Abstract
The prevalence and morbidity of asthma, a chronic inflammatory airway disease, is increasing. Animal models provide a meaningful but limited view of the mechanisms of asthma in humans. A systems-level view of asthma that integrates multiple levels of molecular and functional information is needed. For this, we compiled a gene expression compendium from five publicly available mouse microarray datasets and a gene knowledge base of 4,305 gene annotation sets. Using this collection we generated a high-level map of the functional themes that characterize animal models of asthma, dominated by innate and adaptive immune response. We used Module Networks analysis to identify co-regulated gene modules. The resulting modules reflect four distinct responses to treatment, including early response, general induction, repression, and IL-13–dependent response. One module with a persistent induction in response to treatment is mainly composed of genes with suggested roles in asthma, suggesting a similar role for other module members. Analysis of IL-13–dependent response using protein interaction networks highlights a role for TGF-β1 as a key regulator of asthma. Our analysis demonstrates the discovery potential of systems-level approaches and provides a framework for applying such approaches to asthma.
Keywords: house dust mite, IL-13, ovalbumin, systems biology, TGF-β
CLINICAL RELEVANCE
In this article, we provide a framework for a systems-level analysis of microarray data and create a global map of asthma. Our insights, the analytic framework, and the accompanying website should be of use to basic and clinical asthma researchers.
Asthma is a chronic lung disease characterized by airway inflammation, hyperresponsiveness, remodeling, and obstruction (1). The lung phenotype in asthma is believed to be determined by the interaction of the environment with the patient's genetic background (2). This interaction leads to a dramatic change in the airway microenvironment that includes activation of inflammatory pathways, recruitment of immune cells that are not usually present in the airway, and a dramatic change in the phenotype of airway resident cells. While individual changes in many of these factors may generate components of the asthmatic phenotype, it is the converging effects of these pathways on recruited and altered cells that determine the patient's disease.
The advent of high-throughput technologies for gene and protein profiling has greatly improved our ability to characterize the behavior of genes and proteins in health and disease. Using animal models of allergic airway disease, investigators applied DNA microarrays to identify potential regulators of asthmatic airway inflammation such as C5 (3), ARG1 (4), ADAM8 (5), SPRR2 (6) as well as to explore the pathways activated by IL13 and STAT6 during the development of allergen-induced lung inflammation (7–9). Transcriptional analysis of the response to IL-13, allergen challenge, syncytial virus infection, and corticosteroids in epithelial cells identified multiple and rarely overlapping genes (10–14). While many of these studies used elegant experimental approaches to dissect pathways and to identify and validate potential novel key regulatory molecules, the majority of their insights were obtained using statistical methods that select individual genes based on their relevance to a specific experimental setup, such as a certain knockout or allergen. These approaches, although successful, tend to reduce the complexity of the data and do not provide a global, systems-level view of the process studied. Furthermore, the dependence on gene-level analysis magnifies the impact of the noise generated by the experimental models and the different technical aspects of microarray sample preparation, hybridization, and scanning. Lastly, such methods are not capable of integrating multiple levels of information. Recently, there has been an increased interest in methods that allow a systems view of the studied process, a view that observes not only the components of a system but also their emergent properties. This includes methods that uncover the functional themes that characterize gene expression profiles (15, 16) as well as methods that use advanced computational algorithms for the integration of multiple levels of information (17–19) and identification of regulatory modules in complex tissue and in disease (20).
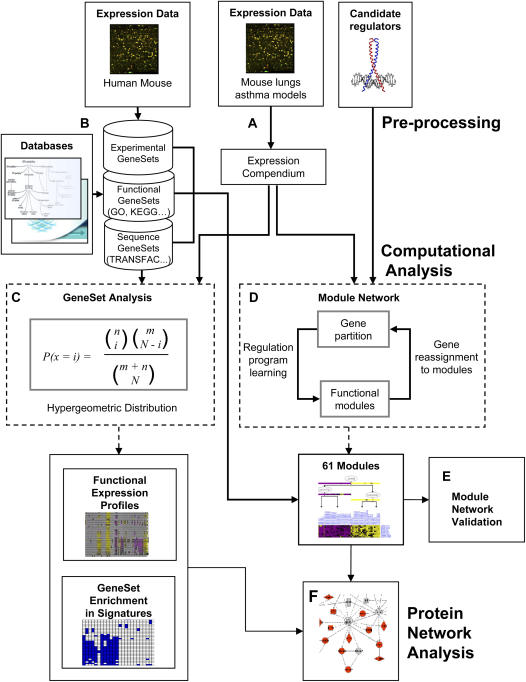
In this study we create a global map of asthma using publicly available gene expression datasets from multiple sources and tools that allow integration of multiple levels of information, such as functional annotations and protein interactions (Figure 1). In addition to providing a comprehensive description of this map, we present examples of novel observations. These observations include individual gene expression heterogeneity of genetically identical animals, a transcriptionally distinct module of known and potentially novel asthma genes, and support for a central role of TGF-β in IL-13–dependent allergic lung inflammation. The map, as well as all datasets, analyses, and additional examples, are available on the interactive AsthmaMap website (http://compbio.cs.huji.ac.il/AsthmaMap).
Figure 1.
Analysis flow. (A) The data were combined, normalized, and filtered, resulting in a unified compendium of 102 experiments and 7,238 genes. (B) A total of 4,305 gene sets were generated from three types of data: experimental, functional, and sequence. Experimental sets are differentially expressed genes from other expression studies in human and mice lungs, and functional and sequence gene sets are derived from multiple databases. (C) Enrichment analysis was performed on experiments and signatures using hypergeometric distribution. Unsupervised reconstruction of transcriptional modules was performed using the Module Networks algorithm (D), and was validated using an independent validation dataset (E). (F) The modules were utilized to generate Protein regulatory network.
MATERIALS AND METHODS
Datasets
We searched NCBI Gene Expression Omnibus (GEO) for all in vivo asthma murine models gene expression datasets, publicly available by June 2006. Five datasets that passed our inclusion criteria (see online supplement) were combined to generate the expression compendium, and an additional dataset published in 2007 was used for independent validation (Table 1). IL-13 (GEO series GSE1301, Wills-Karp and coworkers): 12 lung samples from IL-13 knockout (IL-13–KO) and BALB/cJ wild-type (WT) mice that were treated with house dust mite (HDM) or with PBS as control. RAG (GEO series GSE483, Wills-Karp and colleagues): 7 lung samples from BALB/cJ mice that were treated with ragweed pollen protein plus Alum or with PBS as control. MAH (Murine Airway Hyperresponsiveness, GEO series GSE3184, Wills-Karp and associates): 20 lung samples taken from asthma-sensitive A/J mice and 2 lung samples taken from resistant C3H/MeJ mice. Lungs were harvested at 6 and 24 hours after challenging the mice with HDM/ovalbumin or with PBS as control. DEA (Dissection of Experimental Asthma [4]): 11 lung samples from BALB/cJ mice that were treated with ovalbumin or aspergillus fumigatus (ASP) allergens, or with saline as control. FTM (Focused Transgenic Modeling [8]): 50 lung and tracheal perfusate (TP) samples from four mice strains, which were treated with ovalbumin or with PBS. The four strains are: (1) IL-13+, Stat6+/−; (2) IL-13+, Stat6−/−; (3) IL-13–Epi (IL-13 overexpresses, STAT6 expressed only in epithelial cells); and (4) BALB/cJ WT. The datasets have been previously described (3–5, 8) and recently reviewed by Rolph and coworkers (21). TEST (GEO series GSE6858 [22]): 16 lung samples taken from BALB/cJ recombinase-activating gene–deficient (RD) mice and from WT mice. Lungs were collected 1 day after challenging with ovalbumin or PBS as control. This dataset was used to as an independent validation set. See Table 1 for description of all datasets.
