Abstract
Ketolides, which represent the newest macrolide antibiotics, are generally perceived to be noninducers of inducible erm genes. In the study described in this paper we investigated the effects of several macrolide and ketolide compounds on the expression of the inducible erm(C) gene by Escherichia coli cells. Exposure to 14-member-ring macrolide drugs and to azithromycin led to a rapid and pronounced increase in the extent of dimethylation of Erm(C) target residue A2058 in 23S rRNA. When cells were incubated with subinhibitory concentrations of ketolides, the extent of A2058 dimethylation was also increased, albeit to a lower level and with kinetics slower than those observed with macrolides. The induction of erm(C) expression by ketolides was further confirmed by using a reporter construct which allows the colorimetric detection of induction in a disc diffusion assay. Most of the ketolides tested, including the clinically relevant compounds telithromycin and cethromycin, were able to induce the reporter expression, even though the induction occurred within a more narrow range of concentrations compared to the concentration range at which induction was achieved with the inducing macrolide antibiotics. No induction of the reporter expression was observed with 16-member-ring macrolide antibiotics or with a control drug, chloramphenicol. The deletion of three codons of the erm(C) leader peptide eliminated macrolide-dependent induction but left ketolide-dependent induction unchanged. We conclude that ketolides are generally capable of inducing erm genes. The narrow range of ketolide inducing concentrations, coupled with the slow rate of induction and the lower steady-state level of ribosome methylation, may mask this effect in MIC assays.
Macrolide antibiotics have a proven record of excellence in the clinical setting. These are safe and efficient drugs which have been utilized in medical practice for many decades. The widespread use of macrolides has created favorable conditions for the selection of resistant strains. Nowadays, macrolide resistance is prevalent among clinical isolates (10, 15, 18). The two main mechanisms of macrolide resistance are drug-efflux membrane pumps and modification of the drug target site in the ribosome. The latter is brought about by the action of Erm-type methyltransferases. These enzymes, which likely originated in macrolide producers, methylate a single adenine in 23S rRNA (16). The target nucleotide residue (A2058 in Escherichia coli) is located in the macrolide binding site, and its mono- or dimethylation disturbs the shape and chemical makeup of the drug binding site, thereby reducing the affinity of the macrolide to the ribosome (7, 27, 34). Dimethylation of A2058 also renders the ribosome resistant to lincosamides and streptogramin B antibiotics, which bind to an overlapping site in the ribosome.
A2058 is located in the nascent peptide exit tunnel, and modification of its chemical structure by Erm methyltransferases apparently is not favorable for the cell unless macrolide antibiotics are present. Because of that, macrolide producers and, later, pathogenic strains targeted by macrolides have acquired an elegant and efficient strategy that enables them to turn on rRNA modification by Erm only when it is absolutely necessary. Cells carrying inducible erm genes express the methylase only upon exposure to antibiotics. This is achieved due to the specific secondary structure of the mRNA (reviewed in reference 37). In the case of inducible erm(C), the gene encoding the methylase, erm(C) is preceded by a short leader cistron [erm(C)L in Fig. 1]. In the absence of macrolides, the ribosome binding site of erm(C) (RBSE) is sequestered in the mRNA secondary structure. Therefore, it is inaccessible to initiating ribosomes and Erm is not expressed. At the same time, the leader open reading frame (ORF) erm(C)L is efficiently translated by the ribosomes. When cells are exposed to low concentrations of inducing macrolide antibiotics, the drug-bound ribosomes engaged in the translation of erm(C)L stall. The stalled ribosome destabilizes the ground-level mRNA secondary structure and shifts the equilibrium to the induced conformation. RBSE becomes accessible, and erm(C) can then be translated by the ribosomes, which are not yet bound by antibiotics. This type of ribosome stalling—and, hence, the induction of erm(C) expression—depends on the amino acid sequence of the nascent peptide and the chemical structure of the antibiotic (21, 22). Although the molecular mechanisms leading to ribosome stalling in this system are essentially unknown, they likely involve specific interactions between the ribosome, the nascent peptide, and the drug.
FIG. 1.
Conformational transition in erm(C) mRNA leading to the induction of erm(C) expression. The amino acid sequence of the leader peptide and the first three amino acids of Erm(C) are indicated. The ribosome binding sites of the leader peptide gene erm(C)L (RBSL) and erm(C) (RBSE) are marked. Complementary segments of mRNA that form two hairpins (ovals 1 and 2 and ovals 3 and 4) in the noninduced structure or one hairpin (ovals 2 and 3) in the induced structure are marked. The position of the ribosome in the leader ORF in the hypothetical stalled complex induced by the interaction of the nascent peptide, ribosome, and antibiotic (AB) and leading to the induction of erm(C) expression is shown.
The widespread occurrence of inducible erm genes as well as other mechanisms of resistance provided a strong incentive for the development of newer macrolide drugs that can evade resistance. The latest version of macrolides, the ketolides, appear to have achieved this goal. Three general features distinguish clinically active ketolides from previous 14-member-ring macrolides: (i) the lack of cladinose at the C-3 position of the lactone ring (in ketolides it is replaced with a keto function), (ii) the presence of an 11,12-linked carbamate, and (iii) the presence of an extended alkyl-aryl side chain (Fig. 2). Some of these features are required for the tight binding of ketolides to the ribosome (12, 39). Notably, ketolides were generally reported to be noninducers of inducible erm genes (1-3).
FIG. 2.
Chemical structures of the macrolide and ketolide antibiotics used in this work.
Notwithstanding the general lack of understanding of the mechanisms leading to drug-dependent ribosome stalling and erm induction, the perceived inability of ketolides to induce erm remains unexplained in molecular terms. In addition, some of the published data suggest that certain ketolides may exhibit from weak to significant induction of erm(B) genes (6, 25, 41).
