Abstract
Recently, the emergence of reduced susceptibility to daptomycin has been linked to the reduced vancomycin susceptibility that occurs after vancomycin exposure in Staphylococcus aureus in vivo and in vitro. This study evaluated this propensity in clinical isolates of S. aureus using an in vitro pharmacokinetic/pharmacodynamic model with simulated endocardial vegetations over 8 days. Five clinical isolates (four methicillin-resistant S. aureus isolates and one methicillin-susceptible S. aureus [MSSA] isolate), all of which were reported to have become nonsusceptible to daptomycin, were evaluated. The following regimens were evaluated: vancomycin 1 g every 12 h for 4 days followed by daptomycin 6 mg/kg of body weight daily for 4 days and daptomycin 6 mg/kg daily for 8 days. If nonsusceptibility was detected, the following regimens were evaluated: no treatment for 4 days followed by daptomycin 6 mg/kg daily for 4 days, vancomycin 1 g every 12 h for 4 days followed by daptomycin 10 mg/kg daily for 4 days, and daptomycin 10 mg/kg daily for 8 days. The emergence of daptomycin nonsusceptibility (12- to 16-fold MIC increase) was detected only with the MSSA isolate with daptomycin 6 mg/kg daily for 4 days after vancomycin exposure. However, the bactericidal activity of daptomycin was maintained and the MIC increases of these isolates, which had no mprF or yycG mutations, were unstable to serial passage on antibiotic-free agar. Subsequent regimens did not demonstrate nonsusceptibility to daptomycin. These findings suggest that reduced daptomycin susceptibility can be a strain-specific and unstable event. Further evaluation of the susceptibility relationship between daptomycin and vancomycin is necessary to understand the factors involved and their clinical significance.
Daptomycin is a newly available lipopeptide antibiotic with potent in vitro and in vivo bactericidal activity against Staphylococcus aureus, including methicillin-resistant S. aureus (MRSA) (3, 23). This agent has been utilized as effective therapy for skin and soft tissue infections, bacteremia, and endocarditis (2, 9). It exerts antibacterial activity by irreversibly binding to the gram-positive cell membrane, causing a rapid depolarization via the leakage of potassium ions, which leads to cell death (28).
The frequency of mutation to daptomycin nonsusceptibility in S. aureus is believed to be low. However, case reports of the emergence of nonsusceptible strains during therapy have now been described (11, 12, 16, 17). While the mechanism of nonsusceptibility is not completely understood, it is believed to be due in part to point mutations that lead to amino acid substitutions in proteins identified as MprF and YycG (10). However, it is still unclear what factors may be involved in the appearance and, ultimately, the expression of these mutations.
Reduced susceptibility to daptomycin has been suggested to be associated with MRSA strains with reduced susceptibility to vancomycin (vancomycin-heterointermediate S. aureus [hVISA] and vancomycin-intermediate S. aureus [VISA] strains) (8, 18, 19, 24, 31). Previous studies have indicated that VISA strains produce a thickened cell wall due to the excess production of peptidoglycan, which prevents the penetration of vancomycin (6, 7). The daptomycin susceptibilities of these strains were found to be reduced compared to those of non-hVISA and non-VISA isolates. Of interest, several investigations have also demonstrated that daptomycin retains its bactericidal activity against these strains (1, 15, 31).
Although a number of studies have demonstrated a relationship between reduced daptomycin susceptibility and increased vancomycin MICs (31), it is unknown if prior exposure to vancomycin induces this phenomenon. In a study by Sakoulas et al. (24), isolates with reduced susceptibility to vancomycin or the potential for developing reduced susceptibility were evaluated for reduced cross-susceptibility. Population analysis profiles of isolates exposed to vancomycin and daptomycin revealed that after vancomycin exposure three of four isolates concomitantly displayed vancomycin and daptomycin heteroresistance in vivo. Other isolates displaying the laboratory-derived glycopeptide-intermediate phenotype demonstrated clear daptomycin heteroresistance but still maintained overall daptomycin susceptibility, with MICs of ≤1 mg/liter (24).
We previously evaluated the in vitro activity of daptomycin before and after vancomycin exposure using a series of clinical isogenic strains (22). Daptomycin displayed activity against isolate S. aureus 616 preexposed to vancomycin. However, daptomycin had reduced activity against the isogenic isolate after vancomycin exposure, with a sixfold increase in the MIC. We hypothesized that prior vancomycin exposure may have played a role in the development of reduced susceptibility to daptomycin in these series of isolates. In addition, new case reports involving reduced daptomycin susceptibility following vancomycin exposure suggest a possible association (27).