TABLE 1.
GENE EXPRESSION COMPENDIUM DATASET SOURCES AND SIGNATURE NAMES
| Name | Source | Strains | Treatment | Platform | Signatures dataset_strain/tissue/time_treatment |
|---|---|---|---|---|---|
| IL-13 | GEO GSE1301 | BALB/cJ Wild type; IL-13 knockout | House Dust Mite | Affymetrix GeneChip Mouse Expression Array 430A and Mouse Genome 430A 2.0 Array | IL13_WT_HDM; IL13_KO_HDM |
| Rag | GEO GSE483 | BALB/cJ | Ragweed Pollen + Alum | Affymetrix GeneChip Murine Genome U74A/B/C Version 1 | RAG_RWP |
| MAH | GEO GSE184 | A/J; C3H/MeJ (C3H) | Ovalbumin at 6 h and 24 h | Affymetrix GeneChip Mouse Expression Array 430A and Mouse Genome 430A 2.0 Array | MAH_OVA; MAH_6_OVA; MAH_24_OVA |
| DEA | (4) | BALB/cJ | Ovalbumin; Aspergillus Fumigatus | Affymetrix GeneChip Murine Genome U74A Version 2 | DEA_OVA; DES_ASP |
| FTM | (8) | IL-13+, Stat+/−(Stat6p); IL-13+, Stat6−/− (Stat6n); hStat6+ (IL13-Epi); BALB/cJ | Ovabumin; Samples taken from Whole Lung and Tracheal Prefusate | UCSF 10Mm Mouse v.2 Oligo Array; UCSF Gladstone 18K Mouse v.2 Oligo Array | FTM_WT_OVA; FTM_Stat6_OVA |
| TEST | (22) | BALB/cJ Wild type; Recombinase activating gene deficient mice | Ovalbumin | Affymetrix GeneChip Mouse Expression Array 430A and Mouse Genome 430A 2.0 Array | TEST_WT_OVA; TEST_RD_OVA |
Compendium Generation
Transcript levels of four datasets (IL13, RAG, MAH, DEA), generated with Affymetrix GeneChip arrays, were determined from their data image files using RMAExpress (23, 24). Transcript levels of FTM were taken from the original article (8), as this is a two-channel array that cannot be processed using RMAExpress. Each dataset was normalized such that the mean expression of every gene is zero. Two compendiums were created. The first (Table E1 in the online supplement), which was used for the supervised analysis, includes the five datasets and consists of 102 samples and 7,238 genes that have at most two absent calls across all samples (6,963 of the 7,238 genes had no absent calls).
For the Module Network analysis, a second compendium was generated from the first four datasets only (Table E2) by choosing all genes that had at most seven absent calls across all samples This unified compendium consists of 8,086 genes and 52 samples (8,084 of the 8,086 genes had no absent calls). FTM dataset was not included in this analysis because its inclusion introduced a bias (see methods portion of the online supplement).
Gene Set Knowledgebase Generation
Three groups of gene sets were used for our analyses.
Functional sets: genes annotated according to their cellular function. These included Gene Ontology, pathway data (KEGG and Superarray), literature-based annotations (Biocarta), and the Genetic Associations Database (25).
Expression datasets: genes that were significantly overexpressed or underexpressed in various microarray studies. Including mouse and human lung resident cells, airways cells, and peripheral blood cells, that were treated with cytokines, growth factors, and stimuli (11, 26, 27).
Sequence sets: genes annotated with their predicted regulatory binding sites taken from TRANSFAC (28) and a comparative study (29), or with their protein domains, taken from InterPro (30). Annotations that originally describe human genes were mapped into mouse genes using Homologous groups as defined in NCBI HomoloGene database (www.ncbi.nlm.nih.gov/entrez/query.fcgi?DB=homologene). See Table 2 for a detailed description of all gene set sources. All gene sets are available at the AsthmaMap website.
TABLE 2.
SOURCES OF GENE SETS
| Name | Type | Source | Description | Date |
|---|---|---|---|---|
| Kegg | Functional Pathway | http://www.genome.jp/kegg | KEGG pathways | January 2005 |
| SA | Functional Pathway | http://www.superarray.com | Superarray pathway annotation | January 2005 |
| Biocarta | Functional | http://www.biocarta.com | BioCarta annotations | January 2005 |
| GO | Functional | http://www.geneontology.org | Gene Ontology. 688 terms up to level 7 In the tree | January 2005 |
| Genetic Association | Functional | http://geneticassociationdb.nih.gov/ | Association of genes with human diseases | January 2005 |
| CSPC | Experimental | (11, 27) | Human Primary cell lines of NHBE, NHLF and BSMC, exposed to cytokines or PBS. Differentially expressed genes (t test P value < 0.05) | 2000 |
| FMC | Experimental | (26) | Lung Fibroblast, exposed to IFN and TGF. Differentially expressed genes (t test PDR = 5%) | 2002 |
| DMM | Experimental | http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE4231 | Different Asthma Mouse Models: Genes that were upregulated or downregulated (t test P value < 0.01) in mouse whole lung after treatments of OVA and Bleo | December 2006 |
| HAH | Experimental | http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3183 | Human Airway Hyperresponsiveness: Genes that were upregulated or downregulated (t test P value < 0.01) in response to IL-13 treatment in airway cells | August 2005 |
| HCL | Experimental | http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE473 | Human CD4+ Lymphocytes: Genes that were differentially expressed (t test P value < 0.01) between cells from patients with asthma and healthy patients with and without atopy. | July 2003 |
| HBE | Experimental | http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE470 | Human Asthma Exacerbatory Factors: Genes that were up-regulated or down-regulated (t test P value < 0.01) in Human airway epithelial cells, after different treatments | July 2003 |
| HAE | Experimental | http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3004 | Human Airway Epithelial: Genes that were upregulated or downregulated (paired t test P value < 0.01) in bronchial epithelium of human subjects before and after an allergen challenge. | August 2005 |
| Promoter | Sequence | http://www.broad.mit.edu/seq/HumanMotifs/ | Discovered promoter motifs, Xie et al. | February 2005 |
| TRANSFAC | Sequence | http://www.gene-regulation.com/pub/databases.html | Binding motif prediction from TRANSFAC version 8.3, P value < 0.01, 1,000 bp upstream | December 2004 |
| InterPro | Sequence | http://www.ebi.ac.uk/interpro/ | InterPro domains | December 2004 |
Data Visualization and Enrichment Analyses
Visualization, heatmap generation, and gene set enrichments were performed using Genomica software (http://genomica.weizmann.ac.il/). Differentially expressed gene signatures were calculated with parametric Student's t test that rejects the null hypothesis if the means are not equal. The t test score was controlled for False Discovery Rate (FDR) (31), using ScoreGenes software (http://compbio.cs.huji.ac.il/scoregenes/). Protein interaction regulatory networks were generated using Ingenuity Pathways Analysis (Ingenuity Systems, Redwood City, CA).