The aim of this work was to get better insights into the effect of ketolides on ribosome stalling and, possibly, the induction of erm(C) expression. We analyzed how the exposure of E. coli cells carrying inducible erm(C) to ketolides affects the extent and kinetics of dimethylation of A2058 in the rRNA of the large ribosomal subunit. We have also constructed a reporter system capable of sensing ribosome stalling during translation of the leader ORF and used this reporter to demonstrate that ketolides do induce the initiation of translation at the start codon of the erm(C) cistron. We conclude that ketolides are generally capable of induction of erm(C) expression. We also show that mutations in the leader peptide sequence differentially affect the induction of erm expression by ketolides and cladinose-containing macrolides.
MATERIALS AND METHODS
Antibiotics, enzymes, chemicals, and kits.
The antibiotics were obtained from Aventis Pharma (telithromycin, roxithromycin, cethromycin, HMR3004, RU69874, RU3562, RU66252, RU56006, RU70645), Sigma (erythromycin, tylosin, josamycin), and the U.S. Pharmacopeia (azithromycin, clarithromycin). The enzymes used for DNA cloning were from Fermentas. [γ-32P]ATP was from MP Biomedicals. Luria-Bertani (LB) broth components and agar were from Difco. Isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were from Fisher Scientific. All oligonucleotide primers were from Integrated DNA Technologies. All other chemicals were from Fisher Scientific or Sigma Aldrich, unless indicated otherwise. Site-directed mutagenesis was carried out with a QuikChange XL mutagenesis kit (Stratagene). Total RNA was isolated from E. coli cells by using an RNeasy kit (Qiagen).
Plasmids and bacterial strains.
The plasmids directly used in this study are listed in Table 1. Plasmid pPOT1AE (31) was used as the vector for the generation of erm-containing plasmids as well as lacZ-containing reporter plasmids. pPOT1AE is based on the pGEX vector (GE Healthcare) and includes the IPTG-inducible Ptac promoter, followed by unique AflII and EcoRI restriction sites and the transcription terminator, the lacIq gene, the ampicillin resistance (Ampr) marker, and the pBR322 origin of replication.
TABLE 1.
Plasmids used in this study
| Plasmid | Basic features | Reference |
|---|---|---|
| pPOT1AE | pGEX-derived vector plasmid containing IPTG-inducible Ptac promoter, tryptophan operon terminator, and unique AflII and EcoRI cloning sites | 31 |
| pERMCT | erm(C) cassette containing the regulatory cistron erm(C)L and methylase gene erm(C) cloned between the AflII and EcoRI sites of the pPOT1AE vector | This work |
| pERMCM | Same as pERMCT but with the plasmid AflII site eliminated and a new AflII site introduced at codons 3 and 4 of erm(C) | This work |
| pERMZα | Reporter plasmid derived from pERMCM in which the entire erm(C), starting from codon 5, was replaced with codons 6 to 60 of the lacZ gene | This work |
All the cloning procedures and most experiments with the engineered constructs were carried out with E. coli strain JM109 (Promega) [F′ traD36 proA+B+ lacIq Δ(lacZ)M15/Δ(lac-proAB) glnV44 e14 negative gyrA96 recA1 relA1 endA1 thi hsdR17]. Macrolide-hypersensitive E. coli TolC− strain ZK796 [F− araD139 Δ(argF-lacU)205 ptsF25 relA1 rpsL150 deoC1 flb-5301 Δ(lacU)169 rpsL (Strr) tolC::Tn10 (Tetr)] (40) was used in some microbiological experiments.
Construction of the pERMZα reporter.
The erm(C) cassette, including the leader ORF and the erm(C) cistron, was PCR amplified from an S. aureus clinical isolate. AflII and EcoRI restriction sites were introduced at the ends of the amplified fragment on the PCR primers. The erm cassette was cloned between the AflII and the EcoRI sites of the pPOT1AE vector to produce plasmid pERMCT (Fig. 3A).
FIG. 3.
Physical maps of the 4,889-bp pERMCT plasmid carrying the inducible erm(C) cassette and the 3,738-bp pERMZα modular reporter plasmid in which the erm(C) cistron was replaced with codons 2 to 60 of the lacZ gene encoding the β-galactosidase α peptide. The β-lactamase gene (bla) and the lac repressor gene (lacIq) are indicated by gray arrows. The Ptac promoter is indicated by a black arrow; the tryptophan terminator is shown by a black triangle. The relevant restriction sites are marked.
The AflII site of plasmid pERMCT was eliminated by cutting the plasmid with the AflII restriction enzyme, filling up the DNA ends with the Klenow fragment of DNA polymerase I in the presence of the four deoxynucleoside triphosphates, and religating the blunt-end linear plasmid. A new AflII restriction site was then introduced at codons 3 and 4 of the erm(C) cistron, and an NdeI site was introduced at the initiator AUG codon of the leader ORF by PCR-based mutagenesis. The resulting plasmid was named pERMCM.
The erm(C) portion in pERMCM was then replaced with the segment of the E. coli lacZ gene encoding the β-galactosidase α peptide (17) (codons 6 to 60 of lacZ) by using the unique restriction sites AflII and EcoRI of pERMCM. The final 3,738-bp reporter construct was named pERMZα (Fig. 3B).
MIC determination.
MIC determinations were carried out by following the protocol recommended by the CLSI (formerly NCCLS) (24).
Kinetics of Erm(C) induction in liquid culture.
