Abstract
The genetic relatedness of ciprofloxacin-resistant Streptococcus pneumoniae isolates collected from 1997 to 2002 (n = 82) and 2003 to 2005 (n = 123) was compared by pulsed-field gel electrophoresis (PFGE). Increased genetic homogeneity among the isolates from 2003 to 2005 (cluster analysis; P < 0.001) appeared to be due to expansion of existing clonal groups and to introduction of new PFGE types.
Although fluoroquinolone resistance in Streptococcus pneumoniae has been slow to emerge, clinical isolates with reduced susceptibility have been described and continue to increase in prevalence in some regions (4, 7, 9, 12). Previous observations provide strong evidence that the spread of penicillin- and multidrug-resistant S. pneumoniae is often driven by the dissemination of a few successful clones (11, 15), and it is has been suggested that further increases in fluoroquinolone resistance will also likely be associated with clonal spread (19).
We recently reported that ciprofloxacin resistance in Canadian S. pneumoniae isolates increased from 0.6% in 1997/1998 to 1.7% in 2000/2001 (1). In 2003, resistance rates increased significantly to 2.3% (P < 0.05) and continued to rise annually to 4.2% by 2005 (1). The objectives of this study were to investigate the role of clonal spread in the rapid increase of ciprofloxacin resistance in S. pneumoniae in Canada since 2003 and to determine whether these isolates are genetically related to each other or to known pandemic clones. In our analysis we compared the genetic relatedness of isolates collected between 1997 and 2002 with isolates collected from 2003 to 2005.
In October 1997, a national surveillance study was initiated to monitor the activity of antimicrobial agents against Canadian respiratory tract isolates of S. pneumoniae. Between October 1997 and December 2005 inclusive, 10,741 clinical isolates of S. pneumoniae were submitted to the coordinating laboratory (Department of Clinical Microbiology, Health Sciences Centre, Winnipeg, Manitoba, Canada) from 25 medical centers from all regions of Canada (1, 20).
Susceptibility testing was performed by broth microdilution in accordance with CLSI (formerly NCCLS) recommendations (17) and MIC interpretive standards were defined according to CLSI breakpoints (5). Resistance to ciprofloxacin was defined as an MIC of ≥4 μg/ml (CIPRO package insert; Bayer Healthcare).
Over the course of the study, 205 ciprofloxacin-resistant S. pneumoniae isolates were identified and characterized by pulsed-field gel electrophoresis (PFGE) (13). Twenty-six representative international pneumococcal clones recognized by the Pneumococcal Molecular Epidemiology Network were included in the analysis (15). PFGE profiles were digitized for analysis with BioNumerics software (version 3.5; Applied Maths, Inc., Austin, TX), and dendrograms were constructed by the unweighted pair group method with arithmetic averages with the Dice coefficient (1.0% optimization, 1.5% position tolerance). Isolates were considered to be genetically related if the Dice coefficient of similarity was ≥80%. For the purpose of data analysis, groups were defined as three or four isolates with ≥80% genetic relatedness and clusters were defined as five or more isolates with ≥80% identity.
Sequencing of the quinolone resistance-determining regions (QRDRs) of parC and gyrA was performed previously (1, 16).