Expression Profile of Gene Sets in Individual Animals
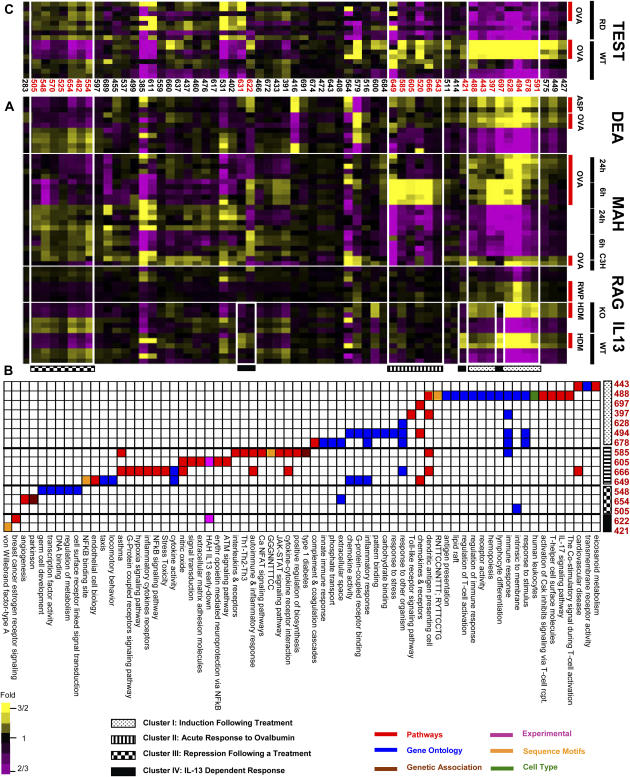
The functional expression profile was generated by calculating for each gene set and each sample the enrichment of genes that are substantially increased (> 2-fold change), compared with all the genes in that sample. For example, if in a certain sample there are X genes, of which x are significantly overexpressed, and there are K genes in a gene set, of which k are significantly overexpressed, the P value is calculated with the hypergeometric distribution over [X,x,K,k] (also known as Fisher's exact test). The same is done for underexpressed genes. We controlled FDR at 5% for all analyses. This strict correction ensures the enrichment is statistically significant. We obtained 258 gene sets enriched with a significant number of substantially changed genes. To address redundancy between gene sets we manually curated them and merged the overlapping ones. The resulting 62 GO and Superarray gene sets are presented in Figure 2.
Figure 2.
Functional profile of individual animals. Gene sets from Gene Ontology (GO) and Superarray (SA) databases that are enriched with substantially expressed genes. Rows correspond to gene sets, columns correspond to experiments, and color indicates the average expression of the genes in each significant gene set. The label of each experiment is presented above the heat map, while red indicates treatment. Rectangles denote function enrichment of general immune response (A, B) and lymphocytes regulation pathways (C). Gene sets were manually curated to eliminate redundancy.
Gene Set Enrichment in Experimental Signatures
Signatures of differentially expressed genes were identified for each dataset independently. The threshold for including a differentially expressed gene in a signature is relatively liberal (t test P value < 0.05). This choice facilitates identification of significant enrichments even when the gene level changes are relatively mild. The enrichment of gene sets in each signature was calculated with the hypergeometric model (FDR < 5%), relative to the enrichment in the complete dataset. We found 281 gene sets enriched in the signatures, of which 161 are presented in Figure 3 after manual curation that merged redundant gene sets.
Figure 3.
Enrichment of gene sets within signatures of differentially expressed genes. The enrichment is determined for each gene set and each signature, compared with the prevalence of that gene set in the complete dataset. One hundred sixty-one gene sets that are significantly enriched (FDR < 5%) within one signature or more are presented. Colored pixels are significantly enriched; color indicates gene set data source. (A, B) Gene sets induced by most treatments. (C, D) Repressed gene sets. (E) Genes induced by IL-13 in human airway cells. (F) IL-13–dependent gene set induction in IL-13–KO and WT mice. (G) Genes induced by TGF-β1 in human airway cells. (H) Genetic association gene sets induced by treatment. Gene sets were manually curated to eliminate redundancy.
Module Networks Analysis
The modules and their regulation programs were automatically detected using Module Networks procedure (32). This method, based on probabilistic graphical models, detects modules of co-expressed genes and their shared regulation programs. The regulation program is a small set of genes, which determines the expression level of the module genes using a decision tree structure (regression tree). Given the expression values and a pool of potential regulator genes, a set of modules and their associated regulation programs are automatically inferred by an iterative procedure. This procedure searches for the best gene partition into modules and for the regulation program of each module, while optimizing a target function. The target function is the Bayesian score, derived from the posterior probability of the model (see Ref. 33 for a detailed description of the algorithm).
We employed the Module Networks on the second compendium, which consists of 52 samples and 8,086 genes. From them, a pool of 1,764 potential regulators was created by choosing all the genes that carry a regulatory role, according to Gene Ontology annotations. The number of modules was determined as the number that achieved the best Bayesian score during the learning (Figure E1). Of the set of potential regulators, 217 regulators were found to regulate at least one module in the inferred network.
Generation of Module Cluster Map
The global view of the modules (Figure 4A) was generated by calculating the average expression over all the genes in each module. In the presented matrix, each column represents the average expression of a single module, and the rows represent experiments. The resulting values were clustered using hierarchical clustering (using Genomica). Four clusters of modules were selected according to their profiles. The gene set enrichment of the modules was calculated with the hypergeometric distribution, and 94 gene sets found to be significant (FDR < 5%); of these, 68 are presented after manual curation that merged redundant sets.
Figure 4.
Module global map. Global view of the 61 modules (columns) that were generated with Module Networks algorithms. (A) For each module, the average expression of its genes in each sample is presented in a heat map. The sample attributes treatment type, strain, and time point appear to the right of the heat map. Clusters of modules with a characteristic profile include: I, induction following a treatment; II, acute response to ovalbumin; III, repression following a treatment; and IV, IL-13–dependent induction. (B) Gene sets enrichment in the modules. Colors indicate gene set source. (C) Module validation (expression patterns of the 61 modules in external TEST data). Note the impressive similarity in module gene expression patterns.
Module Network Validation
We used TEST dataset to independently validate our analysis. This recently published dataset was generated on Affymetrix GeneChip Mouse Genome 430 2.0 Arrays. Cell files were downloaded and normalized using RMAExpress. To measure how well the modules predict the expression of their genes in the new dataset, we measured the correlation of the modules between the new dataset and the old compendium. Pearson correlation was measured for 1,025 genes that participate in the four clusters. Their average expression in the WT treated mice in the TEST data was compared with the average expression in three sample groups from the compendium: the average of OVA-DEA samples, the average of OVA 24-hour BALB-MAH samples, and the average of all treated WT samples.