A 3-ml overnight culture of ZK796 cells transformed with erm(C)-containing plasmid pERMCT was diluted 1:100 in fresh LB broth containing 100 μg/ml ampicillin and 0.5 mM IPTG. The culture was incubated at 37°C for 1.5 to 2 h and split into two flasks. Erythromycin or telithromycin was added to final concentrations of 0.8 μg/ml and 0.4 μg/ml, respectively. One-milliliter aliquots were removed at the desired time points, and RNA was isolated and used as the template for the primer extension analysis of A2058 dimethylation.
Primer extension.
The deoxyoligonucleotide primer GTAAAGGTTCACGGGGTC was used to assess the degree of dimethylation of A2058 in E. coli 23S rRNA. Ten picomoles of the primer was labeled with 32P at the 5′ terminus by incubation in a final volume of 10 μl with 10 μCi [γ-32P]ATP (6,000 Ci/mmol) and 10 units of polynucleotide kinase in a 1× kinase buffer (Fermentas). The reaction mixture was incubated at 37°C for 30 min and then for 2 min at 90°C.
The labeled primer (0.5 pmol) was combined with 1.5 μg of total cellular RNA in 4.5 μl of hybridization buffer (50 mM HEPES-KOH, pH 7, 100 mM KCl). The reaction mixtures were incubated at 90°C for 1 min and then cooled to 45°C over 10 min. The annealed reaction mixtures were then centrifuged for 15 s to collect any condensation.
An extension reaction premix contained 0.65 μl of 10× reaction buffer (1.3 M Tris-HCl, pH 8.5, 100 mM MgCl2, 100 mM dithiothreitol), 1.5 μl of a deoxynucleoside triphosphate-dideoxy CTP (ddCTP) mix (for dideoxycytosine termination; 1 mM dGTP, 1 mM dATP, 1 mM dTTP, and 1 mM ddCTP), 1.75 μl of RNase-free water, and 0.1 μl of avian myeloblastosis virus reverse transcriptase (25 units/ml; Seikagaku America) per reaction sample. The amount of premix was adjusted according to the number of samples to be analyzed. The premix (4.1 μl) was added to each reaction tube with the annealed primer-RNA complex. Samples were incubated at 42°C for 20 min. Following incubation, 120 μl of stop buffer (300 mM sodium acetate, pH 5.5, 70% ethanol, 0.8 mM EDTA) was added to each tube; the samples were incubated on ice for 5 min, followed by centrifugation at 21,000 × g for 15 min in a refrigerated desktop centrifuge. After complete removal of the supernatant, the pellets were air dried for 5 min and resuspended in 5 μl of formamide loading dye (26). Samples (2 μl) were loaded onto a 8-mm-wide slot of a 12% polyacrylamide gel (20 cm by 20 cm by 0.25 mm). The gel was run at 20 W until the bromophenol blue dye reached the bottom of the gel. The gel was transferred to Whatman 3MM filter paper, dried, and exposed on a phosphorimager screen. The screen was scanned on a Molecular Dynamics PhosphorImager, and the band intensities were quantified with ImageQuant software.
Disc-based assay of the pERMZα reporter induction.
A 3-ml culture of cells transformed with the pERMZα reporter plasmid was incubated overnight in LB broth containing 100 μg/μl ampicillin. The culture was diluted 1:100 in fresh LB broth containing ampicillin (100 μg/μl) and IPTG (0.5 mM) and incubated for 2 h. Cells (A600, 0.3; 1.5 × 108 CFU) were added to 5 ml of 0.6% LB agar at 50°C. After a brief mixing of the components of the mixture, the cell suspension was poured on top of a 1.5% LB agar plate containing 100 μg/μl ampicillin, 0.5 mM IPTG, and 80 μg/ml X-Gal. Once the soft agar had solidified, 5-mm-diameter Whatmann 3MM paper discs were placed on Parafilm and wetted with 5 to 10 μl of a solution of the appropriate antibiotic. Antibiotic-containing discs were placed on top of the plates, and the plates were inverted and incubated for 18 to 24 h at 37°C.
RESULTS
Activation of Erm(C)-dependent ribosome methylation by ketolides.
This work represents part of an extended study of drug-dependent ribosome stalling which utilizes some of the unique advantages of the E. coli system. Therefore, the studies reported in this paper were carried out with E. coli as opposed to a gram-positive organism. Given the high degree of conservation of the ribosome elements involved in the interaction with the drug and the nascent peptide, the results obtained in the E. coli system are believed to be applicable to other bacteria as well. To study the effect of ketolide antibiotics on the expression of erm(C), an inducible erm(C) cassette from a clinical S. aureus isolate was cloned into an E. coli plasmid, pPOT1AE (31), under the control of the IPTG-inducible promoter Ptac. The resulting plasmid, pERMCT (Fig. 3A), was transformed into E. coli tolC strain ZK796 (40). Introduction of the inducible erm(C) increased the erythromycin resistance eightfold, whereas no obvious changes in resistance to ketolides (telithromycin or cethromycin) or to a peptidyltransferase inhibitor (chloramphenicol) were observed (Table 2). This result was in agreement with the previous assertion that gram-positive bacteria carrying inducible erm remain sensitive to ketolides (1, 2, 19).
TABLE 2.
