Abstract
Apoptosis has been extensively studied in Drosophila by both biochemical and genetic approaches, but there is a lack of knowledge about the mechanisms of apoptosis regulation in other insects. In mosquitoes, apoptosis occurs during Plasmodium and arbovirus infection in the midgut, suggesting that apoptosis plays a role in mosquito innate immunity. We searched the Aedes aegypti genome for apoptosis-related genes using Drosophila and Anopheles gambiae protein sequences as queries. In this study we have identified eleven caspases, three inhibitor of apoptosis (IAP) proteins, a previously unreported IAP antagonist, and orthologs of Drosophila Ark, Dnr1, and BG4 (also called dFadd). While most of these genes have been previously annotated, we have improved the annotation of several of them, and we also report the discovery of four previously unannotated apoptosis-related genes. We examined the developmental expression profile of these genes in Ae. aegypti larvae, pupae and adults, and we also studied the function of a novel IAP antagonist, IMP. Expression of IMP in mosquito cells caused apoptosis, indicating that it is a functional pro-death protein. Further characterization of these genes will help elucidate the molecular mechanisms of apoptosis regulation in Ae. aegypti.
Introduction
Apoptosis is a key pathway involved in normal processes such as development, tissue homeostasis, and DNA damage responses, as well as pathological processes including cancer, ischemia, neurological diseases, and defense against pathogens like viruses (Vaux and Korsmeyer, 1999; Opferman and Korsmeyer, 2003; James and Green, 2004; Clem, 2005). The process of apoptosis is largely carried out by a family of cysteine proteases called caspases. These enzymes are expressed as zymogens and are activated by multiple stimuli. There are two types of caspases, initiator and effector, which carry out different functions in apoptosis. A death insult first results in activation of one or more members of the initiator class, which then cleave and activate members of the effector class. The effector caspases cleave many cellular targets and dismantle the cell. All caspases consist of a prodomain and large and small catalytic domains, which are freed from each other by cleavage at aspartate residues. The large and small catalytic domains form a dimer, and two of these heterodimers associate to form the active caspase molecule. Initiator caspases contain long prodomains, which are involved in interaction with adaptor proteins, while effector caspases contain short prodomains.
Most of what is known about the molecular mechanisms of apoptosis in insects comes from study of the fruitfly Drosophila melanogaster (reviewed in Hay and Guo, 2006). There are seven caspases encoded in the Drosophila genome, including the three initiator caspases Nc (also known as Dronc), Dredd, and dream (also known as Strica) and the four effector caspases Ice (also known as Drice), Dcp-1, decay, and Damm (also known as Daydream). Among the initiator caspases found in Drosophila, Nc appears to be the most important in carrying out apoptosis, while Dredd is important in the immune response, and dream is relatively uncharacterized. Among the effector caspases, Ice appears to be the most important for apoptosis, with Dcp-1 playing a supportive role.
Activation of initiator caspases by intrinsic signals involves the formation of a large protein complex called the apoptosome. Homologs of an adaptor protein that is an integral part of the apoptosome have been found in the nematode Caenorhabditis elegans (CED-4), in mammals (APAF-1), and in Drosophila (Ark). In mammals, cytochrome c binding to APAF-1 is required for apoptosome formation, but cytochrome c does not appear to be required for apoptosome formation in Drosophila (Zimmermann et al., 2002; Means et al., 2006; Yu et al., 2006; Bao and Shi, 2007).
Multiple gene products regulate caspases, either positively or negatively. Among the most important negative caspase inhibitors are the IAP (Inhibitor of Apoptosis) proteins. IAP proteins were first discovered in baculoviruses (Crook et al., 1993), but are now known to exist in cellular genomes ranging from yeast to mammals, where they play important roles in regulating apoptosis and cell division (Vaux and Silke, 2005). In Drosophila, the IAP protein that is most important in regulating apoptosis is thread (also known as DIAP1). Thread was first identified in an enhancer screen for apoptosis-regulating genes (Hay et al., 1995). Overexpression of thread inhibits apoptosis, while loss of thread leads to spontaneous apoptosis, both in the developing fly embryo and in cultured Drosophila cells (Hay and Guo, 2006). Thread has the ability to directly bind and inhibit effector caspases. It also can bind to Nc and causes its degradation via the ubiquitin-proteasome pathway (Wilson et al., 2002).
In addition to IAPs, a second type of negative caspase inhibitor was recently reported in Drosophila. The Dnr1 (defense repressor 1) protein was first identified in a cell-based screen for innate immunity, and was found to inhibit Dredd activity (Foley and O’Farrell, 2004). More recently, Dnr1 has been shown to inhibit apoptosis by causing a reduction in the level of Nc protein (Primrose et al., 2007).
In Drosophila, apoptosis depends on the expression of a set of proteins collectively referred to as IAP antagonists or RHG proteins (Hay et al., 2004). Four of these proteins, rpr (reaper), grim, W (also known as wrinkled or hid), and skl (sickle), are encoded by genes that are in a chromosomal region called the H99 interval (White et al., 1994; Grether et al., 1995; Chen et al., 1996). The RHG genes rpr and grim encode small proteins that are transcriptionally upregulated in cells that are destined to die (Kumar and Cakouros, 2004), while the W gene encodes a larger protein that is regulated post-translationally by phosphorylation (Bergmann et al., 1998). The protein encoded by skl is less well characterized. The RHG proteins each physically interact with thread through a short motif at their amino termini (Vucic et al., 1997; Vucic et al., 1998), and this interaction plays an important role in determining whether a cell lives or dies.
Apoptosis has been established as a component of the innate immune response in baculovirus infections of lepidopteran insects (Clem, 2005). In addition, cross-talk exists between innate immunity pathways and apoptosis pathways in insects. In Drosophila, Dredd (Elrod-Erickson et al., 2000; Leulier et al., 2000; Stoven et al., 2000), Iap2 (Gesellchen et al., 2005; Kleino et al., 2005), BG4 (also known as dFADD) (Zhou et al., 2005b) and Dnr1 (Foley and O’Farrell, 2004) have already been shown to play roles in innate immunity. At one day post infection with Sindbis virus, the midgut of Aedes aegypti exhibited an increase in expression of the Ae. aegypti ortholog of Dif, which is part of the Toll pathway in Drosophila (Sanders et al., 2005). In Drosophila, it has been shown that the protein MyD88 is a component of the Toll pathway, and MyD88 was shown to bind to BG4 (dFADD) and Dredd (Horng and Medzhitov, 2001). In mammals, FADD plays a role in activation of caspases through the extrinsic pathway (Chinnaiyan et al., 1996).
In mosquitoes, there are reports that arbovirus infection causes pathology resembling apoptosis in the midgut and salivary glands. This pathology has been found in Ae. aegypti infected with Semliki Forest virus (Mims et al., 1966), in Culiseta melanura infected with eastern equine encephalitis virus (Weaver et al., 1988), and in Aedes albopictus infected with Sindbis virus (Bowers et al., 2003). Long-term West Nile virus infection has also been shown to induce cell death in the salivary glands of Culex pipiens quinquefasciatus (Girard et al., 2005) and the same group later suggested that this late pathology affected transmission rates (Girard et al., 2007). Recently it has been shown that a lab-derived strain of Culex pipiens pipiens was refractory to infection with West Nile virus, and that infection with this virus caused extensive cell death in the midgut epithelial cells of these mosquitoes (Vaidyanathan and Scott, 2006). Besides viral infection, a number of apoptosis-related genes, as well as other immune response genes, are expressed in hemocytes of Ae. aegypti and Armigeres subalbatus infected with bacterial pathogens (Bartholomay et al., 2004). In addition, Plasmodium can elicit pathology resembling apoptosis in mosquito vectors. Plasmodium berghei infection causes apoptosis in midgut cells of Anopheles stephensi (Han et al., 2000) and An. gambiae (Vlachou et al., 2004), and activation of Ancaspase 7 in midgut cells of An. stephensi (Abraham et al., 2004). With P. gallinaceum and Ae. aegypti, ookinete infection of midgut cells has also been reported to activate caspases (Zieler and Dvorak, 2000).
Even though numerous reports suggest that cell death might play a role in certain infections of mosquitoes, knowledge of the basic mechanics of apoptosis in mosquito vectors is lacking. There have been reports of IAP1 genes in Aedes triseriatus (Blitvich et al., 2002) and Ae. albopictus (Li et al., 2007). A recent study suggests that these genes are regulated by an alternative splicing mechanism (Beck et al., 2007). The initiator caspases Dredd and Dronc have been reported in Ae. aegypti (Cooper et al., 2007a; Cooper et al., 2007b). There has also been a report of an IAP-antagonist related to the rpr protein from Drosophila found in mosquitoes called Michelob_x (Zhou et al., 2005a). In addition, a number of caspases and IAP proteins were recently annotated in the Ae. aegypti genome (Waterhouse et al., 2007). We independently identified a number of apoptosis-related genes, using the available Ae. aegypti genome sequence. In most cases, our results agreed with these previous annotations, but we have made improvements to the annotation of several of these genes, and we have identified four additional apoptosis-related genes that were not previously annotated in Ae. aegypti.