Ingenuity Protein Network Analysis
Interaction networks were generated by analyzing genes in distinct module clusters using Ingenuity Networks analysis. Examples of significant networks and canonic pathways for every cluster are presented on the AsthmaMap webiste. In the case of IL-13–dependent cluster presented below, eight significant networks (P value < 0.01) were algorithmically generated based on their connectivity. The two highest scored networks (P value < 1e-43) were merged to generate the network presented in Figure 5B. Here, genes are represented as nodes, and the biological relationship between two nodes (e.g., protein–protein interaction) is represented as an edge. All edges are supported by at least one reference from the literature, from a textbook, or from canonical information stored in the Ingenuity Pathways Knowledge Base. Edges which are supported only by co-expression evidence are not presented. Human, mouse, and rat orthologs of a gene are represented as a single node in the network.
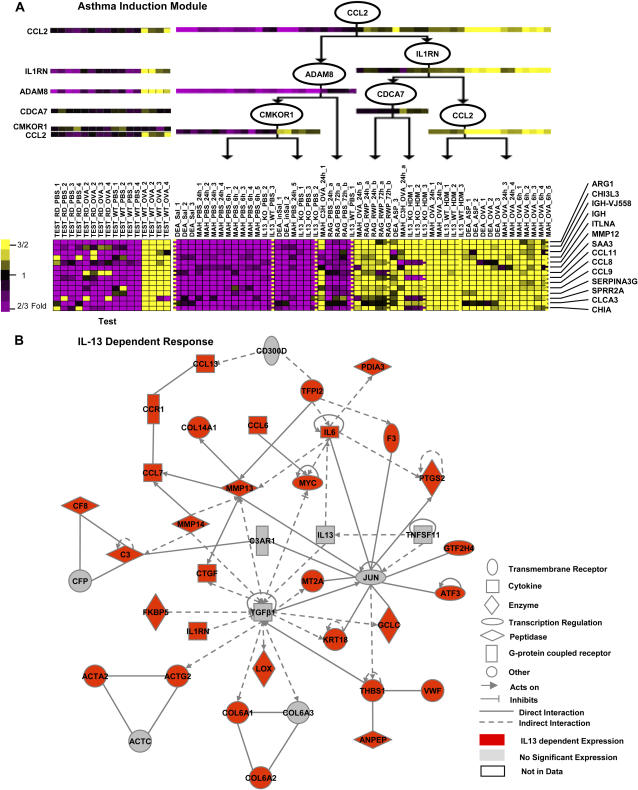
Figure 5.
Protein interaction regulatory networks within module clusters. (A) Module 494, globally induced following treatment, along with its regulation program. The rows are genes, and the tree represents the regulation program. The left panel shows the expression of the genes and regulators in the independent validation dataset TEST. (B) Protein interaction network within IL-13–dependent cluster of modules. Nodes represent proteins, edges indicate all direct or indirect interactions besides co-expression. Red color indicates IL-13–dependent expression (i.e., genes that are induced by treatment only at the presence of IL-13).
AsthmaMap Website
The complete map, containing all the pre-processed data (expression compendium, 4,305 gene sets, and new gene lists reported here), is available on the interactive AsthmaMap website (http://compbio.cs.huji.ac.il/AsthmaMap). The website allows visualizing the sets along with the expression patterns of their genes. The sets can be downloaded in a format applicable for Genomica software. In addition, user-defined gene lists can be uploaded and analyzed for enrichment in respect to any of the sets available in this study.
RESULTS
Expression Compendium and Gene Knowledgebase
We combined five different studies of pulmonary gene expression from mouse models of asthma. After normalization and filtering (see Materials and Methods), we had a unified compendium describing the expression of 7,238 genes in 102 samples (Table E1). The sample set includes treatments of various mouse strains and cell types with different inducers of asthma (Table 1). We analyzed the compendium with a gene knowledgebase that includes 4,305 gene sets from three types of information sources (Table 2): functional annotations derived from different annotation databases such as Gene Ontology, pathway analyses, and disease association; sequence annotations (genes sharing the same predicted cis-regulatory motif in their promoters or genes encoding the same protein domain); and experimental annotations (genes that were differentially expressed between two conditions in other DNA microarray studies). We then identified modules of co-regulated genes using Module Networks analysis (32), and validated them with a new independent validation gene expression dataset. Finally, we proposed potential regulatory networks within these modules, by projecting the discovered modules to protein interaction networks. The complete map, containing all the pre-processed data (expression compendium, 4,305 gene sets, and new gene lists reported here), is available on the interactive AsthmaMap website (http://compbio.cs.huji.ac.il/AsthmaMap).
Gene Set Expression Profiles Reveal Diverse Response in Individual Animals
To examine the functional profile of individual animals based on their gene set expression, we inspected the expression profiles of gene sets across all samples. Specifically, we looked for gene sets that were enriched (FDR < 5%) for genes substantially changed (> 2-fold) in one sample or more. Out of 4,305 gene sets, only 258 were found to be significant, of which 62 GO and Superarray gene sets are presented in Figure 2.
We can see from the expression profiles that most treatments cause an impressive homogenous increase in multiple gene sets, including general immune response, cytokines, chemotaxis, and G protein–coupled receptor signaling (Figures 2A and 2B). This increase can be seen across all animals with an intact IL13-STAT6 pathway. On the other hand, increase in regulation of T cell and B cell activation, or antigen processing and presentation gene sets, seemed to vary among individual animals (Figure 2C). Interestingly, in some treated animals none of the gene sets are increased. Assuming that the experimental annotations are correct and that these are genetically similar to their experimental peers, we can only speculate that this lack of response may suggest a technical cause or an overlooked biological cause, of which experimentalists should be aware.
Emergent Gene Set Enrichment in Experimental Signatures
To obtain a better global view of the themes in the data, we looked at the gene set enrichment in differentially expressed gene signatures that distinguish between each treatment and its control experiments (Figure 3). Unlike the previous analysis, this global view shows themes with moderate response to treatment and it masks individual variance.
The most immediate conclusion from this view is that the functional profile of genes induced by treatments is drastically different from those repressed. As expected, Gene Ontology gene sets related to antigen processing, hemopoiesis, cytokines, and response to stimulus are induced by all treatments (Figure 3A). Similarly, pathway gene sets like inflammation, cytokines, chemokines, and receptors dominate induced genes across all treatments (Figure 3B). Among the genes decreased, growth factors and development-related genes are dominant (Figures 3C and 3D).
Reassuringly, IL-13–regulated gene sets identified in human hyperresponsive airway cells (HAH) stimulated by IL-13 were enriched in most models, but not in IL-13–KO mice (Figure 3E). Protease inhibitor activity and eicosanoid metabolism (arachdionic acid pathway) characterize HDM-induced signature in WT but not IL-13–KO mice (Figure 3F).
Recently, there has been an increased interest in the role of TGF-β, a master regulator of fibrosis that suppresses inflammatory response, in asthma (34, 35). Although several TGF-β pathway genes were repressed after treatment and TGF-β1 itself was unchanged, we found that experimental gene sets of genes induced by TGF-β in lung fibroblasts, were enriched in most treated animals with an intact IL-13 pathway (Figure 3G), supporting the notion that indeed TGF-β may regulate gene expression patterns associated with acute inflammatory response.
To assess whether the genes induced in every model were enriched with genes known to be associated with human disease we generated gene sets based on “Genetic Association” database (http://geneticassociationdb.nih.gov/). Indeed, sets of genes known to be associated with rheumatoid arthritis, inflammation, and asthma were all enriched in signatures increased by treatment (Figure 3H; see AsthmaMap website for detailed lists).