The objective of this study was to evaluate vancomycin exposure followed by daptomycin exposure in clinical isolates of S. aureus by using an in vitro pharmacokinetic (PK)/pharmacodynamic (PD) model with simulated endocardial vegetations (SEVs) over 8 days to determine the activity of daptomycin and the propensity for the development of nonsusceptibility.
MATERIALS AND METHODS
Bacterial strains.
Five clinical isolates of S. aureus were evaluated. Four isolates (three MRSA isolates and one methicillin-susceptible S. aureus [MSSA] isolate) were reported to demonstrate daptomycin nonsusceptibility following vancomycin exposure. The remaining MRSA isolate was not exposed to vancomycin but became daptomycin nonsusceptible after the suboptimal dosing of daptomycin (22, 27). Of the MRSA isolates, one displayed characteristics of community-associated MRSA isolates (USA-300, Panton-Valentine leukocidin positive, staphylococcal chromosomal cassette mec type IV) and the others were hospital derived (USA-100, Panton-Valentine leukocidin negative, staphylococcal chromosomal cassette mec type II) (27).
Antibiotics.
Daptomycin analytical-grade powder was provided by the manufacturer (Cubist Pharmaceuticals, Lexington, MA). Vancomycin analytical-grade powder was purchased commercially (Sigma Chemical Company, St. Louis, MO). Stock solutions were freshly prepared and were used at the beginning of each day throughout the experiment.
Media.
Mueller-Hinton broth (Difco Laboratories, Detroit, MI) supplemented with calcium titrated to physiologic levels (1.1 to 1.3 mM) and magnesium (12.5 mg/liter) (SMHB) was used for all in vitro PD models and susceptibility testing. Protein binding was accounted for by the addition of 3.5 to 4 g/dl albumin (American Red Cross, Detroit, MI) to all model media and by the high protein content of the SEVs (see “SEVs” below). Colony counts were determined with tryptic soy agar (TSA; Difco) plates.
Susceptibility.
MICs were determined by broth microdilution in SMHB or by the Etest methodology according to the guidelines of the Clinical and Laboratory Standards Institute (5). Minimum bactericidal concentrations were defined as a 99.9% killing of the starting inoculum and were determined according to the guidelines of the Clinical and Laboratory Standards Institute by plating 5 μl from the broth microdilution MIC and all other wells with organism growth inhibition onto agar growth media.
SEVs.
Organism inocula were prepared by spreading isolates onto six TSA plates and incubating the plates for 18 to 24 h. The resulting growth was collected and placed into 9 ml SMHB. SEVs were prepared in 1.5-ml siliconized Eppendorf tubes by mixing 50 μl of this organism suspension (final inoculum, 109 CFU/g) with 0.5 ml of human cryoprecipitate antihemolytic factor from human volunteer donors (American Red Cross) and 2.5 μl of platelet suspension (platelets mixed with normal saline; 250,000 to 500,000 platelets per clot). After these components were mixed and a monofilament line was added to each mixture, 0.05 ml bovine thrombin (5,000 units/ml) was added to each tube. The resulting SEVs were then removed from the Eppendorf tubes with a sterile 21-gauge needle and inserted into the model. This methodology results in SEVs of approximately 3 to 3.5 g/dl of albumin and 6.8 to 7.4 g/dl of total protein (1).
In vitro PD infection model.
An in vitro infection model consisting of a 250-ml glass apparatus with ports containing the suspended SEVs was utilized for all simulations. The apparatus was prefilled with medium, and the antibiotics were administered over an 8-day period as boluses into the central compartment via an injection port. The model apparatus was maintained in a 37°C water bath throughout the procedure, and a magnetic stir bar was placed in the medium for thorough mixing of the drug in the model. Fresh medium was continuously supplied and removed from the compartment along with the drug via a peristaltic pump (Masterflex; Cole-Parmer Instrument Company, Chicago, IL) at a rate set to simulate the half-lives of the antibiotics. The pH was monitored throughout all experiments with daptomycin due to the possible effects of pH on its activity. All model experiments were performed in duplicate to ensure reproducibility.
Simulated antimicrobial regimens.
Two regimens were initially evaluated, including vancomycin 1 g every 12 h (maximum concentration [Cmax], 30 mg/liter; minimum concentration [Cmin], 10 mg/liter) for 4 days followed by daptomycin 6 mg/kg of body weight every 24 h (Cmax, 98.6 mg/liter; Cmin, 6.2 mg/liter) for 4 days and daptomycin 6 mg/kg every 24 h for 8 days. If these regimens resulted in reduced susceptibility to daptomycin, model experiments of no vancomycin treatment for 4 days (growth control) were followed by treatment with daptomycin 6 mg/kg every 24 h for 4 days to confirm a possible relationship to vancomycin. Regimens of vancomycin 1 g every 12 h for 4 days followed by daptomycin 10 mg/kg every 24 h (Cmax, 164.3 mg/liter; Cmin, 10.3 mg/liter) for 4 days and daptomycin 10 mg/kg every 24 h for 8 days were also evaluated.
