Abstract
Human macrophage inflammatory protein 3α (MIP-3α), also known as CCL20, is a 70-amino-acid chemokine which exclusively binds to chemokine receptor 6. In addition, the protein also has direct antimicrobial, antifungal, and antiviral activities. The solution structure of MIP-3α was solved by the use of two-dimensional homonuclear proton nuclear magnetic resonance (NMR). The structure reveals the characteristic chemokine fold, with three antiparallel β strands followed by a C-terminal α helix. In contrast to the crystal structures of MIP-3α, the solution structure was found to be monomeric. Another difference between the NMR and crystal structures lies in the angle of the α helix with respect to the β strands, which measure 69 and ∼56.5° in the two structures, respectively. NMR diffusion and pH titration studies revealed a distinct tendency for MIP-3α to form dimers at neutral pH and monomers at lower pH, dependent on the protonation state of His40. Molecular dynamics simulations of both the monomeric and the dimeric forms of MIP-3α supported the notion that the chemokine undergoes a change in helix angle upon dimerization and also highlighted the important hydrophobic and hydrogen bonding contacts made by His40 in the dimer interface. Moreover, a constrained N terminus and a smaller binding groove were observed in dimeric MIP-3α simulations, which could explain why monomeric MIP-3α may be more adept at receptor binding and activation. The solution structure of a synthetic peptide consisting of the last 20 residues of MIP-3α displayed a highly amphipathic α helix, reminiscent of various antimicrobial peptides. Antimicrobial assays with this peptide revealed strong and moderate bactericidal activities against Escherichia coli and Staphylococcus aureus, respectively. This confirms that the C-terminal α-helical region of MIP-3α plays a significant part in its broad anti-infective activity.
Chemokines are chemotactic cytokines which are small, positively charged proteins that regulate immune responses. They are involved in a number of biological processes, including trafficking of various subpopulations of leukocytes (15). They are released at sites of inflammation and form a chemotactic gradient that leukocytes, which possess the corresponding receptor, can recognize and follow. In humans, the chemokine family is comprised of approximately 46 members which interact with about 20 different G-protein coupled receptors expressed on chemokine target cells (78). Interactions with receptors are thought to take place in a two-step process (7). First, the region between two conserved N-terminal cysteines and the first β strand of the chemokine, called the N loop, contacts the N-terminal region and other extracellular domains on the receptor. Collectively, this is known as the binding groove, which forms a pocket with part of β strand 3. This is followed by an interaction between the helical center of the receptor with the extreme N-terminal flexible region of the chemokine to activate the receptor (1, 7). Chemokines may form homodimers under various conditions; however, it has been shown that the monomeric form is fully functional and usually binds to receptors more strongly (1, 28, 43, 58). The role and function of chemokine dimers are unclear and are still the topic of much debate and research (3, 17, 62). It has been shown that chemokines can dimerize when they bind to cell surface glycosaminoglycans (GAGs), which increases the local concentration of chemokines and is thought to contribute to cell migration (26).
Human macrophage inflammatory protein 3α (MIP-3α) is a 70-amino-acid, 8-kDa protein of the CC chemokine family that contains two disulfide bonds. The protein is also known as CCL20 or LARC (liver and activation-regulated chemokine). MIP-3α has been linked to a variety of diseases, including cancer (38), rheumatoid arthritis (64), and diseased periodontal tissues (29). It attracts memory T cells and natural killer cells to sites of inflammation, as well as immature dendritic cells (12). CC chemokine receptor 6 (CCR6) is the only receptor for MIP-3α and, in turn, is bound only by this chemokine (46). In contrast, most chemokines and their receptors are quite promiscuous and usually bind to a variety of partners (1). Although not members of the chemokine family, human β-defensins 1, 2, and 3 (HBD1 to HBD3, respectively) have been shown to bind to and activate CCR6, in addition to MIP-3α, and are capable of inducing chemotaxis (74, 77). This role reversal appears in the opposite direction, too, as MIP-3α and many other chemokines have pronounced antimicrobial activity (9, 76). In fact, it was shown that MIP-3α possesses higher antimicrobial activity than HBD1 and HBD2 against Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 29213 (27). Thereby, MIP-3α presents an interesting link between the adaptive and innate immune responses in humans. Moreover, it has been shown that several chemokines, including MIP-3α, can also have direct antiviral (37, 56) and antifungal (76) activities. It was shown recently that a 12-residue peptide from the C terminus of MIP-3α possesses antimicrobial activity (23). In addition, another chemokine, interleukin-8 (IL-8/CXCL8), is not antimicrobial itself, but a peptide corresponding to its C-terminal α-helical region is in fact bactericidal (6). This raises the question of whether the bulk of the anti-infective properties of MIP-3α can be linked to its highly cationic C-terminal α helix.
Here we present the three-dimensional solution structure of human MIP-3α as determined by two-dimensional homonuclear proton nuclear magnetic resonance (NMR). In addition, NMR diffusion studies and multiple molecular dynamics (MD) simulations were performed to investigate the structural and potentially functional changes upon MIP-3α dimerization. Finally, we elucidated the solution structure of a peptide corresponding to the 20-most C-terminal amino acids of MIP-3α and report on its antibacterial activity.
MATERIALS AND METHODS
Materials.
MIP-3α was obtained from Peprotech (Rocky Hill, NJ) and was further purified by high-pressure liquid chromatography (HPLC). The protein showed one band during mass spectrometry, isoelectric focusing, and gel electrophoresis experiments. A peptide corresponding to the 20 C-terminal residues of MIP-3α was synthesized by 21st Century Biochemicals (Marlboro, MA) and was shown to be >95% pure by HPLC and mass spectrometry (see Fig. 1B for the sequence).
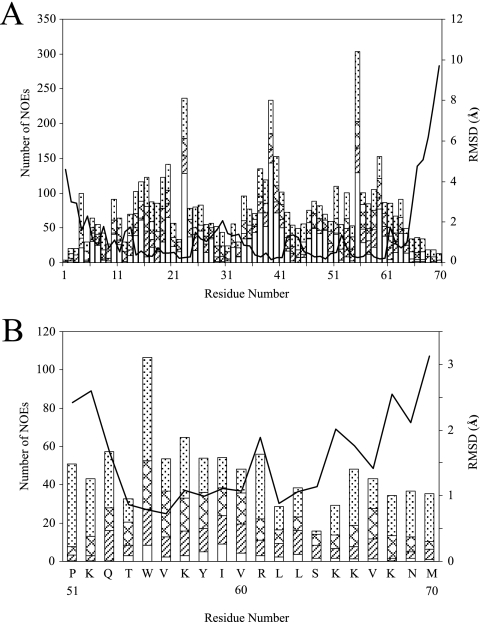
FIG. 1.
Number of NOEs per residue and RMSD plot for the MIP-3α protein (A) and its 20-residue C-terminal peptide (B). Intraresidue (dotted bars), sequential (zigzagged bars), medium-range (striped bars), and long-range (white bars) NOEs are shown. The lines represent the RMSDs per residue for the 20 lowest-energy structures.
NMR spectroscopy.
Approximately 2 mg of purified MIP-3α was dissolved in 500 μl of 90:10 H2O-D2O. The solution was unbuffered and was adjusted to a pH of 4.2. To determine the protein concentration, the UV absorbance was measured in solution at 280 nm by using an extinction coefficient of 8,490 M−1 cm−1, as determined by ProtParam (19). The concentration was found to be 0.87 mM. The two-dimensional nuclear Overhauser effect (NOE) spectroscopy (NOESY), total correlation spectroscopy (TOCSY), and double quantum filtered correlation spectroscopy (DQF-COSY) spectra for the full-length protein were acquired on both Bruker Avance 500- and 700-MHz spectrometers at 25°C. The Bruker Avance 500-MHz spectrometer was equipped with a 5-mm TXI cryoprobe with a z gradient. The 700-MHz spectrometer was equipped with a 5-mm TBI triple-axis-gradient probe. The mixing times for the full-length chemokine were 250 ms and 120 ms for the NOESY and TOCSY experiments, respectively, and in both experiments 2048 × 600 complex data points were acquired in the F2 and F1 dimensions. The spectral sweep widths were 12 ppm. The DQF-COSY spectrum was acquired with 4096 × 512 complex data points. The spectral sweep widths were 12 ppm. The peptide sample was prepared by dissolving it in 90:10 H2O-D2O and adding d25-sodium dodecyl sulfate (SDS) to a final concentration of 100 mM and pH 4.2. The concentration was established to be 2.3 mM by the use of the UV absorbance at 280 nm and an extinction coefficient of 6,990 M−1 cm−1, as determined by ProtParam (19). A simple aqueous solvent was not used for the MIP-3α peptide because circular dichroism experiments showed the absence of structure in aqueous solution (see Fig. S1 in the supplemental material). For the MIP-3α peptide, all spectra were collected on the 700-MHz spectrometer. The NOESY spectrum was collected with 4096 × 400 complex data points and a 120-ms mixing time, while the TOCSY spectrum (mixing time, 100 ms) had 4096 × 320 complex data points and the DQF-COSY spectrum was composed of 4096 × 250 complex data points. Solvent suppression was achieved by excitation sculpting (32). All spectra were referenced internally against a dimethyl silapentane sulfonate standard.
To investigate the relative rate of amide proton exchange, the lyophilized intact protein was redissolved in 99.9% D2O and a two-dimensional TOCSY spectrum was acquired in 1 h immediately after the protein was redissolved. To obtain the true intensity of correlations around the residual solvent peak, two-dimensional TOCSY and NOESY spectra were collected at both field strengths with the protein dissolved in D2O and with no solvent suppression. The data were processed with NMRPipe (version 3.4) software (11). The two-dimensional spectra were zero filled once in each dimension and multiplied by a shifted sine-bell squared function before Fourier transformation.
Structure calculations.
The two-dimensional spectra were analyzed by NMRView (version 5.0.4) software (34). The assignment of proton chemical shifts and spin systems was done by well-established, standard methods (75). Very broad Φ dihedral angles were used to restrain the angles to the favorable regions of the Ramachandran space, with the exception of those for glycines and prolines. In the peptide, Φ and ϕ dihedral restraints were loosely constrained to the helical portion of the Ramachandran spectrum, based on α-H chemical-shift index values (73). Structure calculations were performed by using the program CNS (version 1.1) (8) within the ARIA (version 1.2) package (with diffusion anisotropy) (48, 57). The CNS program uses both MD and a simulated annealing protocol to create structures of the lowest energy with the fewest constraint violations possible. Default ARIA program parameters were used in all runs except the final run, where the number of structures calculated in the seventh and eighth iterations were increased to 40 and 100 structures, respectively, and to 40 and 150 structures, respectively, for the peptide. In addition, the 20 lowest-energy structures were kept in the final iteration for statistical analysis, and 35 structures were created with water refinement for the intact protein. The structures calculated were visualized by using MOLMOL (version 2k.1) (39) and were analyzed by using PROCHECK (version 3.2) (42).
pH titrations and NMR diffusion.
The pH titration studies were carried out with the MIP-3α protein dissolved in D2O as described above. One-dimensional spectra were acquired on a Bruker Avance 700-MHz spectrometer at 25°C with 32,000 data points and 400 scans. The pH was adjusted in small increments from pH 7.0 down to pH 4.4 by using dilute DCl. The pH was measured immediately before and after NMR experiments and was not adjusted for the deuterium isotope effect.
For the diffusion experiments, the protein was lyophilized and redissolved in D2O. The sample was monitored in relation to an internal dioxane reference. The dioxane was prepared as a 1% solution in D2O, of which approximately 5 μl was added to provide an internal standard. The results of pulsed-field-gradient diffusion experiments were collected by use of the PG-SLED sequence (54). The data were acquired on the 700-MHz spectrometer by collecting 56 scans of 16,000 data points at each gradient amplitude and incrementing the gradient strength in 64 steps from 1.25% to 80% of the maximum output of the linear gradient amplifier. Fourier transformation and data analysis were performed by use of the Bruker XWINNMR software package (version 3.0). Calculations of theoretical hydrodynamic radii (RH) were performed by using the empirical equation for folded proteins, (4.75 ± 1.11) × 0.29N ± 0.02, where N is the number of residues (72). The experimental RH of MIP-3α was calculated according to the relationship (Dref/Dprotein) × RH(ref), where RH(ref) is the hydrodynamic radius of dioxane and D is the measured diffusion coefficient of MIP-3α or dioxane (35). The effective hydrodynamic radius of dioxane was taken to be 2.12 Å (35).
MD simulations.
One MD simulation was conducted with the MIP-3α dimer, obtained from the crystal dimer structure of MIP-3α with accession code 1M8A (27). Three MD simulations were set up for monomeric MIP-3α: one for the NMR solution structure and one each for the monomeric components of the MIP-3α crystal dimer. From here on, dimA and dimB refer to monomer A and monomer B in the dimer simulation, respectively, while monA and monB refer to the same proteins but in their simulations as monomers. Because the X-ray structure contained the coordinates only for residues 5 to 65, the N and C termini were added to the pdb file in a solvent-exposed orientation by using MOLMOL (39). The protein structures were inserted into a rhombic dodecahedron box fully solvated with simple-point-charge water. The overall charge in the simulation box was neutralized by introducing chloride ions, resulting in concentrations between 40 and 120 mM. The simulation boxes measured approximately 8 by 8 by 5 nm and contained between 8,000 and 12,000 water molecules.
All MD simulations were conducted with version 3.2.1 of the GROMACS program (5, 47) with the GROMOS96 (version 43a2) force field (67). The neighbor search was conducted by the grid method, with the neighbor list updated every three steps and a neighbor list cutoff of 0.9 nm. All MD simulations were carried out by using periodic boundary conditions. Constant temperature and pressure were maintained by using the Berendsen algorithm (4). The temperature was held at 300 K, with a τT = 0.1 ps. The pressure was kept at 1 bar with isotropic temperature coupling and with a coupling constant τP = 1.0 ps and compressibility of 4.5 × 10−5 bar−1. Bond lengths were restrained using the LINCS method (24). Coulomb energies were calculated according to fast-particle-mesh Ewald electrostatics (10) with a coulomb cutoff of 0.9 nm. Van der Waals energies were collected with a twin range cutoff of 0.9/1.4 nm. To allow for a 5-fs time step, fast motions were removed by treating certain hydrogen atoms as dummy hydrogens (16). The simulation was conducted for 10 million steps, amounting to 50 ns of simulation time for each MD simulation.
Antibacterial assays.
The MICs and minimal bactericidal concentrations (MBCs) were determined for the MIP-3α C-terminal peptide. E. coli ATCC 25922 and S. aureus ATCC 25923 strains were grown in Bacto Peptone medium and were diluted to 2 × 106 CFU/ml in 96-well plates. The bacteria were exposed to various peptide concentrations, and the cell turbidity was monitored by use of the absorbance at 540 nm. MBCs were obtained by pipetting an overnight culture on paper disk antibiotic sensitivity medium II and further incubation. This procedure has previously been described in detail (69).
In a second assay, the MBCs were established for S. aureus 42D and E. coli ML35. Bacterial suspensions of 2 μl at 107 CFU/ml were suspended in 80 μl of 10 mM phosphate buffer, pH 7.0, supplemented with 1% (vol/vol) tryptic soy broth. This solution was incubated with the MIP-3α peptide in a total volume of 200 μl. Aliquots of 10 μl and 20 μl were then placed on blood agar plates at 0 and 2 h after incubation at 37°C under agitation. The concentration of peptide that killed >99.9% of the inoculum at 2 h after incubation is the MBC.
Protein Data Bank accession numbers.
The atomic coordinates for the human MIP-3α protein have been deposited in the Protein Data Bank (access code 2jyo), Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ. The chemical shifts and table of assignments have been deposited in the BioMagResBank (University of Wisconsin—Madison) under access code 15596.
RESULTS
NMR assignments.
The two-dimensional NOESY spectrum of MIP-3α displayed excellent chemical shift dispersion for a protein of 70 amino acids due to the presence of both α-helical and β-sheet segments. Although there was some peak overlap in the spectra of the peptide in SDS micelles due to broad line widths, the resolution was sufficient for peak assignment (see Fig. S2 in the supplemental material). The TOCSY and COSY spectra yielded largely conclusive spin system assignments for both the intact chemokine and the MIP-3α peptide, allowing a nearly complete determination of the proton chemical shift. In the full-length protein, residues 1 and 69 could not be specifically assigned in the D2O data due to similar α- and β-proton shifts. The TOCSY spectrum acquired during D2O exchange showed 26 amide protons that exchanged slowly in MIP-3α. Slow exchange occurs with amide protons that are either involved in hydrogen bonding and/or not readily accessible to bulk solvent.
Structure calculations.
No NOEs that would result from intermonomer contacts (e.g., from residue 21 of one monomer to residue 25 of the adjacent monomer across the dimer interface, as seen in the crystal structures) were found in the MIP-3α NOESY spectrum, indicating the presence of a monomer under our experimental conditions. Therefore, all NOESY cross peaks were used for monomer structure determination. The NOEs were well distributed over the entire protein sequence, adding up to ∼2,200 NOEs obtained from the H2O and the D2O NOESY spectra for the intact protein. By using a structure consistent with the initial NOE constraint information, hydrogen bond and broad dihedral angle information was added to the ARIA calculations. Lastly, additional distance information from the D2O NOESY experiment was incorporated. The MIP-3α peptide structure was determined in a similar manner, although no hydrogen bonding restraints were used and dihedral restraints were based on the α-H chemical-shift index. A summary of the number of NOEs used and structural statistics are provided in Fig. 1 (see also Table S1 in the supplemental material).
Solution NMR structure of MIP-3α.
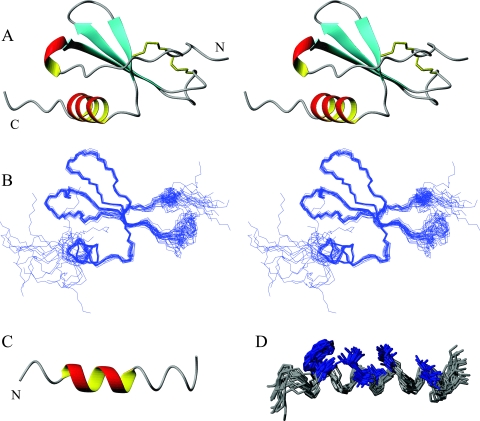
A backbone overlay of the 20 lowest-energy structures is provided in Fig. 2B. The 20 lowest-energy structures of MIP-3α show a short 310 helix followed by three β strands running in an antiparallel fashion and a C-terminal α helix (Fig. 2A). This secondary structure pattern is similar to that observed in other chemokines (36, 51). The N-terminal region is largely flexible, constrained mainly by the two disulfide bonds connecting the N-terminal region with the β-sheet portion of the protein (Fig. 1A and 2A). The arrangement of disulfide bonds was supported by NOEs found in the NOESY spectrum. The amide group of Cys6 shows strong correlations to both the α and β hydrogens of Cys32. The disulfide bonds are followed by a random coil and a short 310 helix, which leads into the first β strand. The three antiparallel β strands run from residues 20 to 26 (β1), 36 to 41 (β2), and 46 to 49 (β3), separated by the 30s and 40s loops (Fig. 2A). The β1 strand contains a β bulge at residues 20 and 21. The 30s loop (residues 27 to 35) bends back onto the β sheet and is quite flexible (Fig. 2B). The 40s loop displays less variability and is much shorter (residues 42 to 44) and therefore lies largely in plane with the β sheet. The α helix extends from residues 54 to 63 and is present in all of the lowest-energy structures, and it displays interactions typical for a helical segment (see Fig. S3A in the supplemental material). The remaining residues (residues 65 to 70) are disordered, similar to the most N-terminal residues. The root mean square deviation (RMSD) for the backbone atoms over the well-defined region of the protein (residues 6 to 65) is 0.49 Å. Further structural statistics are given in Table S1 in the supplemental material.
FIG. 2.
(A) Stereo ribbon diagram of the water-refined, lowest-energy structure of MIP-3α. The two disulfide bonds are represented in yellow, and the N and the C termini are labeled. (B) Stereo diagram showing an overlay of the protein backbone of the 20 lowest-energy structures. The structures were superimposed over the backbone atoms of residues 6 to 65 and are shown in the same orientation as the stereo ribbon diagrams in panel A. The two termini and the 30s loop (on right-hand side) display the largest amount of disorder. (C) Ribbon diagram of the lowest-energy structure of the MIP-3α C-terminal peptide. (D) Representation of the 20 lowest-energy conformers of the MIP-3α peptide fit to the backbone atoms of residues 4 to 18. The central region is well defined, while the termini are more disordered. Hydrophobic residues Trp, Val, Ile, and Leu (highlighted in blue) assemble on one side of the peptide to form its hydrophobic face. The figure was prepared with the MOLMOL program (39).
Solution NMR structure of the MIP-3α peptide.
The MIP-3α peptide, which consists of the 20-most C-terminal residues in the chemokine, consistently folds into a partial α-helical conformation (Fig. 2C). The NOESY spectrum displays most of the medium-range NOE interactions classically associated with an α helix (see Fig. S3B in the supplemental material). Looking at the 20 lowest-energy structures, the α helix can be seen to span almost the entire sequence of the peptide, although only in portions in the individual structures. The α helix is also seen over residues 4 to 13, which corresponds to the residues in a helical conformation in the intact protein (i.e., residues 54 to 63). The RMSD of the α-helical region in the intact protein compared to that of the α-helical region in the peptide is 0.587 Å over the backbone atoms. The last five residues in the peptide show the largest degree of variation, although they are predominantly in a helical arrangement (Fig. 1B and 2D). There are only a few observable NOEs for Ser14, which disconnects the C-terminal end from the rest of the peptide, giving rise to more variation in that region (Fig. 1B). The side chains in the structured region covering residues 2 to 18 show a clear separation between hydrophobic and polar/charged residues (Fig. 2D). The separation is emphasized by the core hydrophobic residues, consisting of Trp5, Val6, Ile9, Val10, Leu12, and Leu13, which are oriented similarly in intact MIP-3α. This is also reflected in the electrostatic surface plot, which shows a distinct separation of the hydrophobic core away from charged residues (see Fig. 4A in the supplemental material). As observed for the intact protein, the backbone dihedral angles of the ensemble of peptide structures predominantly fall into the most favored region of the Ramachandran plot, which is shown along with other structural statistics in Table S1 in the supplemental material.
FIG. 4.
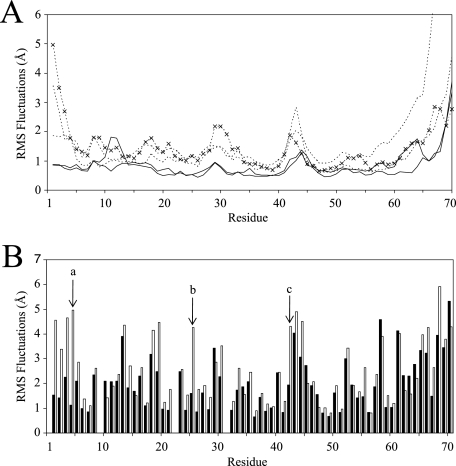
RMSF plots of the MIP-3α MD simulations shown for each residue. (A) Backbone RMSF plots for each MIP-3α monomer. Notice the elevated backbone fluctuations of the monomeric simulations (dotted lines) throughout the protein compared to the fluctuations of the dimer simulation (solid lines). ×, plot from the simulation conducted with the NMR solution structure. (B) Side chain RMSF plots averaged over the last 30 ns of the monomer (empty bars) and dimer (filled bars) simulations. The far N-terminal residues most noticeably display the largest decreases in fluctuations between the monomeric and the dimeric MIP-3α simulations. The side chain fluctuations of Phe4, Arg25, and Lys42 are marked a, b, and c, respectively.
Antibacterial activity of the MIP-3α peptide.
Like the C-terminal region of IL-8, the final helical region of MIP-3α possesses a large number of cationic charges and forms an amphipathic α helix in solution, similar to many well-known antimicrobial peptides (AMPs). Furthermore, intact MIP-3α has been shown to possess potent antimicrobial activity (27, 76). To evaluate what role the C-terminal α helix plays in MIP-3α's anti-infective properties and to see whether it can act as an AMP on its own, the peptide was synthesized and evaluated for its activity against E. coli and S. aureus by two separate methods. The data indicate that the MIP-3α peptide possesses significant bactericidal activity (Table 1). These values for antibacterial activity lie within the same range and, particularly against E. coli, exceed the values for the activities of other AMPs evaluated under the same conditions (Table 1) (66, 68, 69).
TABLE 1.
Antibacterial activities of MIP-3α, its C-terminal peptide, and other antimicrobial peptides determined by various methods
| Peptide or protein |
E. coli
|
S. aureus
|
Reference(s) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml) | MBC (μg/ml) | LD50d (μg/ml) | LD90 (μg/ml) | Lethal concn (μg/ml) | MIC (μg/ml) | MBC (μg/ml) | LD50 (μg/ml) | LD90 (μg/ml) | ||
| MIP-3α peptide51-70a | 12.5 | 15.0, ≤1.1b | 63.0 | 63.0, 4.7b | ||||||
| MIP-3α proteinb | 0.07-0.53c | 0.3 | 0.3 | 10-40c | 23, 27, 76 | |||||
| MIP-3α59-70b | 3.2 | 23 | ||||||||
| Bovine lactoferricina | 50 | 100 | 69 | |||||||
| Indolicidina | 20 | 20 | 10 | 10 | 66 | |||||
| Magainin 2a | 20 | >100 | 68 | |||||||
| Tritrpticina | 20 | 20 | 10-20 | 20 | 66 | |||||
| IL-8 peptide53-72 | 147.4c | 6 | ||||||||
Determined by a microdilution assay.
Determined by a colony count assay conducted under low-salt conditions and with 10 mM potassium phosphate, which commonly yield higher activity values.
Some of these numbers had to be interpolated from figures.
LD50, 50% lethal dose.
NMR diffusion and pH titrations.
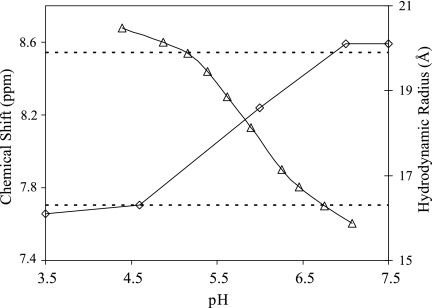
NMR diffusion studies were conducted on the 700-MHz spectrometer by using dioxane as an internal standard (35). Various groups have recently employed this method to study protein hydrodynamic radii (20, 31, 63). It has also been used successfully to study the dimerization of the chemokine stromal cell-derived factor 1 (SDF1/CXCL12) (71). The diffusion results for MIP-3α over a variety of pH values are presented in Fig. 3. The theoretical hydrodynamic radii for spherical proteins similar in size to monomeric and dimeric MIP-3α are 16.3 Å and 19.9 Å, respectively (72). According to these standards, the protein moves from a monomeric state at lower pHs to a dimeric form at higher pHs. The hydrodynamic radii indicate the presence of monomers at pH 3.5 and 4.6 (16.1 and 16.3 Å, respectively), a transitional or mixed species at pH 6.0 (18.6 Å), and finally, dimers at pH 7.0 and 7.5 (20.1 Å). These results suggest that titration of a group with a pKa of ∼6.5 governs the equilibrium, and this group is likely a His side chain. Hence, we performed pH titration experiments with intact MIP-3α. The chemical shift of the Hɛ1 proton on His40 was monitored from pH 4.4 to 7.1, moving from 8.8 ppm at pH 4.4 to 7.6 ppm at pH 7.1, which represents the change in the protonation state of the His40 side chain (Fig. 3). The inflection point of the His40 pH titration curve clearly corresponds to the transition point from the monomeric to the dimeric form of MIP-3α (Fig. 3).
FIG. 3.
NMR diffusion and pH titration results. The chemical shifts of the Hɛ1 proton on His40 decrease distinctively at higher pH values (triangles). This matches the increase in the hydrodynamic radius (Å) obtained from the NMR diffusion experiment (diamonds), and the pH of the transition is in excellent agreement for both experiments. The dotted lines represent the theoretical hydrodynamic radii of completely monomeric (bottom) and completely dimeric (top) MIP-3α.
MD simulations of MIP-3α.
To date only very few MD simulations of chemokine proteins have been reported, and most of these were relatively short, up to a few nanoseconds. Recently, Cui and colleagues have performed longer simulations of the chemokine lymphotactin; however, these were predominantly used to study salt-protein association effects (18, 49). All of our simulations with MIP-3α were conducted for 50 ns in an explicit solvent. In each of the simulations, the protein secondary structure was preserved and the protein ran stably without any unfolding. Relative to their starting structures, the RMSD values steadily increased over a period of approximately 20 ns, after which they leveled out from 0.30 to 0.35 nm, which is normal behavior for proteins (see Fig. S5 in the supplemental material). Because the RMSD values took a significant amount of simulation time to stabilize, certain plots and analyses are presented only for the last 30 ns. A partial movement of the C-terminal-most residues from a solvent-exposed state to a buried conformation underneath the β sheet is seen in all of the monomeric MIP-3α simulations. The transition of the C-terminal region does not appear to have any effect on the rest of the protein, as there are no other movements associated with this transition.
The root mean square fluctuations (RMSFs) of the MIP-3α simulations are lower in the dimeric simulations throughout the protein. The largest fluctuations are seen in the termini as well as in the 30s and 40s loops (Fig. 4). This behavior parallels that determined from the RMSD values obtained from the NMR structures (Fig. 1). The fluctuations of the N-terminal end are not elevated in all the trajectories; in particular, the dimA and dimB trajectories show very little N-terminal fluctuations, whereas the monomeric simulations all show elevated fluctuations at the N terminus. Reduced fluctuations of the N termini in the dimer can be attributed to stabilizing dimer contacts, which also drastically reduce fluctuations in other regions of the dimer interface. For example, residues such as Arg25 in the β1 strand display markedly dampened fluctuations compared to those in the MIP-3α monomer simulations due to hydrogen bonding and decreased solvent exposure (Fig. 4B).
MD simulations: monomer versus dimer simulations.
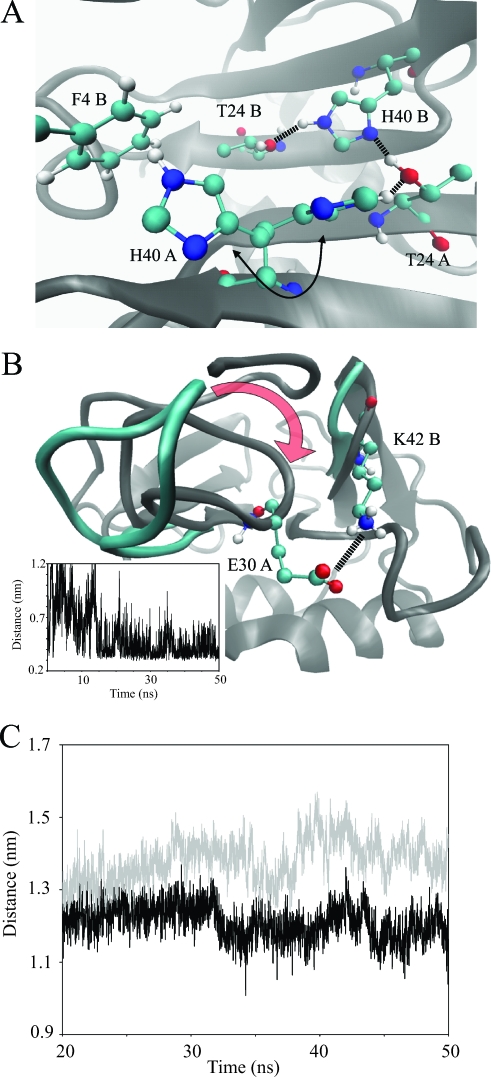
Substantial interactions between the N terminus and the 30s loop of the same monomer of MIP-3α are seen in the simulations; this would not be expected from the high-resolution structures, as increased flexibility is observed in these regions. The far N-terminal region interacts and remains close to the 30s loop of MIP-3α in both the dimer proteins and also in the monA trajectory. The association between these regions of MIP-3α does not appear to be random, since many similar contacts are observed in the three trajectories (i.e., dimA, dimB, and monA). In particular, hydrogen bonding between the backbone NHs of Asn3 and Phe4 and the carbonyl oxygen of Gly31 are crucial for the continued association of the N terminus with the 30s loop and are exclusively seen when these regions associate. In each simulation the N terminus sits on top of the 30s loop in the same manner, with the loop bent toward the face of the β sheet. On average, a total of four hydrogen bonds are formed between the N terminus and the 30s loop to maintain a stable interaction. Although these regions associate in the monA trajectory as well, the behavior of the protein in these parts is not entirely the same. Both the backbone and the side chain fluctuations are much higher in the monA simulation than in the dimer simulation; specifically, the side chain of Phe4 shows only small fluctuations around 1.0 Å in the dimer simulation, while its fluctuations in the monomer simulations are significantly elevated throughout, ranging from 4.0 to 5.8 Å (Fig. 4B). Phe4 is held in place in the MIP-3α dimer through intermonomer contacts, thereby burying the hydrophobic Phe side chain (Fig. 5A). The solvent-accessible surface area (SASA) of Phe4 increases from below 25 Å2 in the dimer simulation to at least 68 Å2 in the monomer simulations. In the crystal dimer structures of MIP-3α, His40 forms an intramonomer hydrogen bond with Thr24 on the upper face of the β sheet (27, 50). These interactions are seen in the dimer simulation as well and involve the side chain of Phe4 (Fig. 5A). At the beginning of the trajectory, the two His40 residues form hydrogen bonds with Thr24 in their respective monomers. As the simulation progresses, the His40 of dimA flips and thereby switches from interacting with the Thr24 of its own monomer to interacting with the Phe4 of dimB, forming close hydrophobic interactions (Fig. 5A). At the same time, His40 in dimB does not change its orientation but instead interacts with the Thr24 side chains of both proteins in the dimer. Therefore, His40 can stabilize the dimer conformation in two ways, both from the intermonomer hydrogen bonding with Thr24 and from hydrophobic interactions with Phe4.
FIG. 5.
(A) Close-up view of the dimer interface and the key interactions observed during the MD simulation. His40 residues interact with each other and participate in hydrogen bonding with Thr24. The His40 of one monomer is shown in two orientations (arrow), as it is observed during the simulation to flip from interacting with Thr24 to interacting with Phe4 of the adjacent monomer. (B) Another view of the dimer interface and, more specifically, the gap between the 30s and 40s loops of adjacent monomers. During the simulation, the 40s loop moves to close the gap to the 30s loop, as indicated by the red arrow. During this process a salt bridge forms between the side chains of Lys42 and Glu30, as shown by the distance plot (see the inset). (C) Average binding groove size measured between the Cαs of Leu15 and Lys43 in the monomeric (gray) and dimeric (black) MD simulations. After initial equilibration, the binding groove is consistently larger in the monomer simulations than in the dimer simulations. Panels A and B were prepared with the VMD program (30).
In the crystal dimer structure of MIP-3α used for the simulation, the distance between the 30s loop from one monomer and the 40s loop of the other is between 12.5 and 12.9 Å (i.e., the distance between Cα residues 31 and 43). These distances decrease significantly to averages of 7.1 and 6.5 Å, respectively, as the simulation progresses, bringing the two loops much closer together than they are in the crystal structure (Fig. 5B). The 30s loop moves into the gap between the two monomers, along with the N terminus, which sits on top of the 30s loop, approaching the 40s loop of the other monomer. As a consequence, Lys42, which is located in the 40s loop of MIP-3α, is highly constrained in the dimer simulation, while it is flexible in the monomer simulations (side chain RMSFs, 19.5 and 43.1 Å, respectively; Fig. 4B). Interestingly, of the three consecutive Lys residues in the residues 40s loop, Lys42 is the only one with restricted side chain fluctuations and a decreased SASA in the dimer simulation (Fig. 4B). The intermonomer interactions of Lys42 involve the formation of a salt bridge between its side chain and Glu30 in the other monomer, which holds the Lys42 side chain in place (Fig. 5B). The simulation thereby reproduces what is also seen in the crystal dimer structures. In both the 2HCI monomers and in one of the 1M8A monomers, the same salt bridge is observed, with distances of about 3 Å in these structures.
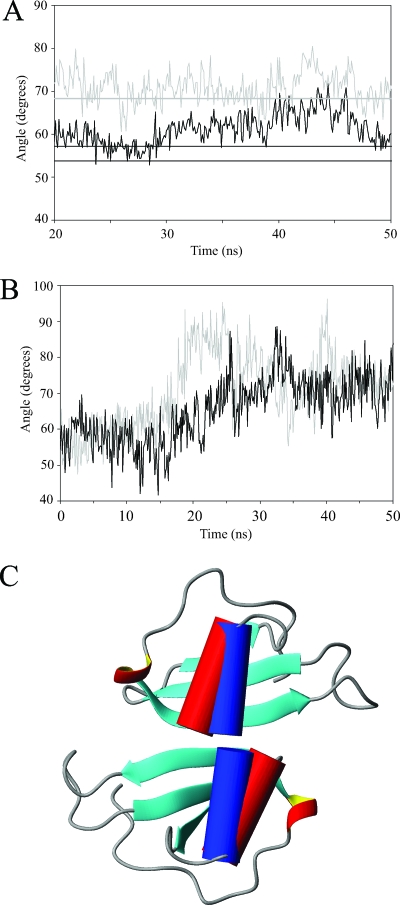
The size of the binding groove in MIP-3α has previously been implicated as an important factor in receptor binding and can be measured by various Cα distances spanning across the cavity (27, 59). For example, the distance between the Cαs of Leu15 and Lys43 consistently produces larger values in the monomeric MIP-3α simulations than in the dimeric simulations. The width of the groove measures, on average, 12.1 Å in the dimer simulations and 13.8 Å in the monomer simulations (Fig. 5C). This discrepancy between the monomeric and dimeric forms of MIP-3α are observed throughout the binding groove, as the measurements of different residues along the groove yield similar results. Another key difference in the behaviors of monomeric and dimeric MIP-3α is seen in the motions of the C-terminal α helix. Similar to the rest of the protein, the α helix is much more stable and is constrained in the protein dimer, whereas greater movement is seen in the MIP-3α monomers. The orientation of the α helix was measured using MOLMOL (39) and is defined as the angle between the axis of the α helix and the axis along the length of the first two β strands. The simulations of MIP-3α in the monomeric conformation produce larger angles than the simulations of MIP-3α in the dimeric conformation. Over the last 30 ns of the trajectory, the average angle of the monomer simulations measures 71°, or about 10° higher than what is seen in the dimer simulation, with an average angle of 61°. Although the angles in the crystal structures are observed to be slightly lower (54 ± 2.4°), the angles of the monomeric simulations are in excellent agreement with the angles derived from our NMR solution structure (68 ± 2.6°) (Fig. 6A). Further evidence to support a difference in helix angles between monomeric and dimeric MIP-3α was obtained from the monA and monB simulations. These simulations, based on the individual monomers of 1M8A in the crystal dimer, showed a clear increase in the helix angle as the simulation progresses (Fig. 6B). Although there was considerable variability in the helix angles for all simulations throughout the trajectories, an obvious trend was visible. Both of the proteins start off with small helix angles, as defined by the starting crystal dimer structure, and subsequently, the helices reorient themselves during the simulation to a more orthogonal position (Fig. 6B). This is similar to what is seen in the monomeric human and murine MIP-3α NMR structures. Since both the monA and the monB trajectories show this progression, it is likely that the helix is in fact held in a different conformation due to chemokine dimerization.
FIG. 6.
(A) Plots of the average angles between the α helix and the β sheet of MIP-3α over the last 30 ns of the MD simulations. The gray curve and the straight line represent the average angle in the monomer simulations and the average angle seen in the NMR structures, respectively. The black curve shows the average angle seen in the MIP-3α dimer simulation. The two black lines represent the helix angles in each of the monomers of the 1M8A crystal dimer structure. (B) Individual helix angles relative to the β strands of the monA and monB simulations, based on the MIP-3α dimer structure, but simulated as monomers. The helix angles increase drastically over the course of the simulation to resemble those observed in the monomeric solution structure and the MD simulation thereof. (C) Ribbon diagram of the MIP-3α crystal dimer (1M8A), with the α helix in cylindrical representation (red cylinders). The blue cylinders represent the orientations of the helices observed in the solution structure. Therefore, in a dimeric conformation, the helix angles observed in the NMR solution structure would result in significant steric clashes. In addition, there is steric overlap from the flexible C terminus that is not shown in these structures.
DISCUSSION
Effect of His40 on dimerization.
Considering the presence of monomers at concentrations as high as 10−4 to 10−3 M, as is the case during NMR data collection, it may not seem reasonable to consider the possibility that dimers readily exist in vivo. However, it has been reported that pH as well as other factors can have significant effects on chemokine dimer formation, showing that under the right physiological conditions MIP-3α can indeed form dimers (3, 27, 71). Our NMR diffusion results support the existence of a changing monomer-dimer equilibrium with a change in pH. Under acidic conditions, the protein is present as a monomer, whereas at higher pHs, close to pH 7.0, the protein clearly appears to dimerize. This suggests that a His side chain may play a key role in the dimerization, and in both crystal dimer structures of human MIP-3α, the two His40 residues face each other directly across the dimer interface on top of the β sheet. The only other His residue in MIP-3α, His16, faces away from the dimer interface and is therefore not likely to be implicated in dimer formation. pH titrations for monitoring of the protonation state of Hɛ1 on His40 were conducted to evaluate its importance in protein dimerization. The unique chemical shifts of the His ring protons have made them attractive as a test subject in similar titration experiments (52, 70). The results of the pH titrations show that the His40 pKa matches well with the diffusion data, as the neutralization of the His40 side chain corresponds to the dimerization event according to the NMR diffusion results (Fig. 3). A study of SDF1 illustrates this concept as well by highlighting the importance of a His residue in the β1 strand on its dimerization (71). The results from our MD simulations further implicate His40 as an important determinant for protein dimerization. Throughout the dimer simulation, the side chain of His40 forms hydrophobic and hydrogen bonding contacts both intra- and intermolecularly, thereby stabilizing the dimeric form of MIP-3α (Fig. 5A). At pHs below the pKa of His40, its side chain is protonated and therefore carries a positive charge. This causes electrostatic repulsion between the two charged His40 side chains, which would prevent them from remaining in close proximity to each other. It is thus highly likely that the dimerization of MIP-3α is mostly dependent on the protonation state of His40. In murine MIP-3α the same His residue is present, and pH-dependent dimerization has also been documented, making it possible that this same mechanism governs its dimerization process (59).
Monomer-dimer properties.
The most pronounced structural difference between the four available MIP-3α structures is the position of the C-terminal α helix relative to that of the β sheet. The murine MIP-3α structure reported the helix to be more orthogonal to the core β strands, measuring about 76 ± 2.0° (59), while in the human crystal structures, the helix lies at a lower angle, between 51 and 57° (27, 50). In our NMR solution structure, the helix is at an intermediate angle relative to the β strands at 69 ± 3°. In addition, our α helix is less curved than the helix in the murine structure, similar to what is seen in the crystal dimer. Other variations between the monomeric murine and human NMR structures of MIP-3α are seen in the 30s and 40s loops, as well as the N-terminal region (see Fig. S6 in the supplemental material). These parts were also identified in a comparison of the murine MIP-3α crystal structure and the human crystal structure and largely fall on the flexible regions of the protein (27). The origins of these differences are difficult to identify, but similar to other changes that have been observed, they may simply stem from species-specific amino acid sequences. In the dimeric crystal structure, the monomers align antiparallel along the first β strand to form a six-stranded β sheet with the α helices stacked close to each other underneath. Although the α helix is unlikely to affect receptor binding since it is thought to face away from the receptor (1), its positioning is crucial when an attempt to form a dimer is made. If the helices lie too perpendicular to the β sheet, as is the case in the monomeric solution structures, the ends of the helices sterically interfere with dimerization (Fig. 6C). Therefore, an adjustment in the helix angle of MIP-3α is necessary before two monomers can arrange to form dimers. This becomes obvious when the solution structure of the MIP-3α monomer is overlaid on the dimeric crystal structure. In that conformation, the α-helices clearly clash, indicating that the helix must shift before dimerization can occur (Fig. 6C). The MD simulations provide further evidence in support of a shift in helix angles between monomeric and dimeric MIP-3α. In both the monA and monB simulations, the helix reorients itself from a shallow angle with respect to the orientation of the β strands into a more perpendicular orientation similar to what is seen in the monomeric solution structures of both human and mouse MIP-3α (Fig. 6B). The ability of MIP-3α to dimerize could therefore be dependent on the range of angles sampled by the α helix. In turn, any factor that influences the position of the helix or its degree of fluctuation will thereby also affect the monomer-dimer equilibrium. For example, the larger angles in the mouse structures may be derived from the increased salt concentration or lower pH that were used during data acquisition. It is also tempting to speculate that GAG binding to MIP-3α may be another contributing factor, especially since GAG binding has been proposed to occur in the C-terminal region of the α helix in certain chemokines (40). In that event, GAG binding could cause the α helix of MIP-3α monomers to stabilize at smaller angles relative to the orientation of the β strands and thereby promote dimer formation.
Receptor binding implications.
MIP-3α displays highly specific and tight binding with its sole receptor, CCR6, which in turn binds to only one chemokine, along with the human β-defensins (61). Such unique and specific binding makes MIP-3α a good candidate for the determination of factors contributing to tight and specific receptor binding. The width of the MIP-3α binding groove has been deemed to be at least partly responsible for MIP-3α's tight receptor binding (27, 59). The distance between the Cαs of Thr11 and Leu45 measures about 12 Å in both the NMR solution structure and the crystal dimer structures of MIP-3α. This is not seen in HBD2, which also binds to CCR6, in which the distance across its similar proposed binding groove measures only 9 Å. Furthermore, all the MD simulations produce distances close to 12 Å, and these virtually never decrease to as low as 9 Å, supporting the idea that the narrow nature of the binding groove could be responsible for the defensin's lower affinity for CCR6. The MD simulations also show that the groove is consistently narrower in dimeric MIP-3α than in the monomer (Fig. 5C). The average size of the groove measures 12.1 Å in the dimer, while it is clearly larger in the monomer, at 13.8 Å. This difference in groove size could be part of the reason why monomeric chemokines are able to bind to their receptors with a greater affinity than the dimeric proteins (17, 62).
Monomeric MIP-3α may also be more adept at activating its receptor due to its more mobile N terminus. In the dimer simulation, the N-terminal region is constrained through its association with its own 30s loop and the opposing monomer's 40s loop. Extensive hydrogen bonding takes place for the duration of the simulation, keeping these parts of the protein in close proximity. In terms of the MIP-3α function, constraining the N terminus may prevent the protein from activating its receptor. Therefore, monomeric MIP-3α, with its freely moving N terminus, is likely to be more proficient at activating CCR6. Should the MIP-3α dimer bind to the receptor, the protein must undergo some conformational change or even dimer dissociation to release the N terminus before it can activate the receptor. This agrees with the hypothesis that chemokine dimers are negative regulators of receptor function, as their N termini are not free to activate the receptor (17, 62). This is similar to what is observed for other CC-type chemokine dimers, e.g., RANTES (CCL5), although the latter has a distinct dimer interface (13). Like most chemokines, MIP-3α dimerizes along its β1 strand, while RANTES associates through its two N-terminal regions.
Comparison of the results of the monomeric and dimeric simulations provides further information about chemokine-receptor interactions. In general, our MD simulations show that the motions in the dimeric form of MIP-3α are suppressed compared to those in the monomeric form. For example, the side chain fluctuations and the SASAs of Lys42 are quite distinct in monomeric and dimeric MIP-3α because of its interaction with Glu30 across the dimer interface. This could prevent a vital electrostatic interaction from forming between the Lys42 of MIP-3α and CCR6. This suggestion is consistent with the findings of other studies that have documented that mutations of lysine or arginine residues in locations close to the 40s loop in MIP-3α decrease receptor binding affinity (22, 33).
Anti-infective properties.
MIP-3α displays a broad array of activities, not only in its classic role as a component of the adaptive immune response but also in innate immunity. Activities have been documented against bacteria such as E. coli and S. aureus (27) as well as against viruses (37), and very moderate activity has been documented against fungi (76). Similar to many AMPs (14), intact MIP-3α shares a highly cationic net charge and an amphipathic surface (see Fig. S7A in the supplemental material). The electrostatic profile is characterized by distinct patches of hydrophobicity and concentrated regions of positive and negative charges. For example, a large portion of the protein surface is cationic, comprised of the 40s loop and the C-terminal α helix, while most of the solvent-exposed negatively charged moieties are concentrated in another region of MIP-3α. The areas of high cationic charge and their separation from negatively charged regions in particular appear to be important for MIP-3α's anti-infective properties. This is reflected in the surfaces of HBD3, IL-8, and MIP-3α (see Fig. S7 in the supplemental material). Although the secondary structure of intact IL-8 is virtually identical to that of MIP-3α, intact IL-8 displays no anti-infective properties. Inspection of their surfaces reveals that IL-8 lacks a highly concentrated cationic region, and its negative charges are not grouped into one general region but are interspersed throughout (see Fig. S7B in the supplemental material). HBD3 displays a drastically different picture, with its surface dominated by positive charges that are interrupted by only two negatively charged species, which would be neutralized by their surrounding Lys and Arg residues (see Fig. S7C in the supplemental material). The high concentration of positively charged residues not only makes HBD3 an antimicrobial agent more efficient than IL-8 and MIP-3α but also makes it more active than HBD1 and HBD2, especially under conditions of high salinity (65). In actuality, the surface profile of HBD3 is very similar in both charge distribution and size to the surface profile of the cationic domain of MIP-3α mentioned above. Furthermore, it has been observed that a number of other antimicrobial chemokines display an analogous, highly cationic region (9, 76). Although all of these chemokines have similar secondary structures, the relative position of the cationic surface area is not consistent throughout. This suggests that merely the presence, rather than the exact placement, of the cationic domain is a requirement for chemokine anti-infective activity. Additionally, the cationic domain has also been implicated in MIP-3α's potency against vaccinia virus and is consistent with the absence of activity against herpes simplex virus type 1, since the viral envelope of only the former and not that of the latter is negatively charged (37, 56).
Within the cationic lobe of MIP-3α, the far C-terminal region is of special interest, as it contains a high concentration of cationic and hydrophobic residues in an amphipathic helical conformation. Similar α-helical regions with flexible cationic tails have been identified as AMPs in other proteins, e.g., lactoferrampin (21). Moreover, this region of the IL-8 chemokine is reported to be antimicrobial (6). The structure of the C-terminal MIP-3α peptide bound to bacterial membrane mimetic SDS micelles shows an amphipathic helix highly similar to what is seen in the intact protein (Fig. 2). This amphipathic nature is thought to be important for an AMP's ability to initially interact with the anionic head groups of bacterial membranes and also the hydrophobic core (14). After initial contact with the membrane, the bactericidal activity can be achieved by membrane disruption or by entry of the peptide into the cell. Comparison of the C-terminal IL-8 peptide and the MIP-3α peptide once again strengthens this concept (see Fig. S4 in the supplemental material). The latter peptide is more highly charged and displays a clearer separation from its hydrophobic residues and consequently seems to be more bactericidal (see Fig. S4 in the supplemental material and Table 1). Both full-length MIP-3α and the C-terminal peptide display similar activity patterns against E. coli and S. aureus, being clearly more active against the former (Table 1). This is consistent with the observation that the C-terminal helix is solely responsible for the antimicrobial activity of MIP-3α. Fragments of MIP-3α spanning residues 1 to 52 and residues 1 to 55 have been found to be inactive as antibacterial agents, while a fragment comprised of residues 1 to 66 is as active as the intact protein (M. Wolf, personal communication). This indicates that the antimicrobial active center of MIP-3α can be pinpointed precisely to the C-terminal α helix. Here we used two methods under conditions with different ionic strengths and similar pHs to validate the antimicrobial activity of the C-terminal MIP-3α peptide (Table 1). pH has a more pronounced effect on the antimicrobial activity of peptides containing one or several histidine residues (45, 53). In this case, the MIP-3α peptide does not contain a histidine, which likely minimizes the effect of pH on its activity in vitro and in vivo (2, 55). The assay with phosphate buffer contained smaller amounts of salt and, as expected, somewhat higher levels of activity, similar to what has been shown for intact MIP-3α and other AMPs (41, 76), were found. Nevertheless, in vivo, MIP-3α is likely to be highly active. For example, tissue samples from patients with psoriasis and contact dermatitis, which are known to have low salt contents, have been shown to have high levels of expression of MIP-3α (25, 76). This would allow MIP-3α and its C-terminal peptide to display antimicrobial activity in these specific tissues. In an environment with higher salt concentrations, activity may occur in concert with the activities of other antimicrobial agents, as a number of AMPs, for example, HBD2, have been shown to have synergistic effects with other antimicrobial species (2, 44).
In conclusion, our solution structure and MD simulations of MIP-3α provide new insights into the properties of monomeric and dimeric chemokines. In a physiological setting, chemokine dimer formation requires contributing elements, such as GAG binding, to compensate for otherwise insufficiently high concentrations. Our data help explain why the monomeric structure, with a wider binding groove, a free N terminus, and overall more flexibility throughout the protein, is better suited for receptor activation. In agreement with other findings published in the literature, it appears that the dimeric chemokine structure serves as a negative regulator of chemokine function and, in addition, increases local concentrations, which help establish a concentration gradient important in cell migration (14). Furthermore, increased local concentrations are also important for the antimicrobial activities of chemokines. At elevated concentrations, MIP-3α may act either as an antimicrobial agent itself or in synergy with other AMPs and chemokines.
Supplementary Material
Acknowledgments
This work was supported by an operating grant from the Canadian Institute for Health Research to H.J.V. H.J.V. holds a senior scientist award from the Alberta Heritage Foundation for Medical Research.
We thank L. T. Nguyen and S. A. J. Zaat for their help with the peptide studies. In addition, we are grateful for the advice of C. M. Shepherd and T. Stockner in regards to the MD simulations and analysis. We thank D. D. McIntyre for his help with the NMR work and D. P. Tieleman for providing us with computing power for the MD simulations.
Footnotes
Published ahead of print on 17 December 2007.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Allen, S. J., S. E. Crown, and T. M. Handel. 2007. Chemokine: receptor structure, interactions, and antagonism. Annu. Rev. Immunol. 25:787-820. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. C., and P. L. Yu. 2005. Factors affecting the antimicrobial activity of ovine-derived cathelicidins against E. coli O157:H7. Int. J. Antimicrob. Agents 25:205-210. [DOI] [PubMed] [Google Scholar]
- 3.Baryshnikova, O. K., and B. D. Sykes. 2006. Backbone dynamics of SDF-1alpha determined by NMR: interpretation in the presence of monomer-dimer equilibrium. Protein Sci. 15:2568-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. Dinola, and J. R. Haak. 1984. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81:3684-3690. [Google Scholar]
- 5.Berendsen, H. J. C., D. van der Spoel, and R. van Drunen. 1995. Gromacs—a message-passing parallel molecular-dynamics implementation. Comput. Phys. Commun. 91:43-56. [Google Scholar]
- 6.Björstad, Å., H. M. Fu, A. Karlsson, C. Dahlgren, and J. Bylund. 2005. Interleukin-8-derived peptide has antibacterial activity. Antimicrob. Agents Chemother. 49:3889-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanpain, C., B. J. Doranz, A. Bondue, C. Govaerts, A. De Leener, G. Vassart, R. W. Doms, A. Proudfoot, and M. Parmentier. 2003. The core domain of chemokines binds CCR5 extracellular domains while their amino terminus interacts with the transmembrane helix bundle. J. Biol. Chem. 278:5179-5187. [DOI] [PubMed] [Google Scholar]
- 8.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. Delano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 9.Cole, A. M., T. Ganz, A. M. Liese, M. D. Burdick, L. Liu, and R. M. Strieter. 2001. Cutting edge: IFN-inducible ELR-CXC chemokines display defensin-like antimicrobial activity. J. Immunol. 167:623-627. [DOI] [PubMed] [Google Scholar]
- 10.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald—an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089-10092. [Google Scholar]
- 11.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. Bax. 1995. NMRPipe—a multidimensional spectral processing system based on Unix pipes. J. Biomol. NMR 6:277-293. [DOI] [PubMed] [Google Scholar]
- 12.Dieu, M. C., B. Vanbervliet, A. Vicari, J. M. Bridon, E. Oldham, S. Aït-Yahia, F. Brière, A. Zlotnik, S. Lebecque, and C. Caux. 1998. Selective recruitment of immature and mature dendritic cells by distinct chemokines expressed in different anatomic sites. J. Exp. Med. 188:373-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duma, L., D. Häussinger, M. Rogowski, P. Lusso, and S. Grzesiek. 2007. Recognition of RANTES by extracellular parts of the CCR5 receptor. J. Mol. Biol. 365:1063-1075. [DOI] [PubMed] [Google Scholar]
- 14.Epand, R. M., and H. J. Vogel. 1999. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta 1462:11-28. [DOI] [PubMed] [Google Scholar]
- 15.Esche, C., C. Stellato, and L. A. Beck. 2005. Chemokines: key players in innate and adaptive immunity. J. Investig. Dermatol. 125:615-628. [DOI] [PubMed] [Google Scholar]
- 16.Feenstra, K. A., B. Hess, and H. J. C. Berendsen. 1999. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J. Comput. Chem. 20:786-798. [DOI] [PubMed] [Google Scholar]
- 17.Fernando, H., C. Chin, J. Rosgen, and K. Rajarathnam. 2004. Dimer dissociation is essential for interleukin-8 (IL-8) binding to CXCR1 receptor. J. Biol. Chem. 279:36175-36178. [DOI] [PubMed] [Google Scholar]
- 18.Formaneck, M. S., L. Ma, and Q. Cui. 2006. Effects of temperature and salt concentration on the structural stability of human lymphotactin: insights from molecular simulations. J. Am. Chem. Soc. 128:9506-9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 20.Guijarro, J. I., M. Sunde, J. A. Jones, I. D. Campbell, and C. M. Dobson. 1998. Amyloid fibril formation by an SH3 domain. Proc. Natl. Acad. Sci. USA 95:4224-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haney, E. F., F. Lau, and H. J. Vogel. 2007. Solution structures and model membrane interactions of lactoferrampin, an antimicrobial peptide derived from bovine lactoferrin. Biochim. Biophys. Acta 1768:2355-2364. [DOI] [PubMed] [Google Scholar]
- 22.Harrison, J. K., A. M. Fong, P. A. W. Swain, S. Z. Chen, Y. R. A. Yu, M. N. Salafranca, W. B. Greenleaf, T. Imai, and D. D. Patel. 2001. Mutational analysis of the fractalkine chemokine domain—basic amino acid residues differentially contribute to CX3CR1 binding, signaling, and cell adhesion. J. Biol. Chem. 276:21632-21641. [DOI] [PubMed] [Google Scholar]
- 23.Hasan, L., L. Mazzucchelli, M. Liebi, M. Lis, R. E. Hunger, A. Tester, C. M. Overall, and M. Wolf. 2006. Function of liver activation-regulated chemokine/CC chemokine ligand 20 is differently affected by cathepsin B and cathepsin D processing. J. Immunol. 176:6512-6522. [DOI] [PubMed] [Google Scholar]
- 24.Hess, B., H. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463-1472. [Google Scholar]
- 25.Homey, B., M. C. Dieu-Nosjean, A. Wiesenborn, C. Massacrier, J. J. Pin, E. Oldham, D. Catron, M. E. Buchanan, A. Muller, R. D. Malefyt, G. Deng, R. Orozco, T. Ruzicka, P. Lehmann, S. Lebecque, C. Caux, and A. Zlotnik. 2000. Up-regulation of macrophage inflammatory protein-3 alpha/CCL20 and CC chemokine receptor 6 in psoriasis. J. Immunol. 164:6621-6632. [DOI] [PubMed] [Google Scholar]
- 26.Hoogewerf, A. J., G. S. V. Kuschert, A. E. I. Proudfoot, F. Borlat, I. Clark-Lewis, C. A. Power, and T. N. C. Wells. 1997. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry 36:13570-13578. [DOI] [PubMed] [Google Scholar]
- 27.Hoover, D. M., C. Boulègue, D. Yang, J. J. Oppenheim, K. Tucker, W. Y. Lu, and J. Lubkowski. 2002. The structure of human macrophage inflammatory protein-3 alpha/CCL20—linking antimicrobial and CC chemokine receptor-6-binding activities with human beta-defensins. J. Biol. Chem. 277:37647-37654. [DOI] [PubMed] [Google Scholar]
- 28.Horcher, M., A. Rot, H. Aschauer, and J. Besemer. 1998. IL-8 derivatives with a reduced potential to form homodimers are fully active in vitro and in vivo. Cytokine 10:1-12. [DOI] [PubMed] [Google Scholar]
- 29.Hosokawa, Y., T. Nakanishi, D. Yamaguchi, K. Takahashi, H. Yumoto, K. Ozaki, and T. Matsuo. 2002. Macrophage inflammatory protein 3 alpha-CC chemokine receptor 6 interactions play an important role in CD4+ T-cell accumulation in periodontal diseased tissue. Clin. Exp. Immunol. 128:548-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33-38. [DOI] [PubMed] [Google Scholar]
- 31.Hunter, H. N., D. B. Fulton, T. Ganz, and H. J. Vogel. 2002. The solution structure of human hepcidin, a peptide hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis. J. Biol. Chem. 277:37597-37603. [DOI] [PubMed] [Google Scholar]
- 32.Hwang, T. L., and A. J. Shaka. 1995. Water suppression that works—excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. A 112:275-279. [Google Scholar]
- 33.Jarnagin, K., D. Grunberger, M. Mulkins, B. Wong, S. Hemmerich, C. Paavola, A. Bloom, S. Bhakta, F. Diehl, R. Freedman, D. McCarley, I. Polsky, A. Ping-Tsou, A. Kosaka, and T. M. Handel. 1999. Identification of surface residues of the monocyte chemotactic protein 1 that affect signaling through the receptor CCR2. Biochemistry 38:16167-16177. [DOI] [PubMed] [Google Scholar]
- 34.Johnson, B. A., and R. A. Blevins. 1994. NMRView—a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4:603-614. [DOI] [PubMed] [Google Scholar]
- 35.Jones, J. A., D. K. Wilkins, L. J. Smith, and C. M. Dobson. 1997. Characterisation of protein unfolding by NMR diffusion measurements. J. Biomol. NMR 10:199-203. [Google Scholar]
- 36.Keizer, D. W., M. P. Crump, T. W. Lee, C. M. Slupsky, I. Clark-Lewis, and B. D. Sykes. 2000. Human CC chemokine I-309, structural consequences of the additional disulfide bond. Biochemistry 39:6053-6059. [DOI] [PubMed] [Google Scholar]
- 37.Kim, B. E., D. Y. M. Leung, J. E. Streib, M. Boguniewicz, Q. A. Hamid, and M. D. Howell. 2007. Macrophage inflammatory protein 3alpha deficiency in atopic dermatitis skin and role in innate immune response to vaccinia virus. J. Allergy Clin. Immunol. 119:457-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleeff, J., T. Kusama, D. L. Rossi, T. Ishiwata, H. Maruyama, H. Friess, M. W. Büchler, A. Zlotnik, and M. Korc. 1999. Detection and localization of Mip-3alpha/LARC/Exodus, a macrophage proinflammatory chemokine, and its CCR6 receptor in human pancreatic cancer. Int. J. Cancer 81:650-657. [DOI] [PubMed] [Google Scholar]
- 39.Koradi, R., M. Billeter, and K. Wüthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51-55. [DOI] [PubMed] [Google Scholar]
- 40.Kuschert, G. S. V., A. J. Hoogewerf, A. E. I. Proudfoot, C. W. Chung, R. M. Cooke, R. E. Hubbard, T. N. C. Wells, and P. N. Sanderson. 1998. Identification of a glycosaminoglycan binding surface on human interleukin-8. Biochemistry 37:11193-11201. [DOI] [PubMed] [Google Scholar]
- 41.Lai, J. R., R. F. Epand, B. Weisblum, R. M. Epand, and S. H. Gellman. 2006. Roles of salt and conformation in the biological and physicochemical behavior of protegrin-1 and designed analogues: correlation of antimicrobial, hemolytic, and lipid bilayer-perturbing activities. Biochemistry 45:15718-15730. [DOI] [PubMed] [Google Scholar]
- 42.Laskowski, R. A., M. W. Macarthur, D. S. Moss, and J. M. Thornton. 1993. Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 43.Laurence, J. S., C. Blanpain, J. W. Burgner, M. Parmentier, and P. J. Liwang. 2000. CC chemokine MIP-1 beta can function as a monomer and depends on Phe13 for receptor binding. Biochemistry 39:3401-3409. [DOI] [PubMed] [Google Scholar]
- 44.Lee, H. Y., A. Andalibi, P. Webster, S. K. Moon, K. Teufert, S. H. Kang, J. D. Li, M. Nagura, T. Ganz, and D. J. Lim. 2004. Antimicrobial activity of innate immune molecules against Streptococcus pneumoniae, Moraxella catarrhalis and nontypeable Haemophilus influenzae. BMC Infect. Dis. 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee, I. H., Y. Cho, and R. I. Lehrer. 1997. Effects of pH and salinity on the antimicrobial properties of clavanins. Infect. Immun. 65:2898-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao, F., R. Alderson, J. Su, S. J. Ullrich, B. L. Kreider, and J. M. Farber. 1997. STRL22 is a receptor for the CC chemokine MIP-3 alpha. Biochem. Biophys. Res. Commun. 236:212-217. [DOI] [PubMed] [Google Scholar]
- 47.Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306-317. [Google Scholar]
- 48.Linge, J. P., S. I. O'Donoghue, and M. Nilges. 2001. Automated assignment of ambiguous nuclear Overhauser effects with ARIA. Methods Enzymol. 339:71-90. [DOI] [PubMed] [Google Scholar]
- 49.Ma, L., and Q. Cui. 2006. The temperature dependence of salt-protein association is sequence specific. Biochemistry 45:14466-14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malik, Z. A., and B. F. Tack. 2006. Structure of human MIP-3alpha chemokine. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62:631-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malkowski, M. G., J. Y. Wu, J. B. Lazar, P. H. Johnson, and B. F. P. Edwards. 1995. The crystal-structure of recombinant human neutrophil-activating peptide-2 (M6L) at 1.9-angstrom resolution. J. Biol. Chem. 270:7077-7087. [DOI] [PubMed] [Google Scholar]
- 52.Markley, J. L. 1975. Observation of histidine residues in proteins by means of nuclear magnetic resonance spectroscopy. Acc. Chem. Res. 8:70-80. [Google Scholar]
- 53.Mason, A. J., C. Gasnier, A. Kichler, G. Prévost, D. Aunis, M. H. Metz-Boutigue, and B. Bechinger. 2006. Enhanced membrane disruption and antibiotic action against pathogenic bacteria by designed histidine-rich peptides at acidic pH. Antimicrob. Agents