Unsupervised Analysis Detects Four Distinct Responses
Analyzing the differentially expressed gene signatures reveals the active functional gene sets, but is of course limited to pre-defined sets. To obtain a refined view of the asthmatic response and to create new gene sets, we employed the Module Networks algorithm (32). This probabilistic method detects modules of co-expressed and co-regulated genes using a Bayesian graphical model, where for each module it reconstructs a regulation program: a set of regulators and combinatorial rules, structured as a regression tree, which determine the expression of the target genes in the module. The regulation programs are the main advantage of Module Network over other clustering methods that detect clusters of genes, but not their potential regulators in the cell.
In this analysis we used only four datasets, as the fifth was hybridized on a different platform, and its inclusion introduced a strong bias (see online supplement). This second compendium describes the expression levels of 8,086 genes in 52 samples (Table E2). Among these genes, we defined 1,764 genes that carry a regulatory role according to Gene Ontology as potential regulators (Table E3). We achieved the best Bayesian score when learning with 61 modules (Figure E1). Examining the average expression of these modules (Figure 4A), we obtained four clusters of modules that could be characterized by their overall response.
Global induction following a treatment (Figure 4A, Cluster I) is characterized by a strong induction by any type of treatment. It is enriched for general components of the immune response: innate and adaptive responses, complement system; chemokines and cytokines, and their receptors (Figure 4B and Table 3). In addition, the cluster of modules is enriched for genes with potentially direct correlation with asthma, such as IL-17 signaling pathway and eicosanoid metabolism.
TABLE 3.
ACTIVE MODULES
| Module Number | Number of Genes | Response | Functional Themes | Root Regulator | Selected Genes |
|---|---|---|---|---|---|
| 443 | 45 | Global induction following treatment | Receptors for interleukins and cytokines; eicosanoid metabolism | CHI3L3: Chitinase 3-like3-3 | PTGS1, PTGES, PTGER4 |
| 488 | 38 | Global induction following treatment | Adaptive immunity: Leukocytes; IL-17 signaling pathway; T-cell co-stimulatory pathways | EMR1: EGF-like hormone receptor with macrophage- restricted expression | CD3D, CD3G, CD2, PTPRC, LCK |
| 494 | 14 | Global induction following treatment | Chemokines and their receptors | CCL2: chemokine (C-C motif) ligand 2 | CHIA, CHI3L3, ARG1, MMP12 |
| 678 | 21 | Global induction following treatment | complement system | FCGR2B: Fc fragment of IgG, low affinity IIb, receptor (CD32) | C3, C3AR1, C1GA, C1GB, C1GC |
| 697 | 15 | Global induction following treatment and IL-13– dependent induction | Innate immunity response | CXCL5: chemokine (C-X-C motif) ligand 5 | CCR1, CCL2, CCL7 |
| 585 | 54 | Acute response to ovalbumin | JAK/STAT pathway | ID3: inhibitor of DNA binding 3 | JAK2, JUNB, IL4, IL10 IL1RA, IL4RA, IL17R, SOCS2 |
| 605 | 35 | Acute response to ovalbumin | Nitric oxide; Extracellular matrix molecules | RGS16: regulator of G protein signaling 16 | SERPINE, FAS, NFKBI |
| 649 | 6 | Acute response to ovalbumin | Chemokines with NF-κB binding site in their promoters. | CMKOR1: chemokine orphan receptor 1 | CXCL1, CXCL2, CXCL5 |
| 666 | 39 | Acute response to ovalbumin | Nitric oxide; NF-κB signaling; interleukins and their receptors | FZD2: frizzled homolog 2 (Drosophila) | IL1A, IL1B, IL1R2, CSF3, CCL3, CCL4, FOS, EGR1, MMP8 |
| 548 | 43 | Repression following treatment | Transcription factor activity and regulation of metabolism. | NUMB: numb gene homolog (Drosophila) | SOX17, SOX18, HOCB5, TOB, SMAD7 |
| 654 | 47 | Repression following treatment | Angiogenesis | IL1RL1: interleukin 1 receptor– like 1 | VEGFA, KDR, ANGPT1, FIGF, FIGF1 |
| 622 | 16 | IL-13–dependent induction | Genes repressed by IL-13 in human airway cells | MAFF: v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) | PTGS2 |
The other three responses are as follows. Acute response to ovalbumin (Figure 4A, Cluster II) enriched for chemokines, cytokines and their receptors, inflammatory and DNA damage signaling pathways (nitric oxide, JAK-STAT, NF-κB, Ca-NFAT, ATM) (Figure 4B and Table 3); repression following a treatment (Figure 4A, Cluster III) enriched for angiogenesis, cell development, and regulation of metabolism (Figure 4B and Table 3); and IL-13–dependent response, characterized by induction only in WT and not in IL-13–KO mice (Figure 4A, Cluster IV). A more detailed analysis of this cluster is described below.
Module Network Validation
To evaluate the module network we used a new dataset (TEST) that was published after the analysis was done (22). The dataset includes lung gene expression measurements, collected from WT and RD mice treated with ovalbumin. We wanted to estimate how well the modules predict the expression of their genes in the new dataset. For this purpose we measured for the four clusters (I–IV), the correlation between the average of treated WT mice samples in TEST data and the average of treated WT mice in the old compendium. Pearson correlation between these two groups is positive: 0.61. When calculating correlation between treated WT mice in TEST data, and ovalbumin-treated WT mice in DEA dataset, the correlation is even higher (Pearson correlation 0.74), and the same is true for ovalbumin-treated WT mice in MAH data (Pearson correlation 0.81). Figures 4C and 5A provide visual demonstration of the impressive similarity of gene expression patterns in modules between gene expression compendium and TEST dataset.
Potential Genes with Novel Role in Asthma Induction
An interesting member of in the global induction cluster is module 494 (Figure 5A). This module, regulated by CCL2, IL1RN, and ADAM8, contains 14 genes that exhibit the most persistent activation after treatment of all modules. Among the regulators, ADAM8 and CCL2 were previously associated with allergic lung inflammation based on microarray data (5, 36). The human gene for IL1RN, an anti-inflammatory cytokine located within interleukin-1 cluster on human chromosome 2q12–2q14, was found to be associated with asthma in several populations (37, 38). Recently, Ramadas and coworkers (39) found differences in IL1RN expression between asthma-susceptible and -resistant mouse strains (A/J and C3H/HeJ, respectively) but not sequence differences.
The genes in the module include several chemokines such as CCL11 (Eotaxin, a known asthma regulator [40]); immunoglobulin-related molecules (IGHA1, IGJ); molecules suggested to be involved in allergic lung inflammation based on array data (ITLNA, CALCA3 [8], ARG1 [4], SPRR2A [6]); and chitinase family members CHIA and CHI3L3. Recently Homer and colleagues (41) demonstrated that both CHIA and CHI3L3 were induced in models of allergic lung inflammation. However, they differed in their distribution—CHIA was expressed in distal airway epithelial cells in which mucus was not expressed, while CHI3L3 expression was limited to central or proximal mid-airways but not distal. While they observed that both were induced by ovalbumin or IL-13 induction, in this module CHIA induction is dependent on an intact IL-13 pathway and CHI3L3 is not. The enrichment of this module with asthma-proven relevant genes should encourage further study of the role of other molecules in this module, such as SERPINA2G (a proteinase inhibitor that regulates cathepsin B activity) and SAA3 (a member of the serum amyloid protein family).