PK analysis.
Samples for PK analysis were obtained in duplicate through the injection port of each model from 0 to 192 h for verification of target antibiotic concentration attainment. In addition, all SEVs were assayed for their antimicrobial concentrations after homogenization, and the SEV concentrations were compared to the model concentrations to determine the percent penetration over time. All samples were stored at −70°C until they were ready for analysis.
Concentrations of daptomycin were determined by a microbioassay with Micrococcus luteus ATCC 9341. Blank 1/4-in. disks were spotted with 20 μl of the standards or samples. Each standard was tested in triplicate by placing disks on Mueller-Hinton agar plates preswabbed with a 0.5 McFarland suspension of the test organism. The plates were incubated for 18 to 24 h at 37°C, and then the zone sizes were measured. Concentrations of 150, 50, 10, 5, and 2.5 mg/liter were used as standards. The standard curves of the zone sizes versus the natural logarithm of the drug concentrations were linear between 2.5 and 150 mg/liter when the standards were prepared in SMHB (r2 = 0.99; between-day coefficients of variation for the high, medium, and low standards, 4.6%, 9.5%, and 1.7%, respectively). This assay has a lower limit of detection of 2.5 mg/liter.
Vancomycin concentrations were determined by a fluorescence polarization immunoassay (TDx; Abbott Diagnostics). This assay has a limit of detection of 2 mg/liter (r2 = 0.99; between-day coefficients of variation for the high, medium, and low standards [75, 35, and 7 mg/liter, respectively], <5%). The half-lives, the areas under the curve (AUC) from 0 to 24 h (AUC0-24), AUC/MIC ratios, and the Cmaxs of daptomycin and vancomycin were determined by the trapezoidal method with PK Analyst software (version 1.10; MicroMath Scientific Software, Salt Lake City, UT).
PD analysis.
Two SEVs were removed from each model at each sample point (for a total of four vegetations) over 0 to 192 h. The SEVs were homogenized and diluted in cold saline and plated onto TSA plates. The plates were then incubated at 35°C for 24 h, at which time the colonies were enumerated and the number of CFU/g was calculated. This method results in a lower limit of detection of 2.0 log10 CFU/g. Antimicrobial carryover was accounted for by serial dilution (10 to 10,000) of the plated samples. The reductions in the log10 CFU/g were determined by plotting time-kill curves on the basis of the number of remaining organisms over 192 h. Bactericidal activity (99.9% killing) was defined as a ≥3-log10-CFU/g reduction in the colony count from the initial inoculum. Bacteriostatic activity was defined as a <3-log10-CFU/g reduction in the colony count from the initial inoculum, while inactivity was defined as no observed reduction from the initial inoculum. The time to achieve a 99.9% bacterial load reduction (a measurement of the killing rate) was determined by linear regression (if r2 was ≥0.95) or by visual inspection.
Nonsusceptibility.
Samples of 100 μl from each time point were plated on Mueller-Hinton agar plates containing 1.5-, 2-, or 3-fold the daptomycin MIC to assess the isolates for the development of nonsusceptibility. The plates were then examined for growth after a 48-h incubation at 35°C. Daptomycin and vancomycin Etest MICs were determined for all model samples, including any growth on the nonsusceptibility screening plates, to confirm changes in the MICs.
DNA sequencing.
The mprF and yycG genes were amplified by PCR, and the products were sequenced in both directions by an automated dideoxy chain termination method by the Applied Genomics Technology Center, Wayne State University. Nucleotide sequence analyses were performed with the DS Gene program (version 1.5; Accelrys, Inc., San Diego, CA) (26).
Statistical analysis.
Changes in the CFU/g from 0 to 192 h along with the time to 99.9% killing (T99.9) for daptomycin and vancomycin were compared by a nonparametric t test or two-way analysis of variance with Tukey's honestly significant different test. A P value of ≤0.05 was considered significant. All statistical analyses were performed with SPSS statistical software (release 14.0; SPSS, Inc., Chicago, IL).
RESULTS
Susceptibility.
The preexposure vancomycin MICs for all MRSA isolates were 2 mg/liter, and the preexposure vancomycin MIC was 1 mg/liter for the MSSA isolate. All isolates were susceptible to daptomycin, with MICs of 0.25 mg/liter for all MRSA isolates and 0.125 mg/liter for the MSSA isolate.
In vitro PKs and PDs.
The PK results for the vancomycin and daptomycin exposures are presented in Table 1. The values of the PK parameters for all regimens were within 10% of the targeted values. In addition, the antibiotic Cmaxs for the SEVs were 61.6 and 67.3% of the Cmaxs achieved in broth for vancomycin and daptomycin, respectively, and are consistent with previously published results obtained with the SEV model (30).
