Abstract
While most Staphylococcus aureus telithromycin-resistant mutants isolated in this study possessed duplications within rplV (encoding ribosomal protein L22), four isolates possessed insertions within rplV that were identical to a portion of the gene rplB (encoding ribosomal protein L2). This novel type of mutation is the result of an apparent gene conversion-like event.
Macrolide antibiotics inhibit bacterial protein synthesis through the binding of 23S rRNA. This binding is thought to block the entry of nascent peptides into the exit tunnel on the 50S ribosomal subunit (11, 13). In the laboratory, spontaneous resistance to macrolides typically occurs by mutations in 23S rRNA (15) or mutations in ribosomal proteins L4 and L22 (4). While alterations in 23S rRNA alter the affinity of macrolides for the ribosome (15), the effects of mutations in L4 and L22, which make contacts within the peptide exit tunnel, are indirect (5, 14). In the case of a small deletion within L22, the entry site of the exit tunnel is widened (5, 14), possibly explaining the mechanism of resistance (5). Resistance in the clinic also occurs through the acquisition of methyltransferases, which are macrolide inducible or constitutively expressed, that modify 23S rRNA, leading to decreased binding of the antibiotic (9). The increasing problem of resistance to macrolides has led to the development of newer molecules that circumvent some of the problems of their predecessors. One example of such molecules is the ketolide telithromycin, which differs from macrolides by virtue of a keto group in the place of a cladinose at position 3 of the 14-member macrolide ring (4, 7). Telithromycin also has an 11,12 carbamate bridge with an alkyl-aryl extension attached. The alkyl-aryl attachment adds additional contacts within domain II of the ribosome, leading to tighter binding, and likely accounts for the observed increased potency against most bacteria (7). An additional feature of telithromycin is that it does not stimulate expression of inducible methyltransferases (7).
The novel binding of telithromycin suggested that laboratory-generated resistance to telithromycin might result from novel mutations in L4 and/or L22. Consequently, resistant mutants were generated in Staphylococcus aureus and characterized. Resistant mutants of S. aureus RN4220 (MIC = 0.03 μg/ml) were selected by spreading 0.3 ml of an overnight culture grown in brain heart infusion broth onto each of 10 Luria-Bertani plates containing 1 μg/ml telithromycin followed by incubation at 37°C for 1 week. After purification on 1 μg/ml telithromycin and MIC determination, the genes encoding ribosomal proteins L4 and L22, rplD and rplV, respectively, were sequenced. As shown in Table 1, each isolate exhibited high MICs to both telithromycin and erythromycin. Sequence analysis indicated that, of the 16 isolates obtained, 14 had mutations in rplV. The remaining two isolates had no alteration in rplD or rplV. No further characterization was performed on these two mutants. Of the 14 isolates that had mutant rplV, 10 had duplications of short stretches of bases in the region encoding the carboxy terminus of L22. The mutations in these 10 isolates are represented by six different types (Table 2). These mutations led to amino acid duplications in the L22 protein (Table 1). Mutants with duplications within the carboxy-terminal region of L22 have been identified in macrolide- and ketolide-resistant isolates of a number of different organisms (1, 2, 3, 4, 6). In S. aureus, identical and very similar mutants were isolated by selection on the streptogramin mixture quinupristin-dalfopristin (8).
TABLE 1.
Alignment of protein sequences and phenotypic properties of S. aureus telithromycin-resistant mutants
| Isolate | Protein sequencea | MIC (μg/ml)b of:
|
Doubling time(min)c | |
|---|---|---|---|---|
| TEL | ERY | |||
| Parent | NYDMNTDELVVKEAYANEG-----PTLKRFRPRAQG---------------RASAINKRTS---------HITIVVS | 0.03 | 0.12 | 30 |
| KT01 | NYDMNTDELVVKEAYANEG-----PTLKRFRPTVRGSVMNPNDHPHGGGEGRASAINKRTS---------HITIVVS | 16 | 32 | 89 |
| KT02 | NYDMNTDELVVKEAYANEG-----PTLKGIRPTVRGSVMNPNDHPHGGGEGRASAINKRTS---------HITIVVS | 8 | 32 | 124 |
| KT04 | NYDMNTDELVVKEAYANEG-----PTLKRFRPRAQG---------------RASAINKRTSHITINKRTSHITIVVS | 16 | 32 | NDd |
| KT05 | NYDMNTDELVVKEAYANEG-----PTLKRFRPRAQG---------------RASAINKRTRSAINKRT-SHITIVVS | 16 | 32 | ND |
| KT06 | NYDMNTDELVVKEAYANEG-----PTLKRFRPRAQG---------------RASAINKRTSAINKRT----SHITIVVS | 4 | 16 | ND |
| KT09 | NYDMNTDELVVKEAYANEG-----PTLKRFRPRAQG---------------RASAINSRASAIN----KRTSHITIVVS | 2 | 8 | ND |
| KT10 | NYDMNTDELVVKEAYANEGPTLEGPTLKRFRPRAQG---------------RASAINKRTS---------HITIVVS | 4 | 4 | ND |
| KT11 | NYDMNTDELVVKEAYANEG-----PTLKRFRPRVRPR-----------AQGRASAINKRTS---------HITIVVS | 8 | 16 | ND |
The start of each protein sequence corresponds to residue 61 of parent L22. Residues in boldface are inserted relative to the parent sequence. Underlined residues are duplicated relative to parent sequence.