Materials and Methods
Cells and insect rearing
ATC-10 (Ae. aegypti) and C6/36 (Ae. albopictus) cells were maintained in L-15 medium (Gibco) supplemented with 20% FBS (ATC-10) or 10% FBS (C6/36) at 25°C. Ae. aegypti mosquitoes (RexD strain) were reared at the Arthropod-Borne and Infectious Disease Laboratory at 26–28°C, 80–82% humidity, under a 10h dark/14h light regime. Adults were maintained on sucrose and naive adult females were collected at 4 days post-eclosion.
Database mining
The Ae. aegypti genome was searched for apoptosis related genes using known proteins from Drosophila (obtained from FlyBase (http://flybase.bio.indiana.edu)) and predicted proteins from the An. gambiae genome as queries in BLAST searches. Supercontigs from the Ae. aegypti genome were obtained and small portions that were identified from the BLAST search were used as queries to search the EST_others database at NCBI using nucleotide-nucleotide BLAST. ESTs from Ae. aegypti identified after the initial BLAST search were then used as queries reiteratively until no new ESTs were obtained. The ESTs were then translated and aligned using Vector NTI to assemble mini-contigs for transcripts. Genscan (http://genes.mit.edu/GENSCAN.html) and fgenesh (http://www.SoftBerry.com) were used to predict genes and complete missing regions of some EST contigs.
Updated annotations for CASPS7, CASPS20, CASPS17, CASPS21, IAP2, DNR1, ARK, and IMP have been submitted to VectorBase (http://www.vectorbase.org) and are visible as part of the “Manual annotation track” in the genome browser. These sequences will be incorporated into the next VectorBase genebuild (Neil Lobo, personal communication).
Protein domain determination
Domains for the proteins were predicted using the programs ExPASY PROSITE (Hulo et al., 2006), SMART (Schultz et al., 1998), and Conserved Domain Database at NCBI (Marchler-Bauer et al., 2005).
Phylogenetic analysis
Amino acid sequences were aligned using the ClustalW algorithm with the default parameters found in MEGA 3.1 (Kumar et al., 2004). Phylogenetic trees were assembled using MEGA 3.1. Trees were built using neighbor end joining, complete deletion, and p-distance. Other parameters were set at default values.
Plasmid construction
Michelob_x cDNA was obtained as a generous gift from L. Zhou (University of Florida). EST clones DV326893, DV369010, DV323242, DV328064, DV330266 were obtained from D. Severson (University of Notre Dame). IMP and Michelob_x cDNAs were inserted into the expression vector pHSP70PLVI+Rpr-epi (Vucic et al., 1997), replacing the Rpr gene using BglII and SpeI, and the constructs were named pHSMichelob_x-c-EpiHisVI+ and pHSIMP-c-EpiHisVI+. This resulted in HA and His6 tags being fused to the C terminus of the inserted coding sequence. The plasmid pHSP70GFPBsu36I, expressing eGFP (enhanced green fluorescent protein), has been previously described (Clarke and Clem, 2002). All other genes were cloned into the pCRII vector (Invitrogen) for sequencing purposes. 5’ RACE was performed as previously published (Beck et al., 2007).
Transfections
C6/36 cells were plated at a density of 2x106 cells per well in six-well plates and incubated overnight. The following day, cells were transfected using FuGENE 6 (Roche) following the manufacturer’s instructions. A total of 6 μg of DNA with 9μl of FuGENE 6 was used for each transfection. Under these conditions, transfection efficiency was approximately 20%.
Annexin V staining and caspase activity measurements
At 24 hours post transfection, cells were harvested and assayed for Annexin V staining and caspase activity. Cells were stained with Annexin V-PE (BD Pharmingen) following manufacturer’s instructions and analyzed by flow cytometry. Caspase activity was measured using N-acetyl-Asp-Glu-Val-Asp-7-amino-4-trifluoromethyl coumarin (Ac-DEVD-AFC) fluorescent substrate as previously described (Muro et al., 2004). The assays were done in triplicate. The caspase assay results include the results from all three assays, while the Annexin V staining results are from one representative assay.
Expression analysis
RNA was isolated from homogenized tissues of pooled individuals or lysed ATC-10 cells by Trizol (Invitrogen) and treated with Turbo DNA-free DNase (Ambion), following manufacturers instructions, to reduce the possibility of genomic DNA contamination. Approximately 2–3 μg of RNA was used to synthesize cDNA using M-MLV RT (Invitrogen) and an oligo dT-20 primer. The resulting cDNA was analyzed for expression of all eighteen genes (Table 1) as well as Michelob_x (GenBank accession number ABD47742) and actin 6 (act-6, GenBank accession number DQ124691). Expression was initially analyzed by semi-quantitative PCR and the amplicons were analyzed by agarose gel electrophoresis to verify their correct size. Real time PCR was performed using the BioRad iCycler Optical Module. Primers were designed by Beacon Designer 3 (Premier Biosoft) and are shown in Table III. Primers were tested by Tm gradient to determine the optimal Tm for each primer. iQ SYBR Green Supermix was used in all cycle threshold (Ct) determinations following manufacturers instructions (BioRad). To determine the concentration of cDNA needed for expression analysis, dilutions of the cDNA were assayed using act-6 primers. For each of the five stages or tissues (L1/L2, L3/L4, pupae, female adult without midgut, and female adult midgut), three cDNA batches were made. All genes were assayed in duplicate using each cDNA batch. The PCR efficiency curves and melt curves were analyzed for each gene in each stage. For each cDNA batch that was analyzed, the RNA from each stage was used to determine the background for each pair of primers. Some primer sets gave a background Ct while others yielded no background Ct values. If the Ct from a certain gene was not below the background Ct, then the gene was considered not to be expressed.
Table 1.
Ae. aegypti apoptotic regulatory genes characterized in this study. Shown are the NCBI predicted proteins (if any), supercontigs the genes reside on, percent amino acid similarity and identity to the closest relative in Drosophila, and whether the confirmed sequence differs from that predicted in NCBI.
| Gene | NCBI protein(s) | Supercontig(s) | %Similarity/Identity | Different from predicted? |
|---|---|---|---|---|
| IAP1 | ABK01289 | 368 | 51.7/39.4 (th) | No |
| IAP2 | EAT41756 | 214 | 46.8/34.3 (IAP2) | Yes |
| IAP5 | EAT33476 | 1049 | 50.6/41 (det) | No |
| DNR1 | EAT48387 | 11 | 45.9/35.1 (Dnr1) | No |
| ARK | EAT48065, EAT48066 | 18 | 30.4/18.8 (Ark) | Yes |
| BG4 | EAT46931 | 46 | 35.1/21 (BG4) | No |
| IMP | EAT44230 | 117 | 10.9/7 (rpr) | No |
| CASPS19 | EAT45302 | 86 | 53.8/37.8 (decay) | No |
| CASPS18 | EAT45303 | 86 | 47.9/37.4 (decay) | No |
| CASPS8 | EAT33369 | 1085 | 47.7/38.3 (Ice) | No |
| CASPS7 | EAT35718 | 791, 659 | 61.1/52.5 (Ice) | Yes |
| CASPS20 | EAT33088 | 1207 | 36.1/28.1 (Ice) | Yes |
| Dredd | EAT33580 | 1019 | 46.6/31.3 (Dredd) | No |
| Dronc | EAT36368 | 589 | 42.4/30.3 (Nc) | No |
| CASPS17 | none | 182 | 35.4/25.3 (Damm) | Yes |
| CASPS21 | none | 182 | 36.9/25 (Damm) | Yes |
| CASPS16 | EAT42502 | 182 | 32.9/23.3 (dream) | No |
| CASPS15 | EAT42503 | 182 | 34.2/24 (dream) | No |
Results
By searching the Ae. aegypti genome, we found numerous genes with homology to known apoptosis-related genes in Drosophila. The majority of these genes have been previously annotated (Waterhouse et al., 2007), but we were able to make improvements to some of these annotations (Table 1). We also identified four additional apoptosis-related genes, including an additional effector-type caspase, orthologs of Drosophila Ark and Dnr1, and an additional IAP antagonist, which we have named IMP (IAP-antagonist Micheob_x-like Protein). In keeping with the previously proposed genetic nomenclature (Waterhouse et al., 2007), we have named the new caspase CASPS21. Table 2 summarizes the bioinformatics analysis for each gene. If the EST overlap method yielded a full-length predicted transcript (as defined in Materials and Methods), and the existence of this transcript was verified by RT-PCR, then information about the gene is found in Table 2 and it is not discussed further here. Those that did not yield a full-length predicted transcript, however, are discussed further below.
Table 2.