TABLE 1.
PKs of vancomycin and daptomycin achieved with each isolate testeda
| S. aureus strain | Antibiotic regimen | Cmax (mg/liter) | Cmin (mg/liter) | Half-life (h) | MIC (mg/liter)
|
AUC0-24/MIC (broth) ratio for D | |
|---|---|---|---|---|---|---|---|
| V | D | ||||||
| MSSA 616 | V 1 g every 12 h | 31.7 ± 0.4 | 10.3 ± 0.2 | 7.4 ± 0.9 | 1 | 0.125 | 536.4 ± 7.8 |
| D 6 mg/kg daily | 106.8 ± 5.4 | 15.9 ± 0.4 | 8.4 ± 1.2 | 10,005.9 ± 248.9 | |||
| D 10 mg/kg daily | 160.2 ± 4.5 | 25.5 ± 5.4 | 9.3 ± 1.6 | 14,480.8 ± 831.6 | |||
| MRSA 675 | V 1 g every 12 h | 32.9 ± 0.4 | 10.4 ± 0.2 | 7.2 ± 0.3 | 2 | 0.25 | 277.1 ± 0.6 |
| D 6 mg/kg daily | 107.2 ± 2.9 | 14.1 ± 0.9 | 8.0 ± 0.7 | 4,990.2 ± 86.7 | |||
| MRSA 36132 | V 1 g every 12 h | 31.4 ± 0.2 | 9.4 ± 2.6 | 6.9 ± 1.0 | 2 | 0.25 | 257.4 ± 9.9 |
| D 6 mg/kg daily | 109.3 ± 3.7 | 17.3 ± 5.3 | 8.8 ± 1.0 | 5,003.6 ± 118.8 | |||
| MRSA WB1 | V 1 g every 12 h | 32.8 ± 0.3 | 9.5 ± 0.6 | 6.6 ± 0.6 | 2 | 0.25 | 271.8 ± 4.4 |
| D 6 mg/kg daily | 98.3 ± 0.2 | 12.7 ± 0.6 | 7.9 ± 0.0 | 4,689.8 ± 19.2 | |||
| MRSA WB2 | V 1 g every 12 h | 33.4 ± 1.5 | 6.8 ± 1.8 | 5.3 ± 0.6 | 2 | 0.25 | 243.3 ± 3.2 |
| D 6 mg/kg daily | 106.1 ± 6.3 | 12.4 ± 3.9 | 7.6 ± 2.0 | 5,643.5 ± 180.0 | |||
V, vancomycin; D, daptomycin.
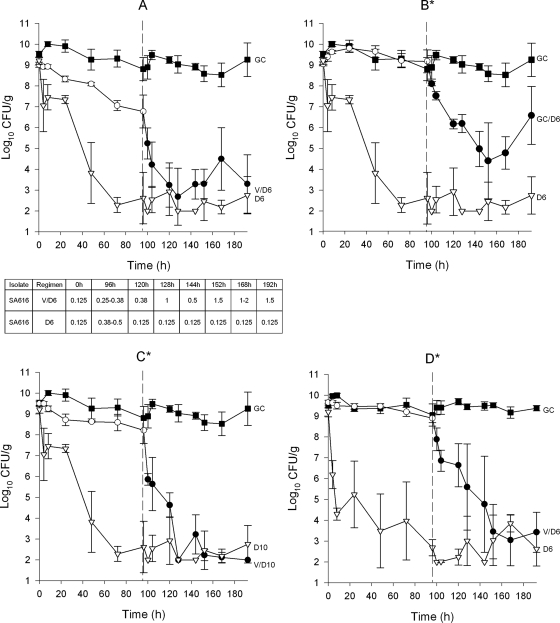
The activities of the daptomycin regimens over the 8 days of the model are displayed in Fig. 1. Daptomycin 6 mg/kg retained its killing activity against all strains following vancomycin exposure. The T99.9 for daptomycin was similar irrespective of prior vancomycin exposure, with mean T99.9s of 4.8 h (range, 1.39 to 11.36 h) for daptomycin regimens without vancomycin exposure and 6.6 h (range, 3.43 to 9.41 h) following vancomycin exposure (P = 0.057). Bactericidal activity was achieved with all daptomycin regimens regardless of prior vancomycin exposure. Overall, daptomycin was more potent than vancomycin regardless of vancomycin preexposure, but daptomycin caused the highest reduction in the numbers of CFU/g if it was administered on day 1 prior to vancomycin exposure. Daptomycin administration on day 1 resulted in an average reduction of 6.1 log10 CFU/g on day 4, whereas the average reduction was only 1.5 log10 CFU/g with vancomycin administration (P < 0.001).