Effect of inducible erm(C) on antibiotic resistance of E. coli
| Antibiotic | MIC (μg/μl)a
|
|
|---|---|---|
| Vector | pERMCT | |
| Erythromycin | 3 | 25 |
| Telithromycin | 1.5 | 1.5 |
| Cethromycin | 1.5 | 1.5 |
| Chloramphenicol | 2 | 2 |
E. coli tolC strain ZK796 was used as the host for the empty vector pPOT1AE or erm(C)-containing plasmid pERMCT.
The resistance conferred by erm induction is the result of a complex sequence of events. The development of the resistance phenotype may depend on the kinetics of erm induction, the “on” and “off” rates of antibiotic binding, the affinity of the drug for the methylated ribosome, and other parameters. We reasoned that MIC data may not accurately reveal the immediate effect of ketolides on the induction of erm expression. Therefore, we investigated more directly whether the exposure of cells that carry inducible erm to ketolide antibiotics leads to the dimethylation of A2058 in 23S rRNA. ZK796 cells transformed with either plasmid pERMCT or the empty vector were incubated for 4 h in the presence of subinhibitory concentrations (ca. 25% of the MIC) of macrolide and ketolide antibiotics. Total rRNA was isolated, and the extent of A2058 dimethylation was analyzed by the method of primer extension arrest (Fig. 4A) (29, 35). A DNA primer complementary to the segment from residues 2961 to 2078 was annealed to the 23S rRNA and was extended with reverse transcriptase in the presence of dATP, dGTP, dTTP, and ddCTP. The dimethylation of A2058 in 23S rRNA prevents reverse transcriptase from incorporating a complementary nucleotide into the cDNA strand and leads to the premature termination of primer extension. The product of primer extension on unmethylated rRNA is 22 nucleotides long, but it is only 20 nucleotides long when 23S rRNA dimethylated at A2058 is used as the template. As can be seen in Fig. 4B, the incubation of erm-carrying cells with the macrolides erythromycin, azithromycin, and RU69874 results in the dimethylation of A2058 in a significant fraction of cellular 23S rRNA. Importantly, exposure of the cells to the ketolide drug telithromycin or cethromycin also led to the dimethylation of A2058, albeit to a lesser extent than exposure to macrolides did.
FIG. 4.
Increase in dimethylation of A2058 in 23S rRNA upon exposure of cells to macrolide and ketolide antibiotics. (A) Principle of detection of A2058 dimethylation by primer extension arrest. In the presence of dATP, dGTP, dTTP, and ddCTP, the 18-nucleotide (nt)-long primer is extended by reverse transcriptase by 4 nucleotides when A2058 is unmodified or by only 2 nucleotides when primer extension is impeded by the dimethylation of A2058. (B) Induction of A2058 dimethylation in cells carrying inducible erm(C) by different antibiotics. Lanes V and E, RNA samples prepared from cells transformed with an empty pPOT1AE vector lacking inducible erm(C) and RNA samples prepared from cells transformed with plasmid pERMCT carrying the inducible erm(C) cassette, respectively. Cells were exposed to antibiotics at approximately one-fourth the MIC for 4 h. (C) Kinetics of induction of A2058 dimethylation by the cladinose-containing macrolide erythromycin or the ketolide telithromycin.
We then investigated the kinetics of erm(C) induction by a macrolide (erythromycin) and a ketolide (telithromycin). After addition of the corresponding drug at subinhibitory concentrations (ca. 25% of the MIC) to the exponential cultures, RNA samples were prepared after defined periods of time and the extent of A2058 dimethylation was assessed by primer extension. Figure 4C shows that upon exposure of cells to erythromycin, a significant fraction (10%) of the 23S rRNA became dimethylated at A2058 after only 30 min of incubation; the fraction of dimethylated rRNA continued to grow rapidly for the next several hours. In contrast, A2058 in cells exposed to a ketolide telithromycin remained essentially unmethylated for at least 2 h, after which time a slow increase in A2058 dimethylation was observed. Even upon prolonged incubation, the level of rRNA methylation in cells incubated with telithromycin remained lower than that in cells exposed to erythromycin. The slow onset of methylation and a lower overall level of A2058 dimethylation upon the exposure of cells to ketolides could be among the reasons why ketolide MICs are not dramatically affected by the presence of inducible erm genes.
Use of pERMZα reporter to assess erm(C) induction by macrolides and ketolides.
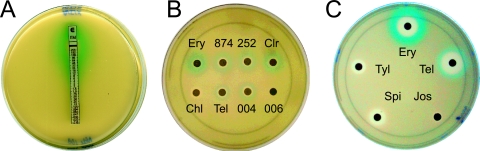
Although the extent of Erm-dependent methylation of 23S rRNA is a more direct readout of erm induction than MIC readings, it is still fairly remote from the act of activation of erm(C) expression. The rate of ribosome assembly, the affinity of the inducing antibiotic for its binding site in the ribosome precursor, and possible competition between the antibiotic and the Erm methyltransferase may affect the rate or the extent of dimethylation of A2058 (4, 5, 8). These factors may complicate the assessment of the immediate effect of the antibiotic on the induction of erm expression. With these considerations in mind, we designed a reporter system which directly monitors the efficiency of initiation of translation from the erm(C) start codon and thus reflects the efficiency of erm induction. To generate such a reporter, we first introduced unique AflII restriction sites at codons 3 and 4 of erm(C) in plasmid pERMCT. This alteration did not affect the extent of induction of erm(C) expression (data not shown). We then fused codons 6 to 60 of the lacZ gene, which encode the α peptide of β-galactosidase (hereafter referred to as lacZα), to the first four codons of this modified erm(C) construct. The final reporter plasmid, pERMZα, had a convenient modular structure in which the regulatory cistron erm(C)L or the reporter gene could be individually manipulated due to the presence of unique sites at the beginning and the end of the corresponding cistrons (Fig. 3B). In E. coli cells capable of α complementation, macrolide-dependent expression of the reporter should turn cells blue on indicator X-Gal plates. Indeed, when an Etest strip (AB Biodisk) with an erythromycin concentration gradient was placed on a lawn of E. coli JM109 cells transformed with pERMZα, an intensive blue halo appeared around the strip (Fig. 5A). Erythromycin concentrations in the range of 1 to 128 μg/ml induced the reporter expression sufficiently so that it was readily detected on the plate after 24 to 36 h of incubation. Cell growth was inhibited at drug concentrations above 128 μg/ml, whereas erythromycin concentrations below 1 μg/ml did not induce reporter expression to the levels sufficient to produce a visible blue color. This result confirmed the previous observations of Weisblum's and Dubnau's laboratories that the induction of erm(C) occurs at drug concentrations much lower than those required to inhibit cell growth (9, 20, 28, 37) and verified the utility of the pERMZα reporter for use in studies of erm(C) induction.