Bioinformatic analysis overview for the genes characterized in this study. Shown are the overlapping ESTs used to determine the transcript (if any), whether the ESTs yielded a full-length transcript, whether the transcript could be verified by RT-PCR from the ATC-10 cell line, and whether gene prediction programs were employed for each gene.
| Gene | ESTs used | ESTs full- length? | Amplify from ATC- 10 cell line? | Gene prediction used? |
|---|---|---|---|---|
| IAP1 | NAa | NA | Yes | No |
| IAP2 | dv383781 | No | Yes | Yes/GenScan |
| eb090402 | ||||
| IAP5 | dv364268 | No | Yes | Yes/GenScan |
| DNR1 | dv237984 | No | Yes | No |
| eg007533 | ||||
| eb089278 | ||||
| dw221134 | ||||
| dw205154 | ||||
| ARK | NA | NA | Yes | No |
| BG4 | dv416615 | Yes | Yes | No |
| dv416614 | ||||
| dv383790 | ||||
| IMP | dv326893 | Yes | NDb | No |
| CASPS19 | dv395012 | Yes | No | No |
| dv330266 | ||||
| CASPS18 | NA | NA | ND | No |
| CASPS8 | dv250139 | Yes | Yes | No |
| dv250137 | ||||
| CASPS7 | dv369010 | No | Yes | Yes/GenScan |
| CASPS20 | dw202685 | Yes | ND | No |
| ee999223 | ||||
| Dredd | dv382387 | Yes | Yes | No |
| dv382385 | ||||
| dw190212 | ||||
| Dronc | dw220447 | Yes | Yes | No |
| dw207394 | ||||
| dv343751 | ||||
| dv356662 | ||||
| CASPS17 | dv323242 | No | No | Yes/fgenesh |
| CASPS21 | NA | NA | ND | Yes/fgenesh |
| CASPS16 | dv241054 | Yes | No | No |
| dv332892 | ||||
| dv328064 | ||||
| CASPS15 | NA | NA | ND | Yes/fgenesh |
NA, not applicable
ND, not determined
Caspases
In NCBI, part of CASPS7 was represented by EAT35718, and this same fragment was also reported by Waterhouse et al. (2007). However, we determined that this was not the full-length transcript using the EST DV369010 (obtained from D. Severson). Portions of DV369010 are found on two supercontigs, 659 and 791. The portion represented by EAT35718, which includes the 3’ end of the transcript, is found on supercontig 659. Gene predictions, using GenScan, predicted that there were additional transcribed sequences further upstream of the gene in supercontig 791. When DV369010 was sequenced further, we found that the gene prediction program was correct, and this was verifed by RT-PCR from ATC-10 cells. This analysis added an additional 76 amino acids to the N-terminus of CASPS7.
CASPS20 bioinformatics analysis yielded a full-length predicted transcript that is represented in NCBI as EAT33088, but the N-termini of the NCBI-predicted protein and the transcript constructed by overlapping ESTs are different. Waterhouse et al. (2007) reported the same sequence that is found in NCBI. 5’ RACE was done on RNA from early stage (L1/L2) larvae and we found that the overlapping EST prediction for the transcript was correct. However, it is possible that there are alternatively spliced forms of CASPS20 present in other stages or tissues.
CASPS17 does not have a predicted protein in NCBI. Waterhouse et al. (2007) identified this gene, but their analysis did not include the full length transcript. CASPS17 is represented by EST DV323242 (obtained from D. Severson). DV323242 has a 3’ stop codon, but further sequencing of DV323242 did not reveal an upstream stop codon in the same reading frame as the start codon. We were able to predict a potential 5’ end of this transcript using the fgenesh gene prediction program, but we were unable to amplify this predicted transcript from ATC-10 cells, meaning that this transcript will require further verification. However, an interesting observation with this gene was that the 3’UTR (represented by DV323242) contained a repetitive DNA element approximately 35 bp in length that is found on multiple supercontigs and ESTs in Ae. aegypti. When this EST is used as a BLAST query against the genome, approximately 100 supercontigs result in hits. The exact nature of this repetitive sequence is unknown at this time.
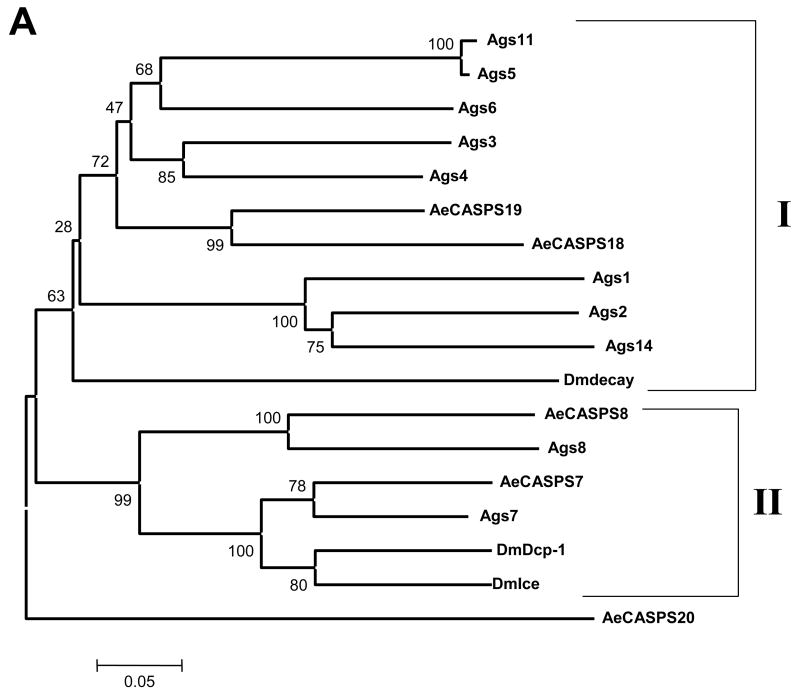
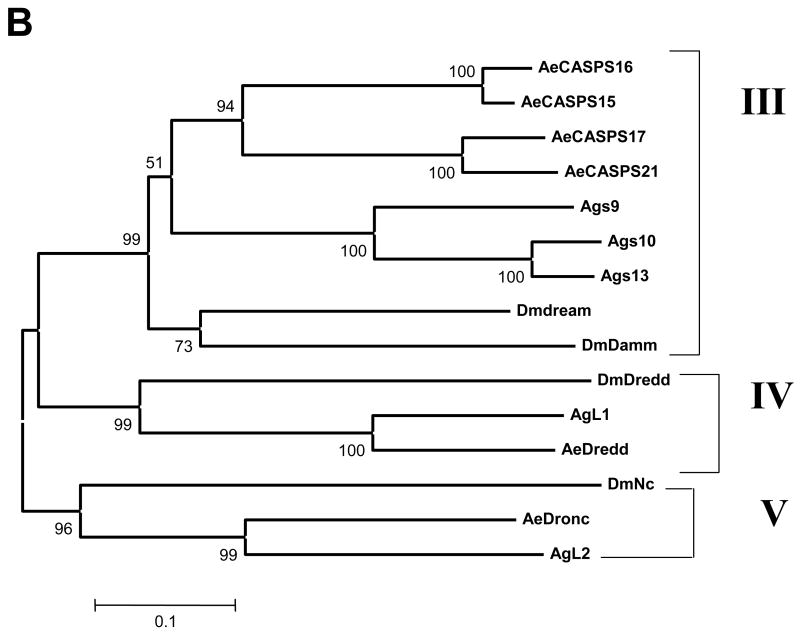
Phylogenetic analysis of the caspases was done using MEGA 3.1 (Kumar et al., 2004), utilizing the full-length amino acid sequences to analyze caspases from Ae. aegypti, An. gambiae and Drosophila. It should be noted that the predicted caspases from An. gambiae have not been confirmed yet, and many are not full length. The results of the phylogenetic analysis are illustrated in Fig. 1A (effector caspases) and B (initiator caspases). Drosophila Damm and its mosquito orthologs, which are predicted to be effector caspases based on their short prodomains, actually aligned more closely with the initiator caspases, and so are included in the phylogenetic analysis with initiator caspases.
Figure 1.
Phylogenetic analysis of caspases in Drosophila, An. gambiae, and Ae. aegypti. Full length amino acid sequences were used to build phylogenetic trees using MEGA 3.1. Panel A includes effector caspases, while panel B includes initiator caspases and caspases related to Damm, which is predicted to be an effector caspase based on its short prodomain, but which groups with initiator caspases. Clades were determined by branching patterns and are represented as vertical lines on the right. Bootstrap values are shown.
The effector caspases (Fig. 1A) are represented by two clades and an outlier caspase from Ae. aegypti, CASPS20. CASPS20 remained an outlier when all of the caspases were included in a single tree (not shown). Clade I includes only one caspase from Drosophila, decay. However, in both mosquito species, and especially in An. gambiae, there has been expansion of this gene. An. gambiae genes in Clade I include s3, s4, s5, s6, and s11, while Ae. aegypti is represented by CASPS19 and CASPS18. Also within Clade I there are additional caspases s1, s2 and s14 from An. gambiae. These are the only caspases in the analysis that have either serine or threonine instead of alanine in the active site sequence QAC(R/Q/G)(G/E) (Vernooy et al., 2000). Clade II includes caspases from all three species analyzed, but we were not able to distinguish clear orthologs for the effector caspases Ice and Dcp-1 from Drosophila, since Ice and Dcp-1 are more closely related to each other than to any of the mosquito caspases. However, CASPS7 and CASPS8 are the closest Ae. aegypti relatives to Drosophila Ice and Dcp-1.