Potential Role of TGF-β1 in IL-13–Induced Allergic Lung Inflammation
One of the limitations of module network analysis is that it is based solely on gene expression of the regulators and target genes. To address regulatory events that are beyond transcriptional regulation we need to add more complete biological information, such as protein–protein interactions. We therefore explored the nature of the interactions of genes between and within modules using Ingenuity Pathways Analysis (Ingenuity Systems). As an example, we subjected the IL-13–dependent cluster of modules to network analysis using Ingenuity. We generated eight potential networks, and merged the two highest scoring networks (P value < 1e-43). The combined network, which contains 40 genes, 31 of which significantly changed, is presented in Figure 5B. This analysis uncovers potential key regulatory effect of genes that do not change significantly in this dataset on genes in this cluster of modules. Surprisingly, although IL-13 is a member of the network, the network's major regulators are TGF-β1 and its downstream transcription factor JUNB (Figure 5B). TGF-β1 regulates eight of the induced network members and is activated or induced by four members, including THBS1, MMP14, and IL-13, suggesting a positive feedback loop. A close look at the network also suggests that at least some of the effects of IL-13 on gene expression in mouse allergic lung inflammation are mediated through TGF-β1.
Additional examples of ingenuity networks found in clusters I, II, and III are available on the AsthmaMap website.
DISCUSSION
The aim of this article is to provide a framework for systems-level analysis of disease-relevant data from multiple sources. Despite inherent difficulties (differences in model parameters, nonideal experimental design, and limited number of animals in experimental subsets), we observe highly meaningful and reproducible patterns and themes that characterize allergic lung inflammation, and are robust to a specific model setup.
One principle applied in this study is gene sets analysis rather than individual gene analysis; here the main benefit is the robustness to noise. Such robustness was critical when uncovering heterogeneity between individual animals, which is not dependent on changes in the levels of single genes (Figure 2). One concern in applying gene expression studies in human clinical research is diversity in genetic background that dictates individual variability. By being less affected by changes in the levels of single genes, gene set profiles allow us to observe changes in global trends and themes in response to a stimulus or an intervention. Such analyses may be highly useful in applying gene expression approaches to guiding human clinical research and potentially, in the future, disease diagnosis and management.
When the gene sets analysis principle is applied to gene expression signatures, a global view of the nature of the genes that are changed during allergic lung inflammation in the mouse lung emerges (Figure 3). This view provides system-level support to previous hypotheses as well as generation of new ones. As an example in this analysis, we found that protease inhibitor activity and eicosanoid metabolism characterize HDM-induced signature in WT but not IL-13–KO mice (Figure 3F). Considering the difference in response to allergen in IL-13–KO and WT mice, the difference in eicosanoid metabolism is predictable: this pathway is often implicated in asthma in humans. More specifically, recently Shim and coworkers (42) demonstrated that ALOX5, a key enzyme in arachidonic acid metabolism, mediates IL-13–induced pulmonary inflammation; Trudeau and colleagues (43) also demonstrated that PTGS2, another key enzyme in this pathway, is regulated by IL-13 in airway epithelial cells. Our findings, which are based on microarray data created years before these articles and on analysis performed independently, provide a systems-level support for the observations by Shim and coworkers and Trudeau and colleagues. In parallel, their results validate and support our analyses. The differences in protease inhibitors activity, however, were not reported so far. These differences suggest that IL-13 effects on airway inflammation and remodeling may also be mediated by modulation of anti-protease activity, a finding that may have important therapeutic implications.
An additional principle used in this study is an unbiased integration of multiple levels of biological information. This principle aims to enhance what the biologists often do, which is to prioritize hypotheses and insights based on their knowledge by using tools that integrate such knowledge. A good example of this approach is the potential regulatory role for TGF-β1 in allergic lung inflammation that we propose. This observation became obvious when we combined gene expression analysis, gene set analysis, and protein–protein interactions information. In fact, the role of TGF-β1 in allergic lung inflammation is not completely understood. Increased levels of TGF-β1 and evidence for TGF-β1 activation have been found in airways, bronchoalveolar lavage, and cells from of patients with asthma (44–46). Polymorphisms in the TGF-β1 promoter have been identified in patients with asthma (47–49) as well as increased levels of TGF-β2 in patients with severe asthma (50). Recently, Leung and coworkers demonstrated that inhibition of TGF-β1 receptor kinase reversed bronchial hyperreactivity in a murine model of allergic lung inflammation (51). Similar results were obtained by Hirano and colleagues with pirfenidone, an antifibrotic agent (52), and by Nakao and coworkers using transgenic mice that overexpress SMAD7, an inhibitor of TGF-β1 signaling (53). Lee and colleagues (54) demonstrated that IL-13–mediated pulmonary fibrosis was mediated through TGF-β1, as did Fichtner-Feigl and coworkers (55). Zhou and colleagues demonstrated synergism between IL-13 and TGF-β1 in TIMP1 induction (56).
In our analysis, lungs of mice after antigen challenge are almost universally enriched with genes induced by TGF-β in airway resident cells (Figure 3G). In addition, protein network analysis demonstrates that IL-13–dependent gene cluster of modules is significantly regulated by TGF-β1 (Figure 5B). Our results indicate that TGF-β1 is induced early in allergic asthmatic response and may play a significant role in all of its stages and not necessarily only in the remodeling phase. Together with the murine TGF-β1 inhibition experiments, these findings suggest that modulating TGF-β1 signaling in the airway may be a potential target for therapeutic intervention in asthma.
One of the interesting questions arising from gene expression data is whether there are a few key molecules that regulate the expression of the rest of the genes. The Module Networks algorithm attempts to address this question by detecting modules of genes that have a similar transcription under some context (context-specific clustering), and a set of regulators and rules that together predict the transcription levels of the target genes under the different contexts.
We presented in details an example of a module (494, Figure 5A) which shows a persistent activation after all types of treatments and consists of many known asthma-related genes, both as module members and as regulators. However, this module also illustrates the limitations of the Module Network approach in gene expression data. It is tempting to hypothesize that the regulators CCL2, IL1RN, and ADAM8 indeed regulate the behavior of the genes in the module. But we cannot rule out that what we obtain is a conditional co-expression that may be driven by regulators outside the data set. Such regulation may occur at the protein level, or by miRNA, or even by simpler mechanisms such as changes in cellular admixtures. Nevertheless, in many cases the transcriptional level reflects a true regulation relationship (32), and the chosen regulators are valid. To address regulatory events that are beyond transcriptional regulation, we need to add more complete biological information, such as physical interactions that support the regulation relationship. The analysis of cluster IV, in which we found that TGF-β may be a regulator of IL-13–dependent genes although its transcriptional levels are not informative, illustrates this point.