FIG. 5.
Induction of expression of the pERMZα reporter in E. coli JM109 cells by various antibiotics. The plates contained ca. 1.5 ×108 cells plated in 0.6% LB agar on top of 1.5% LB agar containing ampicillin, IPTG, and X-Gal. (A) Induction by erythromycin. The Etest strip (AB Biodisk) contains a gradient of erythromycin concentrations from 0.016 to 256 μg/ml. (B) Induction by 14-member-ring macrolides and ketolides. Antibiotic discs were impregnated with the cladinose-containing macrolides erythromycin (Ery; 4.5 mg/ml), RU69874 (874; 10 mg/ml), RU66252 (252; 10 mg/ml), and clarithromycin (Clr; 8 mg/ml) and the cladinose-containing ketolides telithromycin (Tel; 1.5 mg/ml), HMR3004 (004; 1.5 mg/ml), and RU56066 (006; 12 mg/ml). Chloramphenicol (Chl; 0.3 mg/ml) was used as a control. The lack of reporter induction and of a growth inhibition zone around the RU56006 disc is likely explained by the low level of activity of this ketolide against E. coli cells. (C) Sixteen-member-ring macrolides josamycin (Jos), spiramycin (Spi), and tylosin (Tyl), all at 10 mg/ml. Erythromycin (Ery) and telithromycin (Tel) were used as controls. All plates were photographed after 36 to 48 h of incubation.
We next used plasmid pERMZα to qualitatively investigate the difference in reporter induction by a number of macrolide and ketolide drugs. Exponential-phase JM109 cells transformed with pERMZα were plated in soft agar on LB agar plates containing ampicillin, IPTG, and X-Gal. Discs impregnated with high concentrations of various macrolide antibiotics were placed on top of the plate. After 36 h of incubation at 37°C, the induction patterns were examined (Fig. 5B and C and 6A). The concentrations of drugs used on the discs were usually chosen in such a way that a clear zone of no growth was observed around the disc. This was done in order to ensure that the complete concentration range of the drug (from the very low one farthest away from the disc to that exceeding the MIC in the vicinity of the disc) is examined for possible induction effects.
FIG. 6.
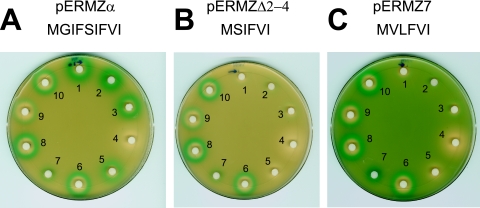
Mutations in the leader ORF affect the spectrum of inducing antibiotics. Plated cells contained the pERMZα reporter with the wild-type leader peptide ORF (A); a mutant lacking codons 2 to 4 of the leader peptide ORF (pERMZΔ2-4) (B); and a third mutant, pERMZ7, with a similar leader truncation and two additional mutations (which changed the nature of amino acid residues 2 and 3) (C). The sequence of the nascent leader peptide in the hypothetical stalled complex is indicated. The discs contained cladinose-containing macrolides (discs 1 to 5) or ketolides (discs 6 to 10) (all at 25 mg/ml): 1, erythromycin; 2, roxithromycin; 3, clarithromycin; 4, azithromycin; 5, RU66252; 6, HMR3004; 7, RU70645; 8, RU3562; 9, cethromycin; 10, telithromycin.
Two qualitative characteristics of the induction mode are worth attention: the intensity of the color of the blue ring, which reflects the extent of induction, and the width of the ring, which correlates with the range of antibiotic concentrations capable of induction of the reporter. A rather consistent pattern emerged when 14-member-ring macrolides and ketolides were compared. All the 3-cladinose-containing compounds (erythromycin, clarithromycin, roxithromycin, RU66252, RU69874) strongly induced the reporter at a broad range of concentrations (Fig. 5B and C and 6A). However, ketolides (telithromycin, cethromycin, RU3004, and RU3562) also induced the expression of the lacZα reporter, although the extent of the induction was lower than that seen with the 3-cladinose-containing macrolides, and the induction occurred within a narrower range of drug concentrations. A difference in the induction pattern afforded by macrolides and ketolides was observed between the drug pairs that differed only in the presence or absence of the C-3-cladinose (telithromycin versus RU69874 or HMR3004 versus RU66252) (Fig. 5B and 6A). Thus, it is the presence of the cladinose residue at position C-3 of the lactone ring that modulates the efficiency or timing of induction.
Notably, telithromycin reproducibly exhibited a stronger inducing activity than cethromycin (compare discs 9 and 10 in Fig. 6A). None of the three 16-member-ring macrolides tested (tylosin, spiramycin, and josamycin) induced reporter expression; this observation agreed with previously reported results (38) (Fig. 5C).
Unexpectedly, exposure to azithromycin, which is known to be a good inducer of erm, did not generate a blue coloration (Fig. 6A, disc 4). The experiment was repeated several times with different batches of azithromycin, but with the same result. Further experiments with JM109 cells transformed with the pUC18 vector constitutively expressing the LacZ α peptide led us to believe that this effect is related to specific inhibition by azithromycin either of β-galactosidase activity or of chromophore generation rather than an inability of the drug to induce the expression of the reporter (data not shown). Investigation of this unexpected effect of azithromycin was beyond the scope of the present work.