Fig. 1B illustrates the phylogeny of the initiator caspases for the three dipteran species analyzed. The initiator caspases fall into three clades, Clades III through V. Clade III includes dream and Damm from Drosophila and their mosquito orthologs. It appears that Drosophila dream and Damm arose from a gene duplication event, which is particularly interesting since they differ in the lengths of their prodomains (dream has a long prodomain, while Damm has a short prodomain). The analysis suggests that these genes have been duplicated within each mosquito lineage. We note that the An. gambiae caspases s9 and s12 are extremely similar (98% nucleotide identity, including intron sequences). Thus we conclude these are alleles of the same gene, rather than two individual genes and so we have excluded s12 from our analysis. In contrast to Clade III, Clades IV and V include clear mosquito orthologs for Drosophila Nc and Dredd.
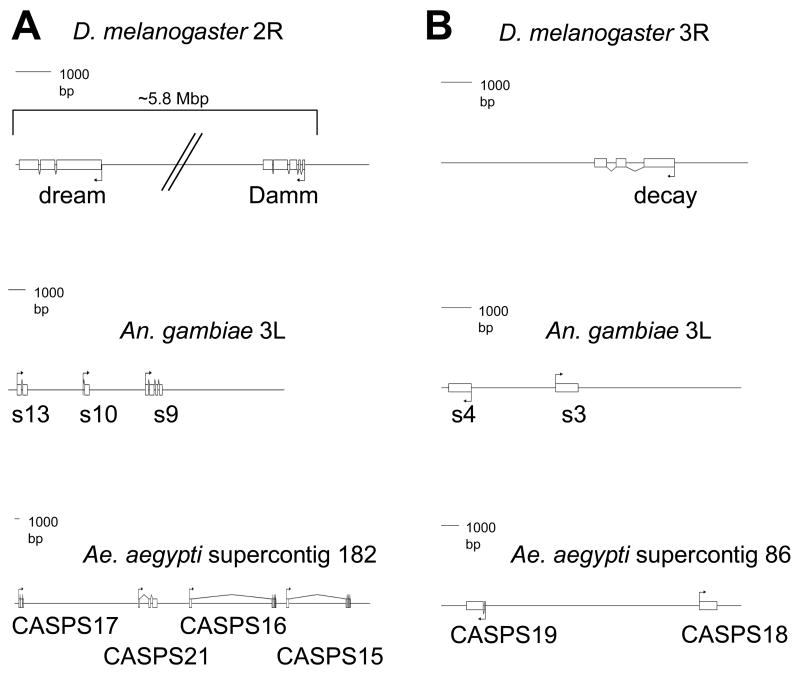
The caspase genes decay, Damm and dream all appear to have undergone duplication events in mosquitoes. Fig. 2 illustrates the genomic architecture for these duplications and how they compare in the three Dipteran species. CASPS15 and CASPS21 were found using gene prediction programs on supercontig 182, close to CASPS16 and CASPS17 (Fig. 2A). CASPS21 is a new caspase that was not previously identified. In Ae. aegypti, the predictions for CASPS16 and CASPS15 yielded introns with sizes of 16 and 11.5 Kbp. This is not surprising given that the Ae. aegypti genome contains a large number of transposable elements in the introns of genes, which has led to intron expansion (Nene et al., 2007). Accordingly, when we performed gene prediction analysis for the Damm and dream orthologs in Ae. aegypti, we found remnants of transposable elements in the introns of some of these genes. Fig. 2B shows the genome arrangements of expanded decay paralogs in mosquitoes.
Figure 2.
Genome architecture of caspase genes within three dipteran species. A, genome organization of Damm and dream in Drosophila and their paralogs in An. gambiae and Ae. aegypti. B, genome organization of Decay in Drosophila and its paralogs in An. gambiae and Ae. aegypti. Genome information was obtained from websites for each organism, as explained in Materials and Methods. The illustrations were produced using GenePalette.
Caspase Regulators
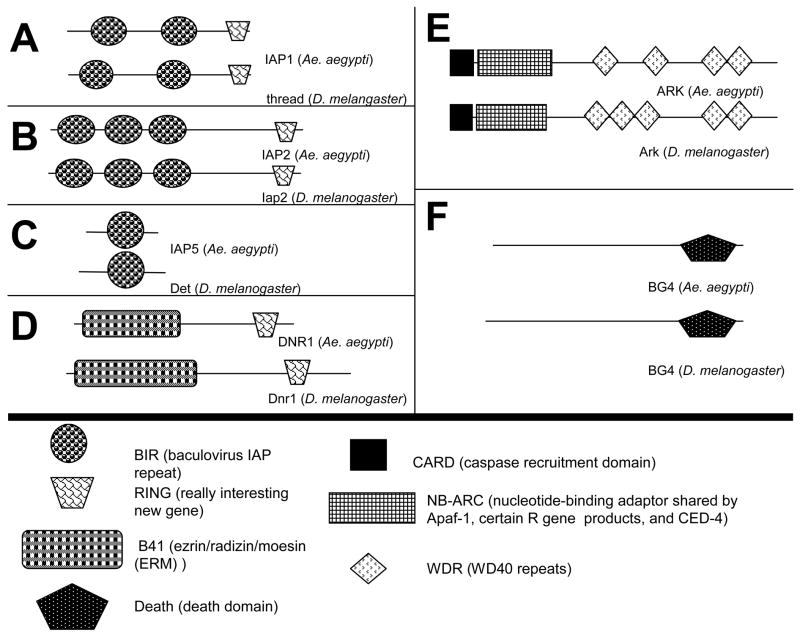
In addition to caspases, we also identified a number of genes that regulate caspases. This group includes proteins that are known in Drosophila to inhibit caspase activity or are involved in caspase activation. Fig. 3 illustrates the protein domains for the predicted Ae. aegypti proteins from the caspase regulator group and their Drosophila orthologs.
Figure 3.
Predicted domain architecture for some of the proteins identified in this study. Protein domains were predicted using SMART. Proteins for Drosophila were obtained from NCBI and FlyBase.
Among the genes in this category that we identified are members of the IAP family. We found five IAP homologs in the Ae. aegypti genome, but we only characterized three. Of the two that we did not characterize, one represents a dBruce ortholog and the other contains a single BIR domain, but determination of its 3’ end was problematic due to the presence of transposable elements. The three IAP proteins that we characterized and their domains are illustrated in Fig. 3A-C. These Ae. aegypti IAPs have a protein architecture similar to that of the Drosophila IAPs. The first, which is designated as IAP1, is the ortholog of thread (th) and is represented in NCBI as ABK1289. The Ae. aegypti ortholog of Iap2 from Drosophila is represented in NCBI as EAT41756, but previous gene predictions do not appear to represent the full transcript because they do not include an initiating methionine. We employed GenScan and found an apparent initiator methionine in supercontig 214. We were able to amplify the predicted transcript from the ATC-10 cell line using RT-PCR, verifying the gene prediction results. This analysis added 90 amino acids to the N-terminus of Ae. aegypti IAP2. IAP5, the ortholog of Drosophila det (also called Deterin), is represented in NCBI as EAT33476. GenScan predictions did not result in additional sequences being included in this transcript.
In addition to members of the IAP family, we also identified the Ae. aegypti ortholog of the caspase inhibitor Dnr1. Ae. aegypti DNR1 has a predicted protein domain architecture similar to that of Dnr1 from Drosophila, as illustrated in Fig. 3D. The ESTs encoding the Ae. aegypti DNR1 did not contain an initiation codon, but did yield 3’ stop codons. However the predicted protein in NCBI (EAT48387) has an initiation codon. We were able to verify the NCBI-predicted transcript by RT-PCR from ATC-10 cells.
We also identified genes that encode activators of caspases. The Ae. aegypti ortholog for Ark was found by using three domains found in Ark, the CARD (caspase recruitment domain), NB-ARC (nucleotide-binding adaptor shared by Apaf-1, certain R gene products and CED-4), and WD-40 domains, in individual BLAST searches against NCBI predicted proteins for Ae. aegypti. Two predicted proteins in NCBI, EAT48065 and EAT48066, contained these domains but were in separate predicted proteins found relatively close to each other. EAT48065 encodes CARD and NB-ARC domains, while EAT 48066 encodes WD-40 repeat domains. Primers were designed from the 5’ end of EAT48065 and 3’ end of EAT48066, and we were able to amplify a single continuous transcript from ATC-10 cells, thus confirming that these domains are all part of a single protein. Fig. 3E illustrates that Ae. aegypti ARK and Drosophila Ark are highly similar in their architecture, except for one fewer WD-40 domain in Ae. aegypti ARK. BG4 is also a known activator of caspases in Drosophila. The Ae. aegypti BG4 ortholog has been previously annotated (Waterhouse et al., 2007). Our bioinformatics analysis yielded a full-length transcript that is represented in NCBI as EAT46931. We were able to verify the full-length Ae. aegypti BG4 transcript by RT-PCR from ATC-10 cells.