FIG. 1.
(A) Growth of MSSA 616 treated with vancomycin 1 g every 12 h followed by daptomycin 6 mg/kg every 24 h, with the corresponding changes in daptomycin MICs; filled squares, growth control; open circles, vancomycin 1 g every 12 h for 4 days, filled circles, daptomycin 6 mg/kg daily for 4 days; open inverted triangles, daptomycin 6 mg/kg daily for 8 days; (B) growth of the MSSA 616 growth control treated with daptomycin 6 mg/kg every 24 h; filled squares, growth control for 8 days; open circles, growth control for 4 days, filled circles, daptomycin 6 mg/kg daily for 4 days; open inverted triangles, daptomycin 6 mg/kg daily for 8 days; (C) growth of MSSA 616 treated with vancomycin 1 g every 12 h followed by daptomycin 10 mg/kg every 24 h; filled squares, growth control; open circles, vancomycin 1 g every 12 h for 4 days, filled circles, daptomycin 10 mg/kg daily for 4 days; open inverted triangles, daptomycin 10 mg/kg daily for 8 days; (D) growth of MRSA WB-1 treated with vancomycin 1 g every 12 h followed by daptomycin 6 mg/kg every 24 h; filled squares, growth control; open circles, vancomycin 1 g every 12 h for 4 days, filled circles, daptomycin 6 mg/kg daily for 4 days; open inverted triangles, daptomycin 6 mg/kg daily for 8 days. *, no increase in MIC throughout the model duration.
The susceptibility results during the in vitro regimens are displayed in Fig. 1A. No change in MIC was detected for any MRSA isolate treated with daptomycin following vancomycin exposure. The daptomycin regimens for 8 days without vancomycin exposure also did not result in the development of nonsusceptible daptomycin isolates. However, prior vancomycin exposure resulted in the appearance of daptomycin MIC elevations of ≥1 mg/liter for the MSSA isolate. The vancomycin MIC increased up to 2 mg/liter (twofold increase) as early as 24 h after the start of the vancomycin regimen and remained elevated throughout the length of the model study. The appearance of increased vancomycin MICs occurred much earlier in the simulation than the appearance of increased daptomycin MICs (24 h and 144 h, respectively). This phenomenon was verified during repeat in vitro PD model experiments with this isolate (a total of four experiments) and the same vancomycin and daptomycin exposure conditions. Of note, daptomycin maintained its bactericidal activity against the MSSA isolate even during the period after vancomycin exposure, and nonsusceptible organisms with T99.9s of 9.46 to 9.91 h with vancomycin preexposure and 10.9 to 11.4 h without vancomycin exposure were determined. These increases in MICs were unstable to five serial passages on TSA, but the MICs remained more than twofold higher than the starting MICs at the end of passage. Sequencing did not demonstrate mutations in mprF or yycG. Furthermore, increases in MICs were not noted when daptomycin was used alone during the 8-day simulation with the MSSA isolate.
Following the appearance of daptomycin nonsuceptibility after vancomycin exposure in this strain, additional daptomycin dosing regimens were evaluated, including 4 days of growth without antibiotic exposure followed by dosing with daptomycin 6 mg/kg daily for 4 days, daptomycin 10 mg/kg daily for 4 days after vancomycin exposure for 4 days, and daptomycin 10 mg/kg alone for 8 days. Daptomycin concentration-dependent killing was displayed by the increased bactericidal activity in the regimens with daptomycin 10 mg/kg both with and without prior vancomycin exposure. Although the activity of daptomycin was significantly reduced following 4 days of growth as a control (T99.9s, 33.4 to 34.9 h) compared to that achieved when daptomycin was dosed for 8 days (T99.9s, 9.46 to 9.91 h) (P < 0.001), there was no emergence of nonsusceptibility without prior vancomycin exposure by the use of this regimen with the MSSA isolate. To identify potential factors that may have contributed to this reduced activity, subsequent studies of biofilm production were performed before and after 4 days of growth from the in vitro PD model. By the use of a previously described method of spectrophotometric analysis of absorbance, the initial isolate, MSSA 616, displayed low levels of biofilm production (mean optical density, 0.02), whereas the isolate obtained after 4 days of growth displayed high levels of biofilm production (mean optical density, 0.38) (29).
DISCUSSION
Reports of a possible relationship between reduced susceptibility to vancomycin and daptomycin have raised hypotheses that prior vancomycin exposure may induce this phenomenon (8, 18-20, 24). This is the first study to expose isolates with an established propensity to lose daptomycin susceptibility to vancomycin and then daptomycin by using human PK/PD simulations.