Altogether, the use of the pERMZα reporter confirmed the notion that ketolides are able to activate the expression of the inducible erm(C) gene and, thus, should generally be viewed as potential inducers.
Alteration of the leader ORF differentially affects induction by macrolides and ketolides.
The C-terminal sequence of the erm(C) leader nascent peptide that resides within the ribosome in the drug-induced stalled complex (…-I-F-V-I) bares some similarity to short resistance-conferring peptides whose expression renders cells resistant to erythromycin (M-X-I/L-F-V) (“E peptides”) (11, 30, 31, 33, 36). However, the size of the erythromycin resistance-conferring peptides is only 4 to 6 amino acids, while the size of the nascent peptide in the stalled erm leader complex is 9 amino acids. We wanted to test whether making the erm leader peptide more similar to E peptides would affect induction. Two constructs were engineered. In one, pERMZΔ2-4, codons 2 to 4 of the erm leader were deleted so that if the stalling site is not changed, the nascent peptide in the stalled complex is expected to be 6 amino acids long (MSIFVI). In the second construct, pERMZ7, the same deletion was combined with two additional mutations, Ser5→Val and Ile6→Leu. These mutations converted the sequence of the nascent peptide in the stalled complex to MVLFVI, which increased its resemblance to one of the most active E peptides, MVLFV (31). When a set of macrolide and ketolide compounds was tested for the ability to induce these new constructs, we observed that in spite of the increased similarity of the “new” leader peptides to E peptides, all the cladinose-containing macrolides lost their ability to induce the expression of the reporter in both plasmids with mutant leader peptide cistrons (Fig. 6B and C). In contrast, all the ketolides retained their ability to induce reporter expression, in spite of the shortening of the leader ORF sequence. This result shows that mutations in the inducible erm can change the spectrum of inducing antibiotics and may eliminate their induction by cladinose-containing macrolides while still maintaining a possibility of induction by ketolides.
DISCUSSION
The purpose of this work was to investigate the effect of ketolides, generally perceived as noninducers of inducible erm genes, on the expression of inducible erm(C). We found that the exposure of E. coli cells carrying inducible erm(C) to ketolide compounds results in increased dimethylation of A2058 in cellular ribosomes. In addition, ketolides could induce the expression of a reporter gene fused to the erm(C) start codon. These observations make us conclude that ketolides are, in fact, capable of inducing erm(C) expression.
The general notion that ketolides do not induce inducible erm genes came primarily from antibiotic susceptibility testing in which cells carrying inducible erm cassettes did not show a marked increase in resistance to ketolide compounds (2). In clinical (MIC) terms, this conclusion is likely to be generally correct. However, the intrinsic limitations of the MIC approach and the multifaceted nature of the development of the resistance phenotype may mask the immediate effect of ketolides on the induction of erm expression. Indeed, studies in which more direct readout systems were implied indicated that ketolides may exhibit some inducing activity on the expression of inducible erm(B) genes (25, 41).
Our data clearly show that ketolides are capable of erm(C) induction in E. coli. There are, however, notable differences in the mode of erm(C) induction afforded by ketolides compared to that afforded by cladinose-containing macrolides. First, the range of drug concentrations over which induction occurs is more narrow in the case of ketolides than in the case of macrolides. With macrolides, the induction ring around the antibiotic disc on a lawn of cells carrying the pERMZα reporter is wide and protrudes far away from the clear zone of inhibition of cell growth, indicating that a broad range of drug concentrations is capable of inducing reporter expression. In contrast, for ketolides the ring of induction is confined to a very narrow zone immediately adjacent to the clear zone of cell growth inhibition (Fig. 5B and C and 6). The implication of this observation is that ketolides can induce the expression of the reporter (and, thus, of erm) only within a narrow range of concentrations approaching the MIC. Exceeding this critical concentration effectively stops protein synthesis and prevents the expression of the inducible erm, whereas at a drug concentration that is too low, induction is inefficient. This is evidently one of the reasons why the general ability of ketolides to induce erm genes does not necessarily result in increased MICs. Second, the induction of A2058 dimethylation by ketolides occurs at a much slower rate than that by macrolides (Fig. 4C). Exposure of erm(C)-containing cells to erythromycin leads to a rapid increase in the level of dimethylation of A2058 in 23S rRNA and the ensuing resistance. In contrast, the onset of A2058 methylation by exposure of cells to telithromycin is significantly delayed compared to the time of onset observed with erythromycin, and the subsequent increase in the fraction of cellular ribosomes dimethylated at A2058 proceeds at a much slower rate. A slow onset of induction and a slow rate of accumulation of A2058-dimethylated ribosomes in the cell may mask the induction of erm(C) expression in conventional MIC testing. One needs to keep in mind that our experiments were carried out with the gram-negative organism E. coli, whereas the erm(C) gene apparently originates in gram-positive bacteria. The expression of inducible erm(C) in E. coli was reported to be “sluggish” compared to that in Staphylococcus aureus, and the extent of rRNA dimethylation in E. coli is lower than that in S. aureus (13, 32). However, the fact that ketolides induce erm(C) expression even in E. coli may mean that in S. aureus and other gram-positive pathogens, the induction of erm(C) by ketolides could be even more pronounced.