Another group of proteins involved in caspase activation are the IAP antagonists, including rpr, W (wrinkled), grim and skl in Drosophila. These proteins do not share significant similarity, except for a small motif called the IAP Binding Motif (IBM) found at the N-terminus. By using Michelob_x as a query, we identified another protein containing an IBM in Ae. aegypti. The EST DV326893 (obtained from D. Severson) contained the transcript with 5’ and 3’ flanking initiation and stop codons. This gene, which we named IMP, is represented in NCBI as EAT44230.
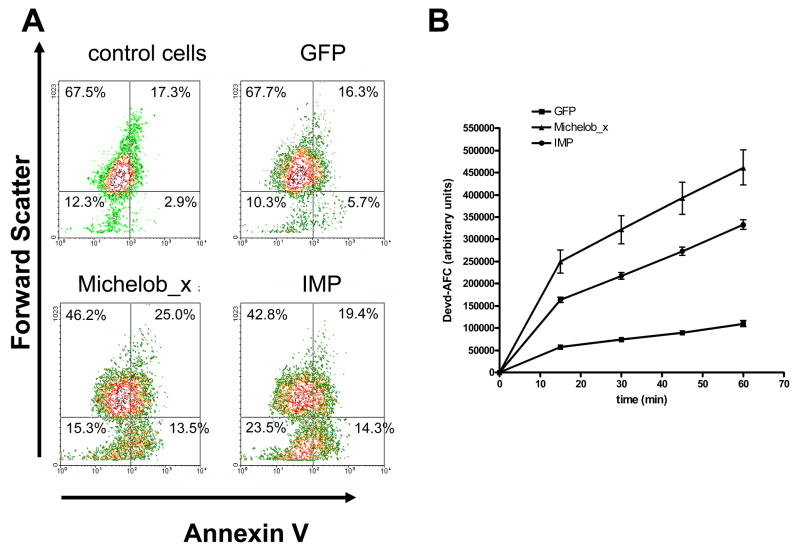
Because of the low level of similarity between IAP antagonists, we sought to verify that IMP encodes a functional pro-apoptotic gene. The IMP cDNA was cloned into an expression vector and expressed in C6/36 cells by transient transfection. Cells overexpressing IMP were analyzed at 24 h post-transfection for Annexin V staining (a marker for early apoptosis) by flow cytometry (Fig. 4A) and for caspase activity by incubating lysate from transfected cells with the (human) caspase-3 substrate Ac-DEVD-AFC and analyzing AFC fluorescence (Fig. 4B). Expression of either Michelob_x or IMP, but not GFP, resulted in higher Annexin V staining (shown by a shift to the right) in a portion of the cells consistent with the level of transfection efficiency routinely observed using these cells (around 20%). There was also a decrease in cell size (shown by a downward shift) upon expression of Michelob_x or IMP, which is also characteristic of apoptotic cells.
Figure 4.
Expression of IMP or Michelob_x causes apoptosis in C6/36 cells. A, AnnexinV staining (X-axis) versus forward scatter (Y-axis) of untransfected (control) cells or cells transfected with constructs expressing GFP, Michelob_x, or IMP, as analyzed by flow cytometry. Percentages are given for each quadrant. Each graph represents analysis of 10,000 cells. B, caspase activity in lysates from cells expressing GFP, Michelob_x, or IMP as determined by liberated AFC fluorescence over 60 min incubation with the caspase substrate Ac-DEVD-AFC. The data shown represent the combined results from three independent transfections.
In support of the flow cytometry results, we also observed increased effector caspase activity in C6/36 cells expressing Michelob_x or IMP. Lysates from cells expressing either Michelob_x or IMP cleaved significantly higher amounts of the effector caspase substrate Ac-DEVD-AFC during a 60 min incubation period than control cells expressing GFP (Fig. 4B). Cells expressing Michelob_x or IMP also exhibited blebbing morphology typical of apoptotic cells (data not shown). Together, these results verify that IMP is a pro-apoptotic protein similar to Michelob_x.
Expression Analysis
To examine the expression of these potential regulators of apoptosis throughout the life cycle of Ae. aegypti, we employed quantitative reverse transcriptase PCR (RT-PCR). We tested early larvae (pooled L1 and L2 larval stages), late larvae (pooled L3 and L4 larval stages), pupae, and adults. For the adults, we analyzed females only, and analyzed midguts separately from the rest of the adult insect.
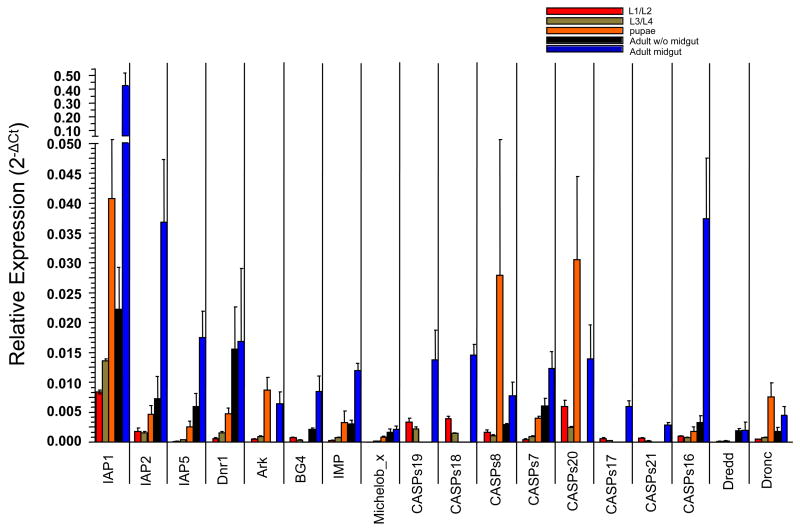
Fig. 5 illustrates the quantitative RT-PCR results for the genes across the five stages and tissues, with the exception of CASPS15. Although we did not detect expression of CASPS15 in any of the samples we assayed, there are ESTs corresponding to CASPS15 in NCBI, indicating that it is an expressed gene. The RT-PCR results were normalized by comparison to act-6 expression and are shown as 2−ΔCt. We observed that expression was highest in the adult female midgut for a majority of the genes, with exceptions being ARK, Dronc, CASPS8 and CASPS20, which showed the highest expression in pupae. The larval stages tended to have the lowest expression for most genes. Caspases were expressed at the highest levels in the midgut. ARK, CASPS17, CASPS21, CASPS20, CASPS19 and CASPS18 did not show Ct values above background in the adult body minus the midgut, while BG4, CASPS17, CASPS21, Dredd, CASPS19 and CASPS18 did not show Ct values above background in pupae. Michelob_x was the only gene that did not show expression above background levels in early larvae (L1/L2). In these cases where Ct values were not above background, it is still possible that these genes are expressed in these stages or tissues, but at low levels. These results suggest that different components of the apoptotic machinery are expressed at varying levels in different developmental stages and tissues in Ae. aegypti.
Figure 5.
Quantitative analysis of transcript levels for the annotated apoptosis regulatory genes in Ae. aegypti throughout the life cycle of the organism. The results of real time RT-PCR analysis in early larvae (L1/L2), late larvae (L3/L4), pupae, adult females without midgut, or adult female midgut are shown. Each data point represents the average 2−ΔCt (+/−SE) obtained from three batches of cDNA made from each stage or tissue.
Discussion
This study reports the annotation of a number of potential apoptosis related genes in Ae. aegypti. To date, our understanding of apoptosis in insects is based almost entirely on studies from Drosophila. This study represents the most thorough attempt to date to identify apoptosis related genes in any other insect. With the exception of IAP1 (Beck et al., 2007), Dredd (Cooper et al., 2007a), Dronc (Cooper et al., 2007b), and Michelob_x (Zhou et al., 2005a), the genes reported here have not been previously verified by cDNA analysis. In silico analysis helps tremendously in annotating genes, but the importance of verifying cDNA sequences is illustrated by the differences we observed between predicted proteins and cDNA comparisons with ARK, CASPS7, CASPS20 and IAP2. We also identified one additional caspase (CASPS21) that has not been previously annotated. The genes identified in this study hold promise for improving our understanding of apoptotic regulation in the important disease vector Ae. aegypti. In addition to elucidating the mechanics of apoptotic regulation, identification of these genes promises to aide in improving our understanding of innate immunity in the yellow fever mosquito. Several of the genes examined in this study are thought to play important roles in Drosophila immunity, including Dredd, Iap2, BG4, and Dnr1. It is likely that these genes are also important in Ae. aegypti immunity, but further study is needed to determine their exact roles in this process.