In conclusion, although many of the observations that we present were found in single datasets or traditional experiments, our global analysis supports the generalizability and reproducibility of these results beyond the specific experimental settings in which they were found. More importantly, by integrating multiple levels of information and complementary analytic approaches, we infer effects of novel regulators that are not necessarily obvious when single datasets are analyzed. Our results demonstrate that the discovery potential in these publicly available datasets is not fully realized. This article and the accompanying AsthmaMap website are a significant step toward realizing this potential.
Supplementary Material
Acknowledgments
The authors thank A. Regev, S. Wenzel, D. Sheppard, M. Selman, A. Choi, and D. A. Thompson for their helpful discussions and productive critiques.
The work of N.K., N.F., and N.N. was in part funded by NIH grant HL 073745. N.K. was also supported by a generous donation from the Simmons Family and by NIH grants HL0793941 and HL0894932. N.N. was also supported by a fellowship from the Maydan Foundation and by a Leibniz Research Center fellowship.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2007-0151OC on October 5, 2007
Conflict of Interest Statement: N.F. serves as a consultant to Agilent Technologies. None of the other authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Busse WW, Lemanske RF Jr. Asthma. N Engl J Med 2001;344:350–362. [DOI] [PubMed] [Google Scholar]
- 2.Castro-Giner F, Kauffmann F, de Cid R, Kogevinas M. Gene-environment interactions in asthma. Occup Environ Med 2006;63:768–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karp CL, Grupe A, Schadt E, Ewart SL, Keane-Moore M, Cuomo PJ, Kohl J, Wahl L, Kuperman D, Germer S, et al. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immunol 2000;1:221–226. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann N, King NE, Laporte J, Yang M, Mishra A, Pope SM, Muntel EE, Witte DP, Pegg AA, Foster PS, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest 2003;111:1863–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King NE, Zimmermann N, Pope SM, Fulkerson PC, Nikolaidis NM, Mishra A, Witte DP, Rothenberg ME. Expression and regulation of a disintegrin and metalloproteinase (ADAM) 8 in experimental asthma. Am J Respir Cell Mol Biol 2004;31:257–265. [DOI] [PubMed] [Google Scholar]
- 6.Zimmermann N, Doepker MP, Witte DP, Stringer KF, Fulkerson PC, Pope SM, Brandt EB, Mishra A, King NE, Nikolaidis NM, et al. Expression and regulation of small proline-rich protein 2 in allergic inflammation. Am J Respir Cell Mol Biol 2005;32:428–435. [DOI] [PubMed] [Google Scholar]
- 7.Follettie MT, Ellis DK, Donaldson DD, Hill AA, Diesl V, DeClercq C, Sypek JP, Dorner AJ, Wills-Karp M. Gene expression analysis in a murine model of allergic asthma reveals overlapping disease and therapy dependent pathways in the lung. Pharmacogenomics J 2006;6:141–152. [DOI] [PubMed] [Google Scholar]
- 8.Kuperman DA, Lewis CC, Woodruff PG, Rodriguez MW, Yang YH, Dolganov GM, Fahy JV, Erle DJ. Dissecting asthma using focused transgenic modeling and functional genomics. J Allergy Clin Immunol 2005;116:305–311. [DOI] [PubMed] [Google Scholar]
- 9.Zimmermann N, Mishra A, King NE, Fulkerson PC, Doepker MP, Nikolaidis NM, Kindinger LE, Moulton EA, Aronow BJ, Rothenberg ME. Transcript signatures in experimental asthma: identification of STAT6-dependent and -independent pathways. J Immunol 2004;172:1815–1824. [DOI] [PubMed] [Google Scholar]
- 10.Lilly CM, Tateno H, Oguma T, Israel E, Sonna LA. Effects of allergen challenge on airway epithelial cell gene expression. Am J Respir Crit Care Med 2005;171:579–586. [DOI] [PubMed] [Google Scholar]
- 11.Lee JH, Kaminski N, Dolganov G, Grunig G, Koth L, Solomon C, Erle DJ, Sheppard D. Interleukin-13 induces dramatically different transcriptional programs in three human airway cell types. Am J Respir Cell Mol Biol 2001;25:474–485. [DOI] [PubMed] [Google Scholar]
- 12.Temple R, Allen E, Fordham J, Phipps S, Schneider HC, Lindauer K, Hayes I, Lockey J, Pollock K, Jupp R. Microarray analysis of eosinophils reveals a number of candidate survival and apoptosis genes. Am J Respir Cell Mol Biol 2001;25:425–433. [DOI] [PubMed] [Google Scholar]
- 13.Kong X, San Juan H, Kumar M, Behera AK, Mohapatra A, Hellermann GR, Mane S, Lockey RF, Mohapatra SS. Respiratory syncytial virus infection activates STAT signaling in human epithelial cells. Biochem Biophys Res Commun 2003;306:616–622. [DOI] [PubMed] [Google Scholar]
- 14.Laprise C, Sladek R, Ponton A, Bernier MC, Hudson TJ, Laviolette M. Functional classes of bronchial mucosa genes that are differentially expressed in asthma. BMC Genomics 2004;5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005;21:3787–3793. [DOI] [PubMed] [Google Scholar]
- 16.Zeeberg BR, Qin H, Narasimhan S, Sunshine M, Cao H, Kane DW, Reimers M, Stephens RM, Bryant D, Burt SK, et al. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID). BMC Bioinformatics 2005;6:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet 2004;36:1090–1098. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu CC, Lin CC, Chen WS, Chen HY, Chang PC, Chen JJ, Yang PC. CRSD: a comprehensive web server for composite regulatory signature discovery. Nucleic Acids Res 2006;34:W571–W577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segal E, Friedman N, Kaminski N, Regev A, Koller D. From signatures to models: understanding cancer using microarrays. Nat Genet 2005;37:S38–S45. [DOI] [PubMed] [Google Scholar]
- 21.Rolph MS, Sisavanh M, Liu SM, Mackay CR. Clues to asthma pathogenesis from microarray expression studies. Pharmacol Ther 2006;109:284–294. [DOI] [PubMed] [Google Scholar]
- 22.Lu X, Jain VV, Finn PW, Perkins DL. Hubs in biological interaction networks exhibit low changes in expression in experimental asthma. Mol Syst Biol 2007;3:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barash Y, Dehan E, Krupsky M, Franklin W, Geraci M, Friedman N, Kaminski N. Comparative analysis of algorithms for signal quantitation from oligonucleotide microarrays. Bioinformatics 2004;20:839–846. [DOI] [PubMed] [Google Scholar]
- 24.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003;4:249–264. [DOI] [PubMed] [Google Scholar]
- 25.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat Genet 2004;36:431–432. [DOI] [PubMed] [Google Scholar]