Even though our finding established that ketolides can induce erm(C) expression, it remains unclear whether the molecular mechanisms of ketolide-dependent induction are the same as those proposed for erythromycin-dependent induction, which imply stalling of the ribosome during the translation of the erm(C)L ORF (37). Some observations argue that these mechanisms may be different. Truncation of the leader peptide ORF by three codons eliminated the induction of reporter expression by cladinose-containing macrolides, although ketolide-dependent induction was still retained (Fig. 6). Similar effects were observed with some point mutations in erm(C)L, where individual amino acid substitutions in the wild-type Erm(C)L sequence eliminated erythromycin-dependent but not telithromycin-dependent induction (N. Vazquez-Laslop and A. S. Mankin, submitted for publication). In addition, toe-printing studies showed that erythromycin but not telithromycin induced the formation of a stable stalled ribosome complex on the wild-type erm(C)L mRNA (Vazquez-Laslop and Mankin, submitted). The latter observation suggests that either the ketolide-induced stalled complex is short-lived or that ketolide-dependent induction operates through a totally different mechanism, one that is possibly independent of ribosome stalling. Further studies will be needed to elucidate the details of the molecular mechanisms of macrolide- and ketolide-dependent induction of inducible erm genes to help design drugs that are true noninducers.
In the course of this work, we have constructed a reporter plasmid, pERMZα, which, when it is present in cells capable of α complementation, provides a fast and facile way to assay the inducing ability of multiple antibiotics. Several reporters of inducible erm expression have been developed in the past by utilizing either the full-size β-galactosidase (LacZ) gene or green fluorescent protein (GFP) gene (6, 14, 23, 25). We believe that our reporter, based on the β-galactosidase α peptide, provides a considerable advantage over these previously described systems. The high background of β-galactosidase activity observed with the constructs carrying the entire lacZ gene prevents the visual detection of induction on indicator plates. Indeed, when the lacZα gene in the pERMZα construct was replaced with the complete lacZ gene, transformed cells formed dark blue colonies even in the absence of the inducing antibiotic, apparently because the translation attenuation system is not strong enough to completely prevent the translation of the downstream cistron in the erm(C) cassette. The use of GFP generally helps to avoid this problem (6), but the slow folding and high degree of stability of GFP may potentially complicate interpretation of the data. We believe that our reporter, which provides an easy readout of inducible erm(C) expression without the necessity for any specialized equipment, may find broad use in studies of the erm-inducing ability of various antibiotics. The only problem that we encountered with our reporter was the unexpected inhibitory effect of azithromycin on β-galactosidase activity. Among the many compounds tested, azithromycin was the only one which exhibited such inhibition. Nevertheless, the potential ability of other drugs to inhibit directly the activity of β-galactosidase must be kept in mind.
Finally, we also noticed that cells carrying the pERMZ7 construct exhibited a bluish hue on indicator plates, even in the absence of an inducing antibiotic (Fig. 6C). This could result from the altered ground-state folding of the mutant mRNA. Another possible explanation is that certain short nascent peptides may stall the ribosome even in the absence of a drug. Such stalling may be used by the cell for the regulation of gene expression and deserves further investigation.
Acknowledgments
We thank Liqun Xiong for help with some experiments, Hiroshi Nokaido for strain ZK796, and Nora Vazquez-Laslop for critical reading of the manuscript.
This work was supported by a grant (grant MCB0515934) from the National Science Foundation.
Footnotes
Published ahead of print on 17 December 2007.
REFERENCES
- 1.Agouridas, C., A. Bonnefoy, and J. F. Chantot. 1997. Antibacterial activity of RU 64004 (HMR 3004), a novel ketolide derivative active against respiratory pathogens. Antimicrob. Agents Chemother. 41:2149-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnefoy, A., A. M. Girard, C. Agouridas, and J. F. Chantot. 1997. Ketolides lack inducibility properties of MLS(B) resistance phenotype. J. Antimicrob. Chemother. 40:85-90. [DOI] [PubMed] [Google Scholar]
- 3.Bryskier, A. 2000. Ketolides—telithromycin, an example of a new class of antibacterial agents. Clin. Microbiol. Infect. 6:661-669. [DOI] [PubMed] [Google Scholar]
- 4.Champney, W. S., and R. Burdine. 1996. 50S ribosomal subunit synthesis and translation are equivalent targets for erythromycin inhibition in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1301-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champney, W. S., H. S. Chittum, and C. L. Tober. 2003. A 50S ribosomal subunit precursor particle is a substrate for the ermC methyltransferase in Staphylococcus aureus cells. Curr. Microbiol. 46:453-460. [DOI] [PubMed] [Google Scholar]
- 6.Clarebout, G., and R. Leclercq. 2002. Fluorescence assay for studying the ability of macrolides to induce production of ribosomal methylase. Antimicrob. Agents Chemother. 46:2269-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douthwaite, S., D. Fourmy, and S. Yoshizawa. 2005. Nucleotide methylations in rRNA that confer resistance to ribosome-targeting antibiotics. Top. Curr. Genet. 12:287-309. [Google Scholar]
- 8.Douthwaite, S., L. H. Hansen, and P. Mauvais. 2000. Macrolide-ketolide inhibition of MLS-resistant ribosomes is improved by alternative drug interaction with domain II of 23S rRNA. Mol. Microbiol. 36:183-193. [DOI] [PubMed] [Google Scholar]
- 9.Dubnau, D. 1985. Induction of ermC requires translation of the leader peptide. EMBO J. 4:533-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felmingham, D., R. Canton, and S. G. Jenkins. 2007. Regional trends in beta-lactam, macrolide, fluoroquinolone and telithromycin resistance among Streptococcus pneumoniae isolates 2001-2004. J. Infect. 55:111-118. [DOI] [PubMed] [Google Scholar]