Apparent gene duplications have occurred with several caspase genes in both Ae. aegypti and An. gambiae. At this time it is not clear why mosquitoes possess more caspase genes than Drosophila. One possibility is that there is more of a need for caspases in regulating the innate immune responses in mosquitoes, since they may be exposed to more potential pathogens because of their hemophagic life style. For Ae. aegypti, these gene duplications are seen in Clade I (CASPS19 and CASPS18) and Clade III (CASPS16, CASPS15, CASPS17 and CASPS21). In Clade I, the Drosophila gene decay is expanded in both mosquito species analyzed. In An. gambiae there are eight homologs of this gene while Ae. aegypti has two homologs. The function of decay has not been studied in detail in Drosophila (Dorstyn et al., 1999), but there may be additional selective pressures for these caspases in mosquitoes. An interesting observation is that CASPS18 has a serine instead of a cysteine in its active site. This makes it unlikely that CASPS18 encodes a functional caspase. Similar levels of expression were observed for both CASPS19 and CASPS18 throughout the life cycle of Ae. aegypti. Thus it is possible that CASPS18 regulates CASPS19 in a dominant-negative manner, similar to what has been shown in humans with caspase-1 being regulated by Pseudo-ICE and ICEBERG (Druilhe et al., 2001).
Clade III includes duplications for both Damm and dream in the mosquitoes. The phylogeny of Damm and dream is interesting, since this appears to be a case where gene duplication occurred and one of the duplicated genes later either acquired or lost a long prodomain sequence. While dream appears to be an initiator caspase, whether Damm is an effector or initiator caspase is not entirely clear. The phylogeny results would suggest that Damm may be an unusual type of initiator caspase, although it is also possible that Damm may not be correctly annotated in any of these insect species. Damm and dream have also not been extensively studied in Drosophila (Doumanis et al., 2001; Harvey et al., 2001), and so studies of these duplicated genes in mosquitoes should not only lead to a better understanding of apoptosis and innate immunity in mosquitoes, but also may help in gaining a better understanding of the role of decay, Damm and dream in Drosophila.
When IMP is used as a BLAST query, a representative for this gene does not appear in other mosquitoes. In Drosophila, the RHG genes share very little similarity, so this is not surprising. However, when Michelob_x from Ae. aegypti is used as a query, one can easily find An. gambiae and Ae. albopictus orthologs of Michelob_x. Thus it appears that IMP is not as well conserved as Michelob_x in mosquitoes. Our results indicate that IMP is a pro-apoptotic gene that likely functions as an IAP antagonist, similar to Michelob_x. In Drosophila, there are at least four IAP antagonists (rpr, grim, W, and skl). Thus it is likely that there are additional IAP antagonists in the genomes of the mosquitoes, but their level of similarity is too low to detect by traditional BLAST searches.
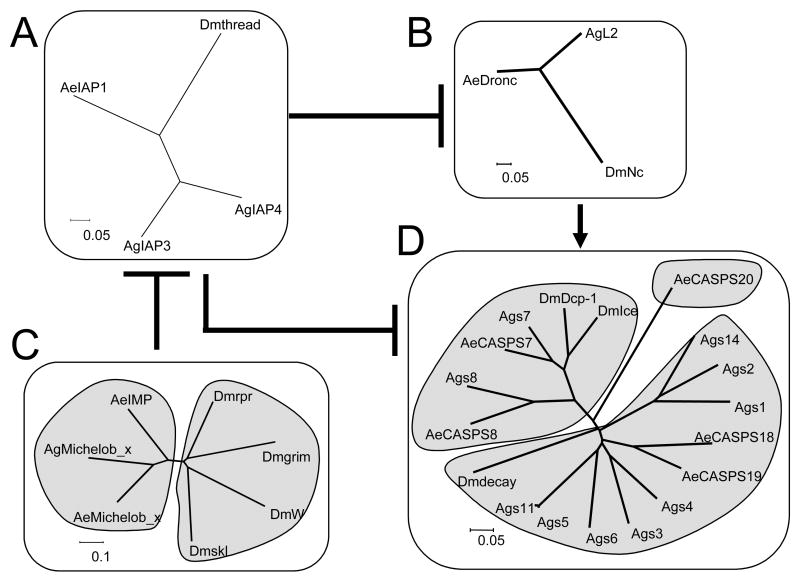
Fig. 6 illustrates the differences between mosquitoes and Drosophila in the numbers of genes involved in the core apoptotic pathway, based on the pathway that has been established in Drosophila. In Drosophila, Nc is the main initiator caspase that activates Ice and Dcp-1, which ultimately leads to death. Thread has been shown to inhibit both Nc and Ice, and IAP antagonist interactions with IAP proteins have also been studied extensively (Chai et al., 2003; Yan et al., 2004). According to this simplified model, thread is required to prevent the accumulation of active Nc, which appears to be constitutively activated in an ARK-dependent manner. IAP antagonists interrupt the interaction between Nc and thread. This interaction frees Nc, which is then able to activate Ice, resulting in apoptosis (Hay and Guo, 2006). Fig. 6C illustrates the high amount of divergence among the IAP antagonists in different insects. However, even though these genes are extremely poorly conserved except for their IBM motifs, the genes still cluster in a species-specific manner. In Fig. 6D we illustrate the expansion that has occurred for the effector caspases in mosquitoes. Another interesting phenomenon is that An. gambiae has two paralogs of thread, while Ae. aegypti has only one version of this gene, as does Drosophila. While the effector caspases and IAP1 genes have duplications in mosquitoes, other genes that make up the core apoptotic pathway have not gone through a duplication event, such as the initiator caspase Nc and the adaptor protein ARK.
Figure 6.
Illustration of the conservation of core apoptosis regulatory genes within Drosophila, An. gambiae and Ae. aegypti. The unrooted trees were built using neighbor end joining, complete deletion, p-distance with full amino acid sequences. The pathway illustrated is based on information obtained from studies using Drosophila. In Drosophila, thread inhibits Nc and Ice, while thread itself is inhibited by the IBM-containing proteins rpr, W, and grim. The initiator caspase Nc is responsible for cleaving and activating effector caspases including Ice and Dcp-1. Nc is activated by Ark, which for simplicity is not shown but which is also conserved in Ae. aegypti.
For a productive infection and transmission cycle to occur, pathogens such as arboviruses and Plasmodium must overcome many barriers in the mosquito vector. These pathogens must establish infection of the midgut epithelium and replicate in these cells, escape from this barrier by passing through the basal lamina, replicate efficiently in other organs, and finally infect the salivary glands and penetrate into the lumen of the salivary glands for transmission (Black et al., 2002). If any one of these barriers is not crossed, the pathogen cannot establish infection and be transmitted from mosquito to a vertebrate host. Interestingly, the midgut showed higher expression for many of the presumptive apoptotic regulating genes as compared to the other tissues or stages. Whether this apparent high level of expression is due to differences in act-6 expression between tissues or stages is not known at this time (since transcript levels were normalized against act-6). In addition, it must be kept in mind that the level of protein may not correspond to the level of transcript. Especially intriguing is that expression of IAP1 was much higher in midgut than in any other tissue or stage analyzed. Based on the known crucial role of thread and SfIAP in preventing spontaneous apoptosis in Drosophila and Spodoptera frugiperda cells, respectively (Igaki et al., 2002; Muro et al., 2002; Zimmermann et al., 2002), it is tempting to speculate that Ae. aegypti IAP1 might have a similar role in regulating apoptosis. This needs to be determined, but if correct, then high levels of IAP1 expression may function to protect midgut cells from spontaneous apoptosis. It has been shown in An. stephensi (Han et al., 2000; Abraham et al., 2004), An. gambiae (Vlachou et al., 2004), and Ae. aegypti (Zieler and Dvorak, 2000) that apoptosis occurs during the establishment of infection in the midgut by different Plasmodium spp. It has also been demonstrated in An. gambiae by microarray analysis that transcript levels of the thread homologs AgIAP3 and AgIAP4 are up-regulated during midgut invasion by P. berghei and down-regulated after invasion (Vlachou et al., 2005). This suggests that, similar to thread and SfIAP, down regulation of AgIAP3 and AgIAP4 by Plasmodium infection may play a role in triggering apoptosis in midgut cells. Depending on the pathogen, regulation of IAP1 levels could possibly play a role in establishment of infection of Ae. aegypti midgut by Plasmodium and/or arboviruses.
Table 3.
