmand F, Vu TH. Identification of p311 as a potential gene regulating alveolar generation. Am J Respir Cell Mol Biol 2006;35:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chua F, Gauldie J, Laurent GJ. Pulmonary fibrosis: searching for model answers. Am J Respir Cell Mol Biol 2005;33:9–13. [DOI] [PubMed] [Google Scholar]
- 33.Dolhnikoff M, Mauad T, Ludwig MS. Extracellular matrix and oscillatory mechanics of rat lung parenchyma in bleomycin-induced fibrosis. Am J Respir Crit Care Med 1999;160:1750–1757. [DOI] [PubMed] [Google Scholar]
- 34.Hoff CR, Perkins DR, Davidson JM. Elastin gene expression is upregulated during pulmonary fibrosis. Connect Tissue Res 1999;40:145–153. [DOI] [PubMed] [Google Scholar]
- 35.Corbel M, Belleguic C, Boichot E, Lagente V. Involvement of gelatinases (MMP-2 and MMP-9) in the development of airway inflammation and pulmonary fibrosis. Cell Biol Toxicol 2002;18:51–61. [DOI] [PubMed] [Google Scholar]
- 36.Aoki F, Kurabayashi M, Hasegawa Y, Kojima I. Attenuation of bleomycin-induced pulmonary fibrosis by follistatin. Am J Respir Crit Care Med 2005;172:713–720. [DOI] [PubMed] [Google Scholar]
- 37.Chilosi M, Poletti V, Zamo A, Lestani M, Piccoli P, Pedron S, Bertaso M, Scarpa A, Maestro R, Semenzato G, et al. Aberrant Wnt/β-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol 2003;162:1495–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, Cohn L, Hamid Q, Elias JA. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science 2004;304:1678–1682. [DOI] [PubMed] [Google Scholar]
- 39.Wills-Karp M, Karp CL. Chitin checking: novel insights into asthma. N Engl J Med 2004;351:1455–1457. [DOI] [PubMed] [Google Scholar]
- 40.Ramanathan M Jr, Lee WK, Lane AP. Increased expression of acidic mammalian chitinase in chronic rhinosinusitis with nasal polyps. Am J Rhinol 2006;20:330–335. [DOI] [PubMed] [Google Scholar]
- 41.Bierbaum S, Nickel R, Koch A, Lau S, Deichmann KA, Wahn U, Superti-Furga A, Heinzmann A. Polymorphisms and haplotypes of acid mammalian chitinase are associated with bronchial asthma. Am J Respir Crit Care Med 2005;172:1505–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fahy JV. Goblet cell and mucin gene abnormalities in asthma. Chest 2002;122(Suppl 6):320S–326S. [DOI] [PubMed] [Google Scholar]
- 43.Ordonez C, Khashayar R, Wong H, Ferrando R, Wu R, Hyde D, Hotchkiss J, Zhang Y, Nokikov A, Dolganov GM, et al. Mild and moderate asthma is associated with airway goblet cell hyperplasia and abnormalities in mucin gene expression. Am J Respir Crit Care Med 2001;163:517–523. [DOI] [PubMed] [Google Scholar]
- 44.Nakanishi A, Morita S, Iwashita H, Sagiya Y, Ashida Y, Shirafuji H, Fujisawa Y, Nishimura O, Fujino M. Role of gob-5 in mucus overproduction and airway hyperresponsiveness in asthma. Proc Natl Acad Sci USA 2001;98:5175–5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu HW, Balzar S, Westcott JY, Trudeau JB, Sun Y, Conrad DJ, Wenzel SE. Expression and activation of 15-lipoxygenase pathway in severe asthma: relationship to eosinophilic phenotype and collagen deposition. Clin Exp Allergy 2002;32:1558–1565. [DOI] [PubMed] [Google Scholar]
- 46.Li H, Romieu I, Sienra-Monge JJ, Ramirez-Aguilar M, Estela Del Rio-Navarro B, Kistner EO, Gjessing HK, Lara-Sanchez Idel C, Chiu GY, London SJ. Genetic polymorphisms in arginase I and II and childhood asthma and atopy. J Allergy Clin Immunol 2006;117:119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King NE, Rothenberg ME, Zimmermann N. Arginine in asthma and lung inflammation. J Nutr 2004;134(Suppl 10):2830S–2836S. [Discussion, 2853S.] [DOI] [PubMed] [Google Scholar]
- 48.Morris CR, Poljakovic M, Lavrisha L, Machado L, Kuypers FA, Morris SM Jr. Decreased arginine bioavailability and increased serum arginase activity in asthma. Am J Respir Crit Care Med 2004;170:148–153. [DOI] [PubMed] [Google Scholar]
- 49.Kuperman DA, Huang X, Koth LL, Chang GH, Dolganov GM, Zhu Z, Elias JA, Sheppard D, Erle DJ. Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat Med 2002;8:885–889. [DOI] [PubMed] [Google Scholar]
- 50.Petersen D, Chandramouli GV, Geoghegan J, Hilburn J, Paarlberg J, Kim CH, Munroe D, Gangi L, Han J, Puri R, et al. Three microarray platforms: an analysis of their concordance in profiling gene expression. BMC Genomics 2005;6:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barker CS, Griffin C, Dolganov GM, Hanspers K, Yang JY, Erle DJ. Increased DNA microarray hybridization specificity using sscDNA targets. BMC Genomics 2005;6:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corry DB, Grunig G, Hadeiba H, Kurup VP, Warnock ML, Sheppard D, Rennick DM, Locksley RM. Requirements for allergen-induced airway hyperreactivity in T and B cell–deficient mice. Mol Med 1998;4:344–355. [PMC free article] [PubMed] [Google Scholar]
- 53.Blackwell TS, Lancaster LH, Blackwell TR, Venkatakrishnan A, Christman JW. Chemotactic gradients predict neutrophilic alveolitis in endotoxin-treated rats. Am J Respir Crit Care Med 1999;159:1644–1652. [DOI] [PubMed] [Google Scholar]
- 54.Koay MA, Christman JW, Wudel LJ, Allos T, Cheng D-S, Chapman WC, Blackwell TS. Modulation of endotoxin-induced NF-κB activation in lung and liver through TNF type I and IL-1 receptors. Am J Physiol Lung Cell Mol Physiol 2002;283:L1247–L1254. [DOI] [PubMed] [Google Scholar]
- 55.Baluk P, Tammela T, Ator E, Lyubynska N, Achen MG, Hicklin DJ, Jeltsch M, Petrova TV, Pytowski B, Stacker SA, et al. Pathogenesis of persistent lymphatic vessel hyperplasia in chronic airway inflammation. J Clin Invest 2005;115:247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kelly BP, Furney SK, Jessen MT, Orme IM. Low-dose aerosol infection model for testing drugs for efficacy against Mycobacterium tuberculosis. Antimicrob Agents Chemother 1996;40:2809–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shinkai K, Mohrs M, Locksley RM. Helper T cells regulate type-2 innate immunity in vivo. Nature 2002;420:825–829. [DOI] [PubMed] [Google Scholar]
- 58.Voehringer D. Type 2 immunity reflects orchestrated recruitment of cells committed to IL-4 production. Immunity 2004;20:267–277. [DOI] [PubMed] [Google Scholar]
- 59.Sawa T, Corry DB, Gropper MA, Ohara M, Kurahashi K, Wiener-Kronish JP. IL-10 improves lung injury and survival in Pseudomonas aeruginosa pneumonia. J Immunol 1997;159:2858–2866. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.