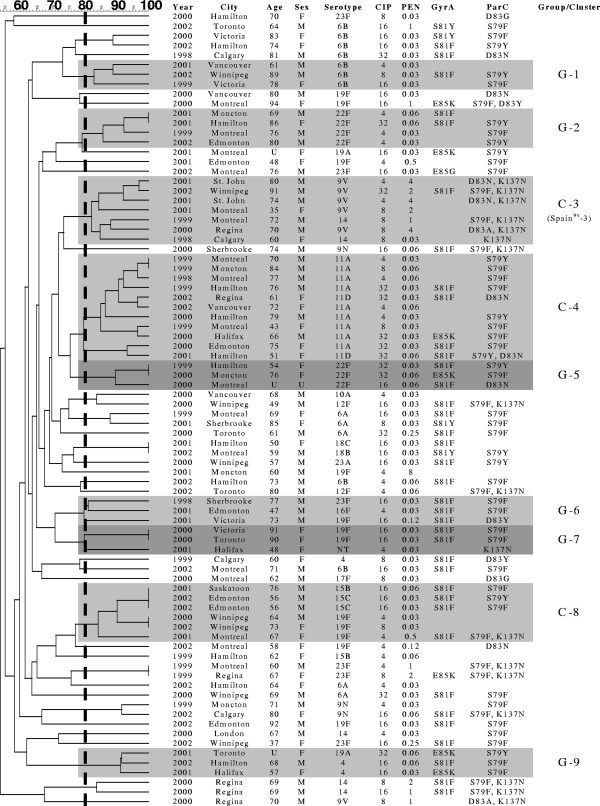
Eighty-two ciprofloxacin-resistant pneumococcal isolates (1.2% [82/6,927]) were collected between October 1997 and December 2002. PFGE analysis revealed six groups of three or four isolates accounting for 19 (23.2%) of the 82 strains (Table 1). Only three clusters of five or more genetically related isolates were observed, containing a total of 24 (29.3%) isolates. Cluster 3 (Fig. 1) included seven isolates, five serotype 9V isolates and two serotype 14 isolates. Comparison to 26 international antibiotic-resistant clones showed that these isolates belonged to PFGE types closely related (≥80%) to the Spain9V-3 clone. The antibiograms of all five serotype 9V isolates were identical to each other and to that of Spain9V-3. Unexpectedly, QRDR mutation patterns were not consistent for isolates in cluster 3.
TABLE 1.
Number of ciprofloxacin-resistant Streptococcus pneumoniae isolates in PFGE groups and clusters
| Parameter | Value for isolates collected during study period
|
P value | |
|---|---|---|---|
| 1997 to 2002 (n = 82) | 2003 to 2005 (n = 123) | ||
| No. of groups (3 or 4 isolates) | 6 | 4 | |
| Total no. of isolates in groups | 19 | 14 | 0.0324 |
| No. of clusters (≥5 isolates) | 3 | 7 | |
| Total no. of isolates in clusters | 24 | 72 | 0.0001 |
| No. of groups and clusters (≥3 isolates) | 9 | 11 | |
| Total no. of isolates in groups and clusters | 43 | 86 | 0.0125 |
FIG. 1.
Genetic relatedness of 82 ciprofloxacin-resistant S. pneumoniae isolates from Canada from 1997 to 2002. The figure shows the genetic relatedness of the isolates (from 60% to 100%) according to the Dice coefficient of similarity and information about the isolates and patients, including the age (in years) and sex (male [M] or female [F]) of the patient, ciprofloxacin MIC (CIP) (μg/ml), penicillin MIC (PEN) (μg/ml), and mutations in GyrA and ParC. The isolates in a group (three or four isolates with ≥80% genetic relatedness) and cluster (five or more isolates with ≥80% genetic relatedness) are indicated by shading. G-1, group 1; C-3, cluster 3.
Cluster 4 contained 11 genetically related isolates belonging to serogroup 11. Five isolates had a ParC mutation only (S79Y/F and/or D83N), and five contained an S81F or E85K mutation in GyrA in addition to the mutation in ParC. Three isolates of serotype 15B/C and three of serotype 19F comprised cluster 8. Serogroup 15 isolates contained mutations at S79 in ParC and S81 in GyrA, while only one of the remaining 19F isolates had QRDR changes. Isolates within these latter two clusters (clusters 4 and 8) were unrelated to the international clones.