- 11.Gaynor, M., and A. S. Mankin. 2003. Macrolide antibiotics: binding site, mechanism of action, resistance. Curr. Top. Med. Chem. 3:949-961. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, L. H., P. Mauvais, and S. Douthwaite. 1999. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol. Microbiol. 31:623-632. [DOI] [PubMed] [Google Scholar]
- 13.Hardy, K., and C. Haefeli. 1982. Expression in Escherichia coli of a staphylococcal gene for resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J. Bacteriol. 152:524-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirsch, D. R., and M. H. Lai. 1984. Regulation of a macrolide resistance-beta-galactosidase (ermC-lacZ) gene fusion in Escherichia coli. J. Bacteriol. 159:381-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klugman, K. P., and J. R. Lonks. 2005. Hidden epidemic of macrolide-resistant pneumococci. Emerg. Infect. Dis. 11:802-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai, C. J., and B. Weisblum. 1971. Altered methylation of ribosomal RNA in an erythromycin-resistant strain of Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 68:856-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langley, K. E., M. R. Villarejo, A. V. Fowler, P. J. Zamenhof, and I. Zabin. 1975. Molecular basis of beta-galactosidase alpha-complementation. Proc. Natl. Acad. Sci. USA 72:1254-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leclercq, R., and P. Courvalin. 1991. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 35:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leclercq, R., and P. Courvalin. 2002. Resistance to macrolides and related antibiotics in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:2727-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayford, M., and B. Weisblum. 1989. Conformational alterations in the ermC transcript in vivo during induction. EMBO J. 8:4307-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayford, M., and B. Weisblum. 1989. ermC leader peptide. Amino acid sequence critical for induction by translational attenuation. J. Mol. Biol. 206:69-79. [DOI] [PubMed] [Google Scholar]
- 22.Mayford, M., and B. Weisblum. 1990. The ermC leader peptide: amino acid alterations leading to differential efficiency of induction by macrolide-lincosamide-streptogramin B antibiotics. J. Bacteriol. 172:3772-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Min, Y. H., J. H. Jeong, Y. J. Choi, H. J. Yun, K. Lee, M. J. Shim, J. H. Kwak, and E. C. Choi. 2003. Heterogeneity of macrolide-lincosamide-streptogramin B resistance phenotypes in enterococci. Antimicrob. Agents Chemother. 47:3415-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7-A6, vol. 23, 6th ed. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 25.Rosato, A., H. Vicarini, A. Bonnefoy, J. F. Chantot, and R. Leclercq. 1998. A new ketolide, HMR 3004, active against streptococci inducibly resistant to erythromycin. Antimicrob. Agents Chemother. 42:1392-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 28.Shivakumar, A. G., J. Hahn, G. Grandi, Y. Kozlov, and D. Dubnau. 1980. Posttranscriptional regulation of an erythromycin resistance protein specified by plasmic pE194. Proc. Natl. Acad. Sci. USA 77:3903-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sigmund, C. D., M. Ettayebi, A. Borden, and E. A. Morgan. 1988. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 164:673-690. [DOI] [PubMed] [Google Scholar]
- 30.Tenson, T., and A. S. Mankin. 2001. Short peptides conferring resistance to macrolide antibiotics. Peptides 22:1661-1668. [DOI] [PubMed] [Google Scholar]
- 31.Tenson, T., L. Xiong, P. Kloss, and A. S. Mankin. 1997. Erythromycin resistance peptides selected from random peptide libraries. J. Biol. Chem. 272:17425-17430. [DOI] [PubMed] [Google Scholar]
- 32.Thakker-Varia, S., A. C. Ranzini, and D. T. Dubin. 1985. Ribosomal RNA methylation in Staphylococcus aureus and Escherichia coli: effect of the “MLS” (erythromycin resistance) methylase. Plasmid 14:152-161. [DOI] [PubMed] [Google Scholar]
- 33.Tripathi, S., P. S. Kloss, and A. S. Mankin. 1998. Ketolide resistance conferred by short peptides. J. Biol. Chem. 273:20073-20077. [DOI] [PubMed] [Google Scholar]
- 34.Tu, D., G. Blaha, P. B. Moore, and T. A. Steitz. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121:257-270. [DOI] [PubMed] [Google Scholar]
- 35.Vester, B., and S. Douthwaite. 1994. Domain V of 23S rRNA contains all the structural elements necessary for recognition by the ErmE methyltransferase. J. Bacteriol. 176:6999-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vimberg, V., L. Xiong, M. Bailey, T. Tenson, and A. Mankin. 2004. Peptide-mediated macrolide resistance reveals possible specific interactions in the nascent peptide exit tunnel. Mol. Microbiol. 54:376-385. [DOI] [PubMed] [Google Scholar]
- 37.Weisblum, B. 1995. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob. Agents Chemother. 39:797-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weisblum, B., M. Y. Graham, T. Gryczan, and D. Dubnau. 1979. Plasmid copy number control: isolation and characterization of high-copy-number mutants of plasmid pE194. J. Bacteriol. 137:635-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong, L., S. Shah, P. Mauvais, and A. S. Mankin. 1999. A ketolide resistance mutation in domain II of 23S rRNA reveals proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31:633-639. [DOI] [PubMed] [Google Scholar]
- 40.Zgurskaya, H. I., and H. Nikaido. 2000. Cross-linked complex between oligomeric periplasmic lipoprotein AcrA and the inner-membrane-associated multidrug efflux pump AcrB from Escherichia coli. J. Bacteriol. 182:4264-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong, P., Z. Cao, R. Hammond, Y. Chen, J. Beyer, V. D. Shortridge, L. Y. Phan, S. Pratt, J. Capobianco, K. A. Reich, R. K. Flamm, Y. S. Or, and L. Katz. 1999. Induction of ribosome methylation in MLS-resistant Streptococcus pneumoniae by macrolides and ketolides. Microb. Drug Resist. 5:183-188. [DOI] [PubMed] [Google Scholar]