TEL, telithromycin, ERY, erythromycin.
Determined in Luria-Bertani medium at 37°C.
ND, not determined.
TABLE 2.
Alignment of rplV sequences of S. aureus telithromycin-resistant mutants
| Isolate | DNA sequencea |
|---|---|
| Parent | ATATGCTAACGAAGGACCAACATTAAAACGTTTCCGTCCACGTGCGCAAGGTCGTGCAAGTGCGATTAACAAACGTACAAGCCACATTACAATCGTCGTAAG |
| KT01 | ATATGCTAACGAAGGACCAACATTAAAACGTTTCCGTCCAACAGTTCGTGGTTCTGTAATGAACCCTAACGATCACCCACACGGTGGTGGTGAAGGTCGTGC |
| KT02 | ATATGCTAACGAAGGACCAACATTAAAAGGTATCCGTCCAACAGTTCGTGGTTCTGTAATGAACCCTAACGATCACCCACACGGTGGTGGTGAAGGTCGTGC |
| KT04 | ATATGCTAACGAAGGACCAACATTAAAACGTTTCCGTCCACGTGCGCAAGGTCGTGCAAGTGCGAT(TAACAAACGTACAAGCCACATTACAAT)2CGTCGT |
| KT05 | ATATGCTAACGAAGGACCAACATTAAAACGTTTCCGTCCACGTGCGCAAGGTCGTGC(AAGTGCGATTAACAAACGTACAAG)2CCACATTACAATCGTCGT |
| KT06 | ATATGCTAACGAAGGACCAACATTAAAACGTTTCCGTCCACGTGCGCAAGGTCGTGCAAG(TGCGATTAACAAACGTACAAG)2CCACATTACAATCGTCGT |
| KT09 | ATATGCTAACGAAGGACCAACATTAAAACGTTTCCGTCCACGTGCGCAAGGTCGTGCGCAAG(GTCGTGCAAGTGCGATTAACA)2AACGTACAAGCCACAT |
| KT10 | ATATGCTAAC(GAAGGACCAACATTA)2AAACGTTTCCGTCCACGTGCGCAAGGTCGTGCAAGTGCGATTAACAAACGTACAAGCCACATTACAATCGTCGT |
| KT11 | ATATGCTAACGAAGGACCAACATTAAAACGTT(TCCGTCCACGTG)2CGCAAGGTCGTGCAAGTGCGATTAACAAACGTACAAGCCACATTACAATCGTCGT |
The start of each sequence corresponds to position 222 of the wild-type rplV gene. Boldface, DNA inserted relative to the parent sequence. Bases in parentheses are duplicated relative to the parent in the mutant.
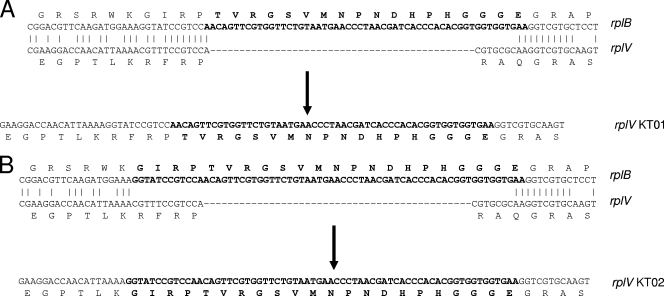
In the remaining four isolates, two similar mutations, resulting in the replacement of amino acids 88 to 90 with the sequence TVRGSVMNPNDHPHGGGE or the replacement of amino acids 84 to 90 with the related sequence GIRPTVRGSVMNPNDHPHGGGE, were identified. A database search with the insertion sequences revealed that they matched a part of ribosomal protein L2 whose gene, rplB, is located upstream of rplV, the two genes being separated by rpsS (encoding ribosomal protein S19). The distance between the insertion point in rplV and the portion homologous with rplB is ca. 790 bp. Comparison of rplB with rplV in the region of the insertion identified limited regions of homology flanking the insertion points of rplV (Fig. 1). It seems likely that this pairing, though limited in length, leads to recombination between the two genes. In an attempt to determine if a reciprocal crossover occurred in rplB, the rplB genes from the four mutants were sequenced and found to be intact. This apparent nonreciprocal transfer of DNA from rplB to rplV is most simply described as gene conversion though it must be noted that a possible gene conversion event can come from gene conversion itself or from a double-crossover event between sister strands that occurs during replication (10, 12). This is the first observation of recombination of any sort between two ribosomal protein genes.
FIG. 1.