Primers utilized for expression analysis of presumptive apoptotic regulators of Ae. aegypti.
| Gene | Forward | Reverse |
|---|---|---|
| IAP1 | CTGAAACTAATGAAGGGCGAAGC | TTGAGATGACTGAAGCGAGGATG |
| IAP2 | CCATTATCGTCGCCGTCTACC | CTTTCAGTCGTTTGTTCTCCTCTTC |
| IAP5 | ACGACAAGGAGGACGAAGAC | TCCAGCAGGTTAAGCATTTCC |
| DNR1 | GAAGATACTGAACTCGGCAAACG | CGGCAGGCGGTAATATGTCC |
| ARK | TGTCTAGCGTTTCGGTCTTGAG | GCGTTGGTTAGCCTGGATAATAATC |
| BG4 | ACTTTGCCTGCTCAATTTCTTTCTC | GATACGCTGTTCTCCCTGTTGG |
| Michelob_x | CAACAGCAAAATCAGAACCAAATCAG | GCACAGCAGACATCGGGAAC |
| IMP | GCTGGACTGAGAACGCCTTC | ACGACTGATGAGAACAACAACAAC |
| CASPS19 | CTCGCCGTGTGACATCATAAC | AAGCAAGGAAGTTCTCGTTTCTC |
| CASPS18 | CTGTCTTGTGGTAGTTGTGATGTC | CGGATGCTTGTGATTCTTCTTCTC |
| CASPS8 | TGGCAAAAGCAAACAGGAAGTC | GGGATGAAGGCGAAGTAATATACG |
| CASPS7 | TTGGCAGAACGCACCGAAAC | CGAAAGTCAGCAGGGTCAGTAG |
| CASPS20 | GCGGATTGCCTGATGGTATTC | ATGCTTGGACTATGAACAACTTCG |
| Dredd | AGAAGTATGTAATATCGTGGAAGAATGC | AGAACAGTGATGCGGCTCAAC |
| Dronc | CAACTTTCCAACTGCCTATAAATTGC | CTCCACCGTATCGTTATTGTTCTTAG |
| CASPS17 | TGCCATTGATGAGAAGAGAATTTGAG | GCCTACTTGTCCCGTGTTACC |
| CASPS21 | CGATTGTAATAAAACGGTTCCTAGTCC | CTATTGACATTTCTGGCATCTCTCTTAG |
| CASPS16 | TCCGCTATCTTCATATTGTATCCTTTG | GACCCGCCACTGTATCTCTG |
| CASPS15 | CCTAACTTGGGTTTGACGATTGC | AATGTCCGCTATCTTCATATTGTATCC |
| Act-6 | AAGGCTAACCGTGAGAAGATGAC | GATTGGGACAGTGTGGGAGAC |
Acknowledgments
We thank David Severson (University of Notre Dame) for providing ESTs from Ae. aegypti, which helped tremendously in determining gene annotation. We also thank Lei Zhou (University of Florida) for Michelob_x cDNA and Lei Zhou and Kristin Michel (Kansas State University) for helpful comments on the manuscript, Ken Jones and Joseph Coolon (Kansas State University) for their expertise in real-time PCR analysis, Scott Bernhardt (Colorado State University) for isolation of life stages and midgut tissues of Ae. aegypti, and Teresa Shippy (Kansas State University) for help with GenePalette. We also thank SuHao Han and Hua Wang for 5’ RACE analysis of CASPS20. This research was supported by NIH grants R21AI067642 (to R.J.C.), R01AI32543 (to C.D.B.), R01AI46435 (to K.E.O.), P20 RR107686 from the National Center for Research Resources (NCRR), P20 RR16475 from the BRIN Program of the NCRR, the Terry C. Johnson Center for Basic Cancer Research, and by the Kansas Agricultural Experiment Station. This is contribution number 08-71-J from the Kansas Agricultural Experiment Station.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham EG, Islam S, Srinivasan P, Ghosh AK, Valenzuela JG, Ribeiro JMC, Kafatos FC, Dimopoulos G, Jacobs-Lorena M. Analysis of the Plasmodium and Anopholes transcriptional repertoire during ookinete development and midgut invasion. J Biol Chem. 2004;279:5573–5580. doi: 10.1074/jbc.M307582200. [DOI] [PubMed] [Google Scholar]
- Bao Q, Shi Y. Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ. 2007;14:56–65. doi: 10.1038/sj.cdd.4402028. [DOI] [PubMed] [Google Scholar]
- Bartholomay LC, Cho WL, Rocheleau TA, Boyle JP, Beck ET, Fuchs JF, Liss P, Rusch M, Butler KM, Wu RC, Lin SP, Kuo HY, Tsao IY, Huang CY, Liu TT, Hsiao KJ, Tsai SF, Yang UC, Nappi AJ, Perna NT, Chen CC, Christensen BM. Description of the transcriptomes of immune response-activated hemocytes from the mosquito vectors Aedes aegypti and Armigeres subalbatus. Infect Immun. 2004;72:4114–4126. doi: 10.1128/IAI.72.7.4114-4126.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck ET, Blair CD, Black WCI, Beaty BJ, Blitvich BJ. Alternative splicing generates multiple transcripts of the inhibitor of apoptosis protein 1 in Aedes and Culex spp. mosquitoes. Insect Biochem Mol Biol. 2007 doi: 10.1016/j.ibmb.2007.07.015. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann A, Agapite J, McCall K, Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–341. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- Black WCI, Bennett KE, Gorrochotegui-Escalante N, Barillas-Mury CV, Fernandez-Salas I, de Lourdes Munoz M, Farfan-Ale JA, Olson KE, Beaty BJ. Flavivirus susceptibility in Aedes aegypti. Arch Med Res. 2002;33:379–388. doi: 10.1016/s0188-4409(02)00373-9. [DOI] [PubMed] [Google Scholar]
- Blitvich BJ, Blair CD, Kempf BJ, Hughes MT, Black WC, Mackie RS, Meredith CT, Beaty BJ, Rayms-Keller A. Developmental- and tissue-specific expression of an inhibitor of apoptosis protein 1 homologue from Aedes triseriatus mosquitoes. Insect Molec Biol. 2002;11:431–442. doi: 10.1046/j.1365-2583.2002.00352.x. [DOI] [PubMed] [Google Scholar]
- Bowers DF, Coleman CG, Brown DT. Sindbis virus-associated pathology in Aedes albopictus (Diptera: Culicidae) J Med Entomol. 2003;40:698–705. doi: 10.1603/0022-2585-40.5.698. [DOI] [PubMed] [Google Scholar]
- Chai J, Yan N, Huh JR, Wu J-W, Li W, Hay BA, Shi Y. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol. 2003;10:892–898. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- Chen P, Nordstrom W, Gish B, Abrams J. grim, a novel cell death gene in Drosophila. Genes Devel. 1996;10:1733–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Chinnaiyan AM, Tepper CG, Seldin MF, O’Rourke K, Kischkel FC, Hellbardt S, Krammer PH, Peter ME, Dixit VM. FADD/MORT1 is a common mediator of CD95 (Fas/APO-1) and tumor necrosis factor receptor-induced apoptosis. J Biol Chem. 1996;271:4961–4965. doi: 10.1074/jbc.271.9.4961. [DOI] [PubMed] [Google Scholar]
- Clarke TE, Clem RJ. Lack of involvement of haemocytes in the establishment and spread of infection in Spodoptera frugiperda larvae infected with the baculovirus Autographa californica M nucleopolyhedrovirus by intrahemocoelic injection. J Gen Virol. 2002;83:1565–1572. doi: 10.1099/0022-1317-83-7-1565. [DOI] [PubMed] [Google Scholar]
- Clem RJ. The role of apoptosis in defense against baculovirus infection in insects. Curr Top Microbiol Immunol. 2005;289:113–130. doi: 10.1007/3-540-27320-4_5. [DOI] [PubMed] [Google Scholar]
- Cooper DM, Pio F, Thi EP, Theilmann D, Lowenberger C. Characterization of Aedes Dredd: a novel initiator caspase from the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2007a;37:559–569. doi: 10.1016/j.ibmb.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Cooper DM, Thi EP, Chamberlain CM, Pio F, Lowenberger C. Aedes Dronc: a novel ecdysone-inducible caspase in the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2007b doi: 10.1111/j.1365-2583.2007.00758.x. in press. [DOI] [PubMed] [Google Scholar]
- Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Read SH, Quinn LM, Richardson H, Kumar S. DECAY, a novel Drosophila caspase related to mammalian caspase-3 and caspase-7. J Biol Chem. 1999;274:30778–30783. doi: 10.1074/jbc.274.43.30778. [DOI] [PubMed] [Google Scholar]
- Doumanis J, Quinn L, Richardson H, Kumar S. STRICA, a novel Drosophila melanogaster caspase with an unusual serine/threonine-rich prodomain, interacts with DIAP1 and DIAP2. Cell Death Differ. 2001;8:387–394. doi: 10.1038/sj.cdd.4400864. [DOI] [PubMed] [Google Scholar]
- Druilhe A, Srinivasula SM, Razmara M, Ahmad M, Alnemri ES. Regulation of IL-1beta generation by Pseudo-ICE and ICEBERG, two dominant negative caspase recruitment domain proteins. Cell Death Differ. 2001;8:649–657. doi: 10.1038/sj.cdd.4400881. [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson M, Mishra S, Schneider D. Interactions between the cellular and humoral immune responses in Drosophila. Curr Biol. 2000;10:781–784. doi: 10.1016/s0960-9822(00)00569-8. [DOI] [PubMed] [Google Scholar]
- Foley E, O’Farrell PH. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2004;2:E203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesellchen V, Kuttenkeuler D, Steckel M, Pelte N, Boutros M. An RNA interference screen identifies Inhibitor of Apoptosis Protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep. 2005;6:979–984. doi: 10.1038/sj.embor.7400530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard YA, Popov V, Wen J, Han V, Higgs S. Ultrastructural study of West Nile virus pathogenesis in Culex pipiens quinquefasciatus (Diptera: Culicidae) J Med Entomol. 2005;42:429–444. doi: 10.1093/jmedent/42.3.429. [DOI] [PubMed] [Google Scholar]