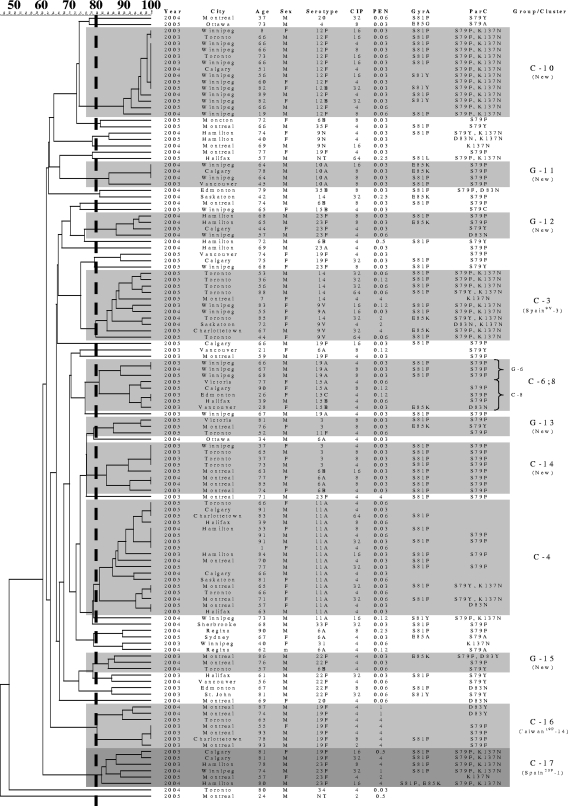
From January 2003 to December 2005, 123 ciprofloxacin-resistant S. pneumoniae isolates (3.2% [123/3,814]) were collected. Dendrogram analysis identified 11 groups and clusters (three or more isolates) accounting for 86 (69.9%) strains (Table 1). The majority (72 [83.7%]) of those isolates belonged to seven clusters containing five or more isolates. Among the 11 groups or clusters, eight new PFGE types were identified (four groups and four clusters) (Fig. 2). In addition, four genotypes (cluster 3, cluster 4, group 6, and cluster 8) seen in S. pneumoniae isolates collected from 1997 to 2002 were also observed in 2003 to 2005 isolates. Interestingly, there appeared to be convergence of group 6 and cluster 8 into a single genotype (designated cluster 6;8). This included three isolates of serotype 19A and five belonging to serogroup 15. Seven (87.5%) of the eight isolates had an S79F or D83N mutation in ParC; only four (50.0%) had a GyrA mutation (S81F or E85K).
FIG. 2.
Genetic relatedness of 123 ciprofloxacin-resistant S. pneumoniae isolates from Canada from 2003 to 2005. The figure shows the genetic relatedness of the isolates (from 60% to 100%) according to the Dice coefficient of similarity and information about the isolates and patients, including the age (in years) and sex (male [M] or female [F]) of the patient, ciprofloxacin MIC (CIP) (μg/ml), penicillin MIC (PEN) (μg/ml), and mutations in GyrA and ParC. The isolates in a group (three or four isolates with ≥80% genetic relatedness) and cluster (five or more isolates with ≥80% genetic relatedness) are indicated by shading. C-10, cluster 10 (a new cluster); G-11, group 11 (a new group).
As with the earlier (1997 to 2002) isolates, the 11 pneumococcal isolates belonging to cluster 3 were genetically related to the Spain9V-3 clone. Approximately 50% were serotype 9V, and 50% were serotype 14. Almost all isolates had identical mutation patterns, including two ParC substitutions and one GyrA change.
Cluster 4 contained 18 isolates belonging to serotype 11A. QRDR mutation patterns were not consistent. Despite being ciprofloxacin resistant, almost half of the isolates had no QRDR mutations. Cluster 10 included 14 isolates that belonged to serotype 12F. All isolates contained S79F and K137N mutations in ParC, and only three did not carry a mutation at S81 in GyrA. These isolates showed the highest degree of genetic similarity (>90%); however, all but three were isolated in Winnipeg, Manitoba, Canada.
Four isolates of serotype 3 and four belonging to serogroup 6 comprised cluster 14. All eight strains had identical resistance profiles and QRDR mutations (S79F in ParC and S81F in GyrA). Neither cluster 10 nor cluster 14 was related to known pneumococcal clones. Isolates in the final two clusters (cluster 16 and cluster 17) were found to be genetically related to the Taiwan19F-14 and Spain23F-1 clones, respectively. All of our ciprofloxacin-resistant S. pneumoniae isolates related to these clones were multidrug resistant. Cluster 16 contained seven serotype 19F isolates while cluster 17 contained a total of six isolates, four serotype 23F isolates and two serotype 19F isolates.
High rates of fluoroquinolone resistance in Hong Kong have been linked to the local dissemination of a strain related to the Spain23F-1 clone (8). Strains related to this and other recognized pneumococcal clones (Spain6B-2, Spain9V-3, Tennessee23F-4, Spain14-5, England14-9, Taiwan19F-14, Taiwan23F-15, Maryland6B-17, Tennessee14-18, Greece6B-22, or Utah35B-24) have also been identified in other countries throughout North America and Europe (2, 3, 6, 10, 14, 18, 19). Despite genetic similarity, the association between clonal spread and fluoroquinolone resistance remains unclear (6). In contrast, genetic heterogeneity among isolates from other regions suggests that clonal spread is not responsible for the observed increases in fluoroquinolone-resistant pneumococci but rather that fluoroquinolone resistance in S. pneumoniae has emerged primarily through de novo mutational events (3, 6, 10, 14, 19).