The emergence of daptomycin nonsusceptibility has been documented in the absence of vancomycin exposure (14). A recent study revealed that subtherapeutic exposures to daptomycin can result in reduced daptomycin binding to whole cells and cytoplasmic membranes and the disappearance of a membrane protein, leading to decreased daptomycin susceptibility and a heteroresistant phenotype in S. aureus (14). Simulating the same subtherapeutic exposures with this isolate using an in vitro PK/PD model, we reproduced this nonsusceptibility but prevented it with a dose of 6 mg/kg every 24 h (21). In the present study, the same initial isolate was exposed to vancomycin for 4 days, followed by 4 days of daptomycin in the PK/PD model. Using the current recommended dose of 6 mg/kg, we were unable to recover daptomycin-nonsusceptible mutants and the activity of daptomycin was maintained following vancomycin exposure.
The impact of prior vancomycin exposure on daptomycin susceptibility has been correlated in vitro by Sakoulas et al. (24). From our previous studies involving a series of clinical isogenic strains of S. aureus 616, we demonstrated that vancomycin may play a role in the development of reduced susceptibility to daptomycin (22). This was further confirmed by the present study with 8-day SEV models, with the development of daptomycin nonsusceptibility (MIC ≥ 1 mg/liter) in S. aureus 616 after 4 days of prior vancomycin exposure in vitro. However, these isolates recovered from the model were not stably daptomycin nonsusceptible and displayed no mutational changes in mprF or yycG. Unstable daptomycin-nonsusceptible isolates have been noted previously in heterogeneous daptomycin-nonsusceptible strains with subpopulations for which the MICs are greater than the daptomycin MIC (14). Of interest, similar to our findings, Pillai et al. have reported that strains with elevated MICs (up to 2 mg/liter) did not possess mutations in mprF (20). Therefore, the previously reported mutational changes are not necessary for reduced daptomycin susceptibility. It is possible that in these types of isolates the pressure of an antibiotic rather than the preexistence of nonsusceptibility, as in wild-type strains, may play a role in the development of reduced susceptibility.
Our results suggest that some strains are more likely than others to lose susceptibility to daptomycin. Only the MSSA isolate (but not the four MRSA isolates) developed reduced daptomycin susceptibility under the vancomycin selection pressure imposed by our model. Upon further testing, all five isolates utilized in the model were found to demonstrate heteroresistance to daptomycin over a concentration range of 0.25 to 6 mg/liter by the use of population analysis (data not shown). Therefore, we could not easily determine if S. aureus 616 was hypermutable. The hypothesis that strains have a proclivity toward the loss of daptomycin susceptibility is further supported by our finding of another MSSA isolate in a patient with prosthetic valve endocarditis deemed inoperable and allergic to penicillin and vancomycin that rapidly lost daptomycin susceptibility during daptomycin therapy. This strain was identical to the MSSA strain evaluated here on the basis of pulsed-field gel analysis, even though it was obtained from a different hospital and the two isolates were recovered over a year apart. Both strains were agr group II (G. Sakoulas, unpublished observations).
The development of the vancomycin-intermediate phenotype has been associated with concentrations of vancomycin below 10 mg/liter (4, 13, 25). The maintenance of vancomycin concentrations above this value (i.e., 15 to 20 mg/liter) may prevent the development of hVISA or VISA strains and, therefore, may potentially minimize the impact of vancomycin on daptomycin therapy. In our in vitro PD model the average vancomycin Cmin of all regimens was 9.0 ± 1.5 mg/liter. However, we were able to find reduced daptomycin and vancomycin susceptibilities (8- to 16-fold and 2-fold increases, respectively) in only one of the five strains tested after vancomycin exposure.
Many unidentified factors may play a role in the development of reduced susceptibility to daptomycin in vivo. Recently, a report described a series of persistent blood isolates that were obtained from a patient with endocarditis receiving vancomycin therapy and that were initially vancomycin susceptible (MIC = 1 mg/liter) but that later became intermediately resistant (MIC = 8 mg/liter) (18). These organisms were evaluated for antimicrobial susceptibility changes and gene mutations. Of note, the patient had also received a single exposure to rifampin and a course of therapy with imipenem. The investigators identified 35 point mutations in 31 loci in these isogenic strains that developed over the time of exposure to vancomycin. Of interest, vancomycin susceptibility was observed to result in a simultaneous 100-fold reduction in daptomycin susceptibility, which coincided with mutations in the rpoC and yyc gene clusters, even though the patient was not treated with daptomycin. In many cases, the isolates used in our study had been exposed not only to vancomycin but also to other antibiotics (nafcillin and gentamicin). The suboptimal exposure to daptomycin in patients with endocarditis, osteomyelitis, and device infections may be a factor in the reduction in susceptibility; notably, the isolates evaluated in our study, which were from patients with complicated infections, including endocarditis, osteomyelitis, and septic arthritis, developed daptomycin nonsusceptibility (14, 22, 27).