- Girard YA, Schneider BS, McGee CE, Wen J, Han VC, Popov V, Mason PW, Higgs S. Salivary gland morphology and virus transmission during long-term cytopathologic West Nile virus infection in Culex mosquitoes. Am J Trop Med Hyg. 2007;76:118–128. [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Han YS, Thompson J, Kafatos FC, Barillas-Mury C. Molecular interactions between Anopheles stephensi midgut cells and Plasmodium berghei: the time bomb theory of ookinete invasion of mosquitoes. EMBO J. 2000;19:6030–6040. doi: 10.1093/emboj/19.22.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey NL, Daish T, Mills K, Dorstyn L, Quinn LM, Read SH, Richardson H, Kumar S. Characterization of the Drosophila caspase, DAMM. J Biol Chem. 2001;276:25342–25350. doi: 10.1074/jbc.M009444200. [DOI] [PubMed] [Google Scholar]
- Hay BA, Guo M. Caspase-dependent cell death in Drosophila. Annu Rev Cell Dev Biol. 2006;22:623–650. doi: 10.1146/annurev.cellbio.21.012804.093845. [DOI] [PubMed] [Google Scholar]
- Hay BA, Huh JR, Guo M. The genetics of cell death: approaches, insights and opportunities in Drosophila. Nat Rev Genet. 2004;5:911–922. doi: 10.1038/nrg1491. [DOI] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Horng T, Medzhitov R. Drosophila MyD88 is an adaptor in the Toll signaling pathway. Proc Natl Acad Sci USA. 2001;98:12654–12658. doi: 10.1073/pnas.231471798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, Langendijk-Genevaux PS, Pagni M, Sigrist CJA. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T, Yamamoto-Goto Y, Tokushige N, Kanda H, Miura M. Downregulation of DIAP1 triggers a novel Drosophila cell death pathway mediated by Dark and Dronc. J Biol Chem. 2002;277:23103–23106. doi: 10.1074/jbc.C200222200. [DOI] [PubMed] [Google Scholar]
- James ER, Green DR. Manipulation of apoptosis in the host-parasite interaction. Trends Parasitol. 2004;20:280–287. doi: 10.1016/j.pt.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Kleino A, Valanne S, Ulvila J, Kallio J, Myllymaki H, Enwald H, Stoven S, Poidevin M, Ueda R, Hultmark D, Lemaitre B, Ramet M. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila. Imd pathway EMBO J. 2005;24:3423–3434. doi: 10.1038/sj.emboj.7600807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Cakouros D. Transcriptional control of the core cell-death machinery. Trends Biochem Sci. 2004;29:193–199. doi: 10.1016/j.tibs.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. The Drosophila caspase Dredd is required to resist Gram-negative bacterial infection. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Li H, Blitvich BJ, Zhang J. The Aedes albopictus inhibitor of apoptosis 1 gene protects vertebrate cells from bluetongue virus-induced apoptosis. Insect Mol Biol. 2007;16:93–105. doi: 10.1111/j.1365-2583.2007.00705.x. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Bryant SH. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means JC, Muro I, Clem RJ. Lack of involvement of mitochondrial factors in caspase activation in a Drosophila cell-free system. Cell Death Diff. 2006;13:1222–1234. doi: 10.1038/sj.cdd.4401821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mims CA, Day MF, Marshall ID. Cytopathic effect of Semliki Forest virus in the mosquito Aedes aegypti. Am J Trop Med Hyg. 1966;15:775–784. doi: 10.4269/ajtmh.1966.15.775. [DOI] [PubMed] [Google Scholar]
- Muro I, Hay BA, Clem RJ. The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J Biol Chem. 2002;277:49644–49650. doi: 10.1074/jbc.M203464200. [DOI] [PubMed] [Google Scholar]
- Muro I, Monser K, Clem RJ. Mechanism of Dronc activation in Drosophila cells. J Cell Sci. 2004;117:5035–5041. doi: 10.1242/jcs.01376. [DOI] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O’Leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman JT, Korsmeyer SJ. Apoptosis in the development and maintenance of the immune system. Nat Immunol. 2003;4:410–415. doi: 10.1038/ni0503-410. [DOI] [PubMed] [Google Scholar]
- Primrose DA, Chaudhry S, Johnson AG, Hrdlicka A, Schindler A, Tran D, Foley E. Interactions of DNR1 with the apoptotic machinery of Drosophila melanogaster. J Cell Sci. 2007;120:1189–1199. doi: 10.1242/jcs.03417. [DOI] [PubMed] [Google Scholar]
- Sanders HR, Foy BD, Evans AM, Ross LS, Beaty BJ, Olson KE, Gill SS. Sindbis virus induces transport processes and alters expression of innate immunity pathway genes in the midgut of the disease vector, Aedes aegypti. Insect Biochem Mol Biol. 2005;35:1293–1307. doi: 10.1016/j.ibmb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoven S, Ando I, Kadalayil L, Engstrom Y, Hultmark D. Activation of the Drosophila NF-kappaB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan R, Scott TW. Apoptosis in mosquito midgut epithelia associated with West Nile virus infection. Apoptosis. 2006;11:1643–1651. doi: 10.1007/s10495-006-8783-y. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Silke J. IAPs, RINGs and ubiquitylation. Nat Rev Molec Cell Biol. 2005;6:287–297. doi: 10.1038/nrm1621. [DOI] [PubMed] [Google Scholar]
- Vernooy SY, Copeland J, Ghaboosi N, Griffin EE, Yoo SJ, Hay BA. Cell Death Regulation in Drosophila: Conservation of Mechanism and Unique Insights. The Jounal of Cell Biology. 2000;150:F69–F75. doi: 10.1083/jcb.150.2.f69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachou D, Schlegelmilch T, Christophides GK, Kafatos FC. Functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium invasion. Curr Biol. 2005;15:1185–1195. doi: 10.1016/j.cub.2005.06.044. [DOI] [PubMed] [Google Scholar]
- Vlachou D, Zimmermann T, Cantera R, Janse CJ, Waters AP, Kafatos FC. Real-time, in vivo analysis of malaria ookinete locomotion and mosquito midgut invasion. Cell Microbiol. 2004;6:671–685. doi: 10.1111/j.1462-5822.2004.00394.x. [DOI] [PubMed] [Google Scholar]
- Vucic D, Kaiser WJ, Harvey AJ, Miller LK. Inhibition of Reaper-induced apoptosis by interaction with inhibitor of apoptosis proteins (IAPs) Proc Natl Acad Sci USA. 1997;94:10183–10188. doi: 10.1073/pnas.94.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucic D, Kaiser WJ, Miller LK. Inhibitor of apoptosis proteins physically interact with and block apoptosis induced by Drosophila proteins HID and GRIM. Mol Cell Biol. 1998;18:3300–3309. doi: 10.1128/mcb.18.6.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, Blandin S, Christensen BM, Dong Y, Jiang H, Kanost MR, Koutsos AC, Levashina EA, Li J, Ligoxygakis P, Maccallum RM, Mayhew GF, Mendes A, Michel K, Osta MA, Paskewitz S, Shin SW, Vlachou D, Wang L, Wei W, Zheng L, Zou Z, Severson DW, Raikhel AS, Kafatos FC, Dimopoulos G, Zdobnov EM, Christophides GK. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Scott TW, Lorenz LH, Lerdthusnee K, Romoser WS. Togavirus-associated pathologic changes in the midgut of a natural mosquito vector. J Virol. 1988;62:2083–2090. doi: 10.1128/jvi.62.6.2083-2090.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Steller H, Meier P. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- Yan SJ, Wu JW, Chai J, Li W, Shi Y. Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nat Struct Mol Biol. 2004;11:420–428. doi: 10.1038/nsmb764. [DOI] [PubMed] [Google Scholar]
- Yu X, Wang L, Acehan D, Wang X, Akey CW. Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J Mol Biol. 2006;355:577–589. doi: 10.1016/j.jmb.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Zhou L, Jiang G, Chan G, Santos CP, Severson DW, Xiao L. Michelob_x is the missing inhibitor of apoptosis protein antagonist in mosquito genomes. EMBO Rep. 2005a;6:769–774. doi: 10.1038/sj.embor.7400473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Silverman N, Hong M, Liao DS, Chung Y, Chen ZJ, Maniatis T. The role of ubiquitination in Drosophila innate immunity. J Biol Chem. 2005b;280:34048–34055. doi: 10.1074/jbc.M506655200. [DOI] [PubMed] [Google Scholar]
- Zieler H, Dvorak JA. Invasion in vitro of mosquito midgut cells by the malaria parasite proceeds by a conserved mechanism and results in death of the invaded midgut cells. Proc Natl Acad Sci USA. 2000;97:11516–11521. doi: 10.1073/pnas.97.21.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann KC, Ricci J-E, Droin NM, Green DR. The role of ARK in stress-induced apoptosis in Drosophila cells. J Cell Biol. 2002;156:1077–1087. doi: 10.1083/jcb.20112068. [DOI] [PMC free article] [PubMed] [Google Scholar]