In our study, we found a greater degree of genetic homogeneity among ciprofloxacin-resistant S. pneumoniae isolates collected from 2003 to 2005 than among ciprofloxacin-resistant strains isolated between 1997 and 2002. Many of the isolates from the 1997 to 2002 study period were genetically unrelated, with only nine PFGE types identified with three or more isolates and only one small cluster of isolates related to the Spain9V-3 clone. Variations in QRDR substitution patterns, serotypes, and antibiotic susceptibilities among some isolates with similar PFGE patterns suggest that new mutational events continue to play a large role in the spread of fluoroquinolone-resistant S. pneumoniae in Canada. However, the observed increase in genetic homogeneity among ciprofloxacin-resistant isolates collected between 2003 and 2005 appears to be the result of a combination of the expansion of existing clonal groups (i.e., Spain9V-3), the appearance and detection of fluoroquinolone resistance among additional international clones (i.e., Spain23F-1 and Taiwan19F-14) in the local pneumococcal population, and the introduction of new PFGE types (clones) containing greater numbers of isolates (seven new clusters of five or more isolates).
Potential limitations of this study include its small sample size (only 205 ciprofloxacin-resistant S. pneumoniae isolates were collected and tested) and the potential bias introduced by segregating isolates based on changes in ciprofloxacin resistance rates. The use of alternate criteria for assignment of strains to sets of isolates for comparison purposes may influence the degree of genetic homogeneity or heterogeneity observed following epidemiological analysis. We also acknowledge that the use of an additional typing scheme, such as multilocus sequence typing, would increase the robustness of our comparisons; however, multilocus sequence typing was beyond the scope of this project.
In summary, genetic relatedness among ciprofloxacin-resistant S. pneumoniae isolates from across Canada increased following a significant increase in ciprofloxacin resistance rates since 2003. Although the development of de novo mutations within the QRDRs of parC and gyrA, presumably as a result of selective pressure from increasing fluoroquinolone use, continues to play a major role in the spread of such resistance, the documentation of ciprofloxacin-resistant strains similar to the Spain23F-1, Spain9V-3, and Taiwan19F-14 international clones among the Canadian pneumococcal population is equally noteworthy. Given the propensity of these strains to acquire resistance to multiple antibiotics and their ability to spread rapidly, ongoing surveillance is essential for monitoring the changing epidemiology of fluoroquinolone resistance in this important pathogen.
Footnotes
Published ahead of print on 7 January 2008.
REFERENCES
- 1.Adam, H. J., K. N. Schurek, K. A. Nichol, C. J. Hoban, T. J. Baudry, N. M. Laing, D. J. Hoban, and G. G. Zhanel. 2007. Molecular characterization of increasing fluoroquinolone resistance in Streptococcus pneumoniae isolates in Canada, 1997 to 2005. Antimicrob. Agents Chemother. 51:198-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alou, L., M. Ramirez, C. Garcia-Rey, J. Prieto, and H. de Lencastre. 2001. Streptococcus pneumoniae isolates with reduced susceptibility to ciprofloxacin in Spain: clonal diversity and appearance of ciprofloxacin-resistant epidemic clones. Antimicrob. Agents Chemother. 45:2955-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canton, R., M. Morosini, M. C. Enright, and I. Morrissey. 2003. Worldwide incidence, molecular epidemiology and mutations implicated in fluoroquinolone-resistant Streptococcus pneumoniae: data from the global PROTEKT surveillance programme. J. Antimicrob. Chemother. 52:944-952. [DOI] [PubMed] [Google Scholar]