Overall, prior exposure to vancomycin produced daptomycin nonsusceptibility in one of the five isolates evaluated, with a result of unstable nonsusceptibility and with the lack of detection of mutations in mprF. Despite increases in the MIC, daptomycin maintained effective killing (T99.9) of this strain. The fact that daptomycin nonsusceptibility following vancomycin exposure in S. aureus remains infrequent is supported by the rare observations of this situation in clinical practice thus far. Further studies are needed to better understand the role that vancomycin may play in the development of reduced susceptibility to daptomycin. What will be the most important will be the determination of the clinical significance of these microbiological observations, since the PK/PD model used here, as well as animal models of endocarditis, is mostly a screening tool for the assessment of antimicrobial therapy. Therefore, the relevance of these observations to the treatment of MRSA infections will need to be verified clinically.
Acknowledgments
This work was supported by a grant from Cubist Pharmaceuticals.
We thank Barbara Robinson-Dunn from William Beaumont Hospital in Royal Oak, MI, for the donation of two of the clinical isolates used in this study.
Footnotes
Published ahead of print on 12 November 2007.
REFERENCES
- 1.Akins, R. L., and M. J. Rybak. 2001. Bactericidal activities of two daptomycin regimens against clinical strains of glycopeptide intermediate-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus faecium, and methicillin-resistant Staphylococcus aureus isolates in an in vitro pharmacodynamic model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 45:454-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbeit, R. D., D. Maki, F. P. Tally, E. Campanaro, and B. I. Eisenstein. 2004. The safety and efficacy of daptomycin for the treatment of complicated skin and skin-structure infections. Clin. Infect. Dis. 38:1673-1681. [DOI] [PubMed] [Google Scholar]
- 3.Cha, R., R. G. Grucz, Jr., and M. J. Rybak. 2003. Daptomycin dose-effect relationship against resistant gram-positive organisms. Antimicrob. Agents Chemother. 47:1598-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charles, P. G., P. B. Ward, P. D. Johnson, B. P. Howden, and M. L. Grayson. 2004. Clinical features associated with bacteremia due to heterogeneous vancomycin-intermediate Staphylococcus aureus. Clin. Infect. Dis. 38:448-451. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; sixteenth informational supplement. M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Cui, L., X. Ma, K. Sato, K. Okuma, F. C. Tenover, E. M. Mamizuka, C. G. Gemmell, M. N. Kim, M. C. Ploy, N. El-Solh, V. Ferraz, and K. Hiramatsu. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui, L., H. Murakami, K. Kuwahara-Arai, H. Hanaki, and K. Hiramatsu. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cui, L., E. Tominaga, H. M. Neoh, and K. Hiramatsu. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:1079-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fowler, V. G., Jr., H. W. Boucher, G. R. Corey, E. Abrutyn, A. W. Karchmer, M. E. Rupp, D. P. Levine, H. F. Chambers, F. P. Tally, G. A. Vigliani, C. H. Cabell, A. S. Link, I. DeMeyer, S. G. Filler, M. Zervos, P. Cook, J. Parsonnet, J. M. Bernstein, C. S. Price, G. N. Forrest, G. Fatkenheuer, M. Gareca, S. J. Rehm, H. R. Brodt, A. Tice, and S. E. Cosgrove. 2006. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N. Engl. J. Med. 355:653-665. [DOI] [PubMed] [Google Scholar]
- 10.Friedman, L., J. D. Alder, and J. A. Silverman. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:2137-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden, M. K., K. Rezai, R. A. Hayes, K. Lolans, J. P. Quinn, and R. A. Weinstein. 2005. Development of daptomycin resistance in vivo in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5285-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirschwerk, D., C. C. Ginocchio, M. Bythrow, and S. Condon. 2006. Diminished susceptibility to daptomycin accompanied by clinical failure in a patient with methicillin-resistant Staphylococcus aureus bacteremia. Infect. Control Hosp. Epidemiol. 27:315-317. [DOI] [PubMed] [Google Scholar]
- 13.Howden, B. P., P. B. Ward, P. G. Charles, T. M. Korman, A. Fuller, P. du Cros, E. A. Grabsch, S. A. Roberts, J. Robson, K. Read, N. Bak, J. Hurley, P. D. Johnson, A. J. Morris, B. C. Mayall, and M. L. Grayson. 2004. Treatment outcomes for serious infections caused by methicillin-resistant Staphylococcus aureus with reduced vancomycin susceptibility. Clin. Infect. Dis. 38:521-528. [DOI] [PubMed] [Google Scholar]