- 26.Gal N, Pardo A, Yakhini Z, Becerril C, Ben-Dor A, Friedman N, Ben-Dov I, Kaminski N. Gene expression analysis of lung fibroblasts derived from idiopathic pulmonary fibrosis patients [abstract]. Am J Respir Crit Care Med 2002;165:A171. [Google Scholar]
- 27.Kaminski N, Lee JH, Allard J, Heller RA, Sheppard D. TGF induces distinct transcriptional programs in airway epithelial and airway smooth muscle cells. Am J Respir Crit Care Med 2000;161:A667. [Google Scholar]
- 28.Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res 2000;28:316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 2005;434:338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res 2001;29:37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Statist Soc Ser B Methodological 1995;57:289–300. [Google Scholar]
- 32.Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet 2003;34:166–176. [DOI] [PubMed] [Google Scholar]
- 33.Segal E, Pe'er D, Regev A, Koller D, Friedman N. Learning module networks. J Mach Learn Res 2005;6:557–588. [Google Scholar]
- 34.Chung KF, Torrego A, Hew M, Sukkar M, Oates T. Expression and activation of TGF-{beta} isoforms in acute allergen-induced remodelling in asthma. Thorax 2007;62:307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makinde T, Murphy RF, Agrawal DK. The regulatory role of TGF-beta in airway remodeling in asthma. Immunol Cell Biol 2007;85:348–356. [DOI] [PubMed] [Google Scholar]
- 36.Fulkerson PC, Zimmermann N, Hassman LM, Finkelman FD, Rothenberg ME. Pulmonary chemokine expression is coordinately regulated by STAT1, STAT6, and IFN-gamma. J Immunol 2004;173:7565–7574. [DOI] [PubMed] [Google Scholar]
- 37.Gohlke H, Illig T, Bahnweg M, Klopp N, Andre E, Altmuller J, Herbon N, Werner M, Knapp M, Pescollderungg L, et al. Association of the interleukin-1 receptor antagonist gene with asthma. Am J Respir Crit Care Med 2004;169:1217–1223. [DOI] [PubMed] [Google Scholar]
- 38.Pattaro C, Heinrich J, Werner M, de Marco R, Wjst M. Association between interleukin-1 receptor antagonist gene and asthma-related traits in a German adult population. Allergy 2006;61:239–244. [DOI] [PubMed] [Google Scholar]
- 39.Ramadas RA, Li X, Shubitowski DM, Samineni S, Wills-Karp M, Ewart SL. IL-1 Receptor antagonist as a positional candidate gene in a murine model of allergic asthma. Immunogenetics 2006;58:851–855. [DOI] [PubMed] [Google Scholar]
- 40.Romagnani S. Cytokines and chemoattractants in allergic inflammation. Mol Immunol 2002;38:881–885. [DOI] [PubMed] [Google Scholar]
- 41.Homer RJ, Zhu Z, Cohn L, Lee CG, White WI, Chen S, Elias JA. Differential expression of chitinases identify subsets of murine airway epithelial cells in allergic inflammation. Am J Physiol Lung Cell Mol Physiol 2006;291:L502–L511. [DOI] [PubMed] [Google Scholar]
- 42.Shim YM, Zhu Z, Zheng T, Lee CG, Homer RJ, Ma B, Elias JA. Role of 5-lipoxygenase in IL-13-induced pulmonary inflammation and remodeling. J Immunol 2006;177:1918–1924. [DOI] [PubMed] [Google Scholar]
- 43.Trudeau J, Hu H, Chibana K, Chu HW, Westcott JY, Wenzel SE. Selective downregulation of prostaglandin E2-related pathways by the Th2 cytokine IL-13. J Allergy Clin Immunol 2006;117:1446–1454. [DOI] [PubMed] [Google Scholar]
- 44.Chakir J, Shannon J, Molet S, Fukakusa M, Elias J, Laviolette M, Boulet LP, Hamid Q. Airway remodeling-associated mediators in moderate to severe asthma: effect of steroids on TGF-beta, IL-11, IL-17, and type I and type III collagen expression. J Allergy Clin Immunol 2003;111:1293–1298. [DOI] [PubMed] [Google Scholar]
- 45.Silverman EK, Speizer FE, Weiss ST, Chapman HA Jr, Schuette A, Campbell EJ, Reilly JJ Jr, Ginns LC, Drazen JM. Familial aggregation of severe, early-onset COPD: candidate gene approaches. Chest 2000;117:273S–274S. [DOI] [PubMed] [Google Scholar]
- 46.Sagara H, Okada T, Okumura K, Ogawa H, Ra C, Fukuda T, Nakao A. Activation of TGF-beta/Smad2 signaling is associated with airway remodeling in asthma. J Allergy Clin Immunol 2002;110:249–254. [DOI] [PubMed] [Google Scholar]
- 47.Mak JC, Leung HC, Ho SP, Law BK, Ho AS, Lam WK, Ip MS, Chan-Yeung MM. Analysis of TGF-beta(1) gene polymorphisms in Hong Kong Chinese patients with asthma. J Allergy Clin Immunol 2006;117:92–96. [DOI] [PubMed] [Google Scholar]
- 48.Nagpal K, Sharma S, B-Rao C, Nahid S, Niphadkar PV, Sharma SK, Ghosh B. TGFbeta1 haplotypes and asthma in Indian populations. J Allergy Clin Immunol 2005;115:527–533. [DOI] [PubMed] [Google Scholar]
- 49.Silverman ES, Palmer LJ, Subramaniam V, Hallock A, Mathew S, Vallone J, Faffe DS, Shikanai T, Raby BA, Weiss ST, et al. Transforming growth factor-beta1 promoter polymorphism C-509T is associated with asthma. Am J Respir Crit Care Med 2004;169:214–219. [DOI] [PubMed] [Google Scholar]
- 50.Balzar S, Chu HW, Silkoff P, Cundall M, Trudeau JB, Strand M, Wenzel S. Increased TGF-beta2 in severe asthma with eosinophilia. J Allergy Clin Immunol 2005;115:110–117. [DOI] [PubMed] [Google Scholar]
- 51.Leung SY, Niimi A, Noble A, Oates T, Williams AS, Medicherla S, Protter AA, Chung KF. Effect of transforming growth factor-beta receptor I kinase inhibitor 2,4-disubstituted pteridine (SD-208) in chronic allergic airway inflammation and remodeling. J Pharmacol Exp Ther 2006;319:586–594. [DOI] [PubMed] [Google Scholar]
- 52.Hirano A, Kanehiro A, Ono K, Ito W, Yoshida A, Okada C, Nakashima H, Tanimoto Y, Kataoka M, Gelfand EW, et al. Pirfenidone modulates airway responsiveness, inflammation, and remodeling after repeated challenge. Am J Respir Cell Mol Biol 2006;35:366–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakao A, Sagara H, Setoguchi Y, Okada T, Okumura K, Ogawa H, Fukuda T. Expression of Smad7 in bronchial epithelial cells is inversely correlated to basement membrane thickness and airway hyperresponsiveness in patients with asthma. J Allergy Clin Immunol 2002;110:873–878. [DOI] [PubMed] [Google Scholar]
- 54.Lee CG, Homer RJ, Zhu Z, Lanone S, Wang X, Koteliansky V, Shipley JM, Gotwals P, Noble P, Chen Q, et al. Interleukin-13 induces tissue fibrosis by selectively stimulating and activating transforming growth factor beta(1). J Exp Med 2001;194:809–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fichtner-Feigl S, Strober W, Kawakami K, Puri RK, Kitani A. IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat Med 2006;12:99–106. [DOI] [PubMed] [Google Scholar]
- 56.Zhou X, Trudeau JB, Schoonover KJ, Lundin JI, Barnes SM, Cundall MJ, Wenzel SE. Interleukin-13 augments transforming growth factor-beta1-induced tissue inhibitor of metalloproteinase-1 expression in primary human airway fibroblasts. Am J Physiol Cell Physiol 2005;288:C435–C442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.