- 4.Chen, D. K., A. McGeer, J. C. de Azavedo, and D. E. Low for The Canadian Bacterial Surveillance Network. 1999. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. N. Engl. J. Med. 341:233-239. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. CLSI/NCCLS M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Davies, T. A., R. Goldschmidt, S. Pfleger, M. Loeloff, K. Bush, D. F. Sahm, and A. Evangelista. 2003. Cross-resistance, relatedness and allele analysis of fluoroquinolone-resistant US clinical isolates of Streptococcus pneumoniae (1998-2000). J. Antimicrob. Chemother. 52:168-175. [DOI] [PubMed] [Google Scholar]
- 7.de la Campa, A. G., L. Balsalobre, C. Ardanuy, A. Fenoll, E. Perez-Trallero, and J. Linares. 2004. Fluoroquinolone resistance in penicillin-resistant Streptococcus pneumoniae clones, Spain. Emerg. Infect. Dis. 10:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho, P. L., W. C. Yam, T. K. Cheung, W. W. Ng, T. L. Que, D. N. Tsang, T. K. Ng, and W. H. Seto. 2001. Fluoroquinolone resistance among Streptococcus pneumoniae in Hong Kong linked to the Spanish 23F clone. Emerg. Infect. Dis. 7:906-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho, P. L., R. W. Yung, D. N. Tsang, T. L. Que, M. Ho, W. H. Seto, T. K. Ng, W. C. Yam, and W. W. Ng. 2001. Increasing resistance of Streptococcus pneumoniae to fluoroquinolones: results of a Hong Kong multicentre study in 2000. J. Antimicrob. Chemother. 48:659-665. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, C. N., W. H. Benjamin, Jr., S. A. Moser, S. K. Hollingshead, X. Zheng, M. J. Crain, M. H. Nahm, and K. B. Waites. 2003. Genetic relatedness of levofloxacin-nonsusceptible Streptococcus pneumoniae isolates from North America. J. Clin. Microbiol. 41:2458-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klugman, K. P. 2002. The successful clone: the vector of dissemination of resistance in Streptococcus pneumoniae. J. Antimicrob. Chemother. 50(Suppl. S2):1-5. [DOI] [PubMed] [Google Scholar]
- 12.Liñares, J., A. G. de la Campa, and R. Pallares. 1999. Fluoroquinolone resistance in Streptococcus pneumoniae. N. Engl. J. Med. 341:1546-1547. [DOI] [PubMed] [Google Scholar]
- 13.McEllistrem, M. C., J. E. Stout, and L. H. Harrison. 2000. Simplified protocol for pulsed-field gel electrophoresis analysis of Streptococcus pneumoniae. J. Clin. Microbiol. 38:351-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGee, L., C. E. Goldsmith, and K. P. Klugman. 2002. Fluoroquinolone resistance among clinical isolates of Streptococcus pneumoniae belonging to international multiresistant clones. J. Antimicrob. Chemother. 49:173-176. [DOI] [PubMed] [Google Scholar]
- 15.McGee, L., L. McDougal, J. Zhou, B. G. Spratt, F. C. Tenover, R. George, R. Hakenbeck, W. Hryniewicz, J. C. Lefévre, A. Tomasz, and K. P. Klugman. 2001. Nomenclature of major antimicrobial-resistant clones of Streptococcus pneumoniae defined by the Pneumococcal Molecular Epidemiology Network. J. Clin. Microbiol. 39:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morrissey, I., and J. George. 1999. Activities of fluoroquinolones against Streptococcus pneumoniae type II topoisomerases purified as recombinant proteins. Antimicrob. Agents Chemother. 43:2579-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. M7-A6. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 18.Pletz, M. W., L. McGee, J. Jorgensen, B. Beall, R. R. Facklam, C. G. Whitney, and K. P. Klugman. 2004. Levofloxacin-resistant invasive Streptococcus pneumoniae in the United States: evidence for clonal spread and the impact of conjugate pneumococcal vaccine. Antimicrob. Agents Chemother. 48:3491-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richter, S. S., K. P. Heilmann, S. E. Beekmann, N. J. Miller, C. L. Rice, and G. V. Doern. 2005. The molecular epidemiology of Streptococcus pneumoniae with quinolone resistance mutations. Clin. Infect. Dis. 40:225-235. [DOI] [PubMed] [Google Scholar]
- 20.Zhanel, G. G., L. Palatnick, K. A. Nichol, T. Bellyou, D. E. Low, and D. J. Hoban. 2003. Antimicrobial resistance in respiratory tract Streptococcus pneumoniae isolates: results of the Canadian Respiratory Organism Susceptibility Study, 1997 to 2002. Antimicrob. Agents Chemother. 47:1867-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]