- 14.Kaatz, G. W., T. S. Lundstrom, and S. M. Seo. 2006. Mechanisms of daptomycin resistance in Staphylococcus aureus. Int. J. Antimicrob. Agents 28:280-287. [DOI] [PubMed] [Google Scholar]
- 15.LaPlante, K. L., and M. J. Rybak. 2004. Clinical glycopeptide-intermediate staphylococci tested against arbekacin, daptomycin, and tigecycline. Diagn. Microbiol. Infect. Dis. 50:125-130. [DOI] [PubMed] [Google Scholar]
- 16.Mangili, A., I. Bica, D. R. Snydman, and D. H. Hamer. 2005. Daptomycin-resistant, methicillin-resistant Staphylococcus aureus bacteremia. Clin. Infect. Dis. 40:1058-1060. [DOI] [PubMed] [Google Scholar]
- 17.Marty, F. M., W. W. Yeh, C. B. Wennersten, L. Venkataraman, E. Albano, E. P. Alyea, H. S. Gold, L. R. Baden, and S. K. Pillai. 2006. Emergence of a clinical daptomycin-resistant Staphylococcus aureus isolate during treatment of methicillin-resistant Staphylococcus aureus bacteremia and osteomyelitis. J. Clin. Microbiol. 44:595-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mwangi, M. M., S. W. Wu, Y. Zhou, K. Sieradzki, H. de Lencastre, P. Richardson, D. Bruce, E. Rubin, E. Myers, E. D. Siggia, and A. Tomasz. 2007. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 104:9451-9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel, J. B., L. A. Jevitt, J. Hageman, L. C. McDonald, and F. C. Tenover. 2006. An association between reduced susceptibility to daptomycin and reduced susceptibility to vancomycin in Staphylococcus aureus. Clin. Infect. Dis. 42:1652-1653. [DOI] [PubMed] [Google Scholar]
- 20.Pillai, S. K., H. S. Gold, G. Sakoulas, C. Wennersten, R. C. Moellering, Jr., and G. M. Eliopoulos. 2007. Daptomycin nonsusceptibility in Staphylococcus aureus with reduced vancomycin susceptibility is independent of alterations in MprF. Antimicrob. Agents Chemother. 51:2223-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rose, W. E., M. J. Rybak, and G. W. Kaatz. 2007. Evaluation of daptomycin treatment of Staphylococcus aureus bacterial endocarditis: an in vitro and in vivo simulation using historical and current dosing strategies. J. Antimicrob. Chemother. 60:334-340. [DOI] [PubMed] [Google Scholar]
- 22.Rose, W. E., M. J. Rybak, and G. Sakoulas. 2006. Daptomycin activity on S. aureus isolates pre and post exposure to vancomycin during the treatment for bacterial endocarditis, abstr. C1-0686. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 23.Safdar, N., D. Andes, and W. A. Craig. 2004. In vivo pharmacodynamic activity of daptomycin. Antimicrob. Agents Chemother. 48:63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakoulas, G., J. Alder, C. Thauvin-Eliopoulos, R. C. Moellering, Jr., and G. M. Eliopoulos. 2006. Induction of daptomycin heterogeneous susceptibility in Staphylococcus aureus by exposure to vancomycin. Antimicrob. Agents Chemother. 50:1581-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakoulas, G., H. S. Gold, R. A. Cohen, L. Venkataraman, R. C. Moellering, and G. M. Eliopoulos. 2006. Effects of prolonged vancomycin administration on methicillin-resistant Staphylococcus aureus (MRSA) in a patient with recurrent bacteraemia. J. Antimicrob. Chemother. 57:699-704. [DOI] [PubMed] [Google Scholar]
- 26.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheth, A., C. F. Carpenter, and B. Robinson-Dunn. 2006. Reduced vancomycin susceptibility and daptomycin nonsusceptibility associated with treatment failure in 2 cases of MRSA bacteremia, abstr. C2-1159. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC.
- 28.Silverman, J. A., N. G. Perlmutter, and H. M. Shapiro. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stepanovic, S., D. Vukovic, I. Dakic, B. Savic, and M. Svabic-Vlahovic. 2000. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 40:175-179. [DOI] [PubMed] [Google Scholar]
- 30.Tsuji, B. T., and M. J. Rybak. 2005. Short-course gentamicin in combination with daptomycin or vancomycin against Staphylococcus aureus in an in vitro pharmacodynamic model with simulated endocardial vegetations. Antimicrob. Agents Chemother. 49:2735-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wootton, M., A. P. Macgowan, and T. R. Walsh. 2006. Comparative bactericidal activities of daptomycin and vancomycin against glycopeptide-intermediate Staphylococcus aureus (GISA) and heterogeneous GISA isolates. Antimicrob. Agents Chemother. 50:4195-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]