Alignment of rplB and rplV at the region surrounding the insertion site of rplV in the telithromycin-resistant mutants. Also shown are the resulting rplV sequences after the gene conversion-like event (arrow). (A) rplV sequence of KT01; (B) rplV sequence of KT02. Boldface, inserted residues and bases. Note the difference in similarity between the 3′ flanks of rplB and rplV among the two types of mutations.
In an effort to determine if rplB-rplV recombination is unique to selection for telithromycin resistance, mutants were selected on 10 μg/ml erythromycin using the same conditions as for the original experiment. Primers specific to rplB and rplV were used to screen colonies for the recombination event by PCR. Of 19 high-level-erythromycin-resistant colonies none showed evidence of the recombination event. Likewise, 27 mutants were selected on 5 μg/ml erythromycin, and none exhibited evidence of recombination. No mutants were recovered when selected on 20 or 50 μg/ml erythromycin. As a control, five additional mutants were selected on 1 μg/ml telithromycin. In this case one of the five isolated mutants exhibited rplB-rplV recombination. The levels used for selection on erythromycin, 5 and 10 μg/ml (40 and 80 times the MIC, respectively), are comparable to the level used for selection of telithromycin resistance (33 times the MIC). From this it appears that the recombination event is specific to telithromycin selection.
Mutants exhibiting the rplB-rplV recombination event grow extremely slow; the doubling times in a rich medium are three and four times longer than that of the parent (Table 1). This growth defect likely indicates a fitness cost, which, in turn, suggests that the mutations are not clinically relevant.
The mechanism by which high levels of telithromycin, but not erythromycin, lead to the recombination event we report here is open to speculation. Because recombination increases upon induction of the SOS response, one possibility is that high levels of telithromycin induce an SOS response while high levels of erythromycin do not.
Footnotes
Published ahead of print on 14 January 2008.
REFERENCES
- 1.Cagliero, C., C. Mouline, A. Cloeckaert, and S. Payot. 2006. Synergy between efflux pump CmeABC and modifications in ribosomal proteins L4 and L22 in conferring macrolide resistance in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 50:3893-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doktor, S. Z., V. D. Shortridge, J. M. Beyer, and R. K. Flamm. 2004. Epidemiology of macrolide and/or lincosamide resistant Streptococcus pneumoniae clinical isolates with ribosomal mutations. Diagn. Microbiol. Infect. Dis. 49:47-52. [DOI] [PubMed] [Google Scholar]
- 3.Farrell, D. J., I. Morrissey, S. Bakker, S. Buckridge, and D. Felmingham. 2004. In vitro activities of telithromycin, linezolid, and quinupristin-dalfopristin against Streptococcus pneumoniae with macrolide resistance due to ribosomal mutations. Antimicrob. Agents Chemother. 48:3169-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franceschi, F., Z. Kanyo, E. C. Sherer, and J. Sutcliffe. 2004. Macrolide resistance from the ribosome perspective. Curr. Drug Targets Infect. Disord. 4:177-191. [DOI] [PubMed] [Google Scholar]
- 5.Gabashvili, I. S., S. T. Gregory, M. Valle, R. Grassucci, M. Worbs, M. C. Wahl, A. E. Dahlberg, and J. Frank. 2001. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell 8:181-188. [DOI] [PubMed] [Google Scholar]
- 6.Hisanaga, T., D. J. Hoban, and G. G. Zhanel. 2005. Mechanisms of resistance to telithromycin in Streptococcus pneumoniae. J. Antimicrob. Chemother. 56:447-450. [DOI] [PubMed] [Google Scholar]
- 7.Lonks, J. R., and D. A. Goldmann. 2005. Telithromycin: a ketolide antibiotic for treatment of respiratory tract infections. Clin. Infect. Dis. 40:1657-1664. [DOI] [PubMed] [Google Scholar]
- 8.Malbruny, B., A. Canu, B. Bozdogan, B. Fantin, V. Zarrouk, S. Dutka-Malen, C. Feger, and R. Leclercq. 2002. Resistance to quinupristin-dalfopristin due to mutation of L22 ribosomal protein in Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2200-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakajima, Y. 1999. Mechanisms of bacterial resistance to macrolide antibiotics. J. Infect. Chemother. 5:61-74. [DOI] [PubMed] [Google Scholar]
- 10.Santoyo, G., and D. Romero. 2005. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol. Rev. 29:169-183. [DOI] [PubMed] [Google Scholar]
- 11.Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 12.Segall, A. M., and J. R. Roth. 1994. Approaches to half-tetrad analysis in bacteria: recombination between repeated, inverse-order chromosomal sequences. Genetics 136:27-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tenson, T., M. Lovmar, and M. Ehrenberg. 2003. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330:1005-1014. [DOI] [PubMed] [Google Scholar]
- 14.Tu, D., G. Blaha, P. B. Moore, and T. A. Steitz. 2005. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 121:257-270. [DOI] [PubMed] [Google Scholar]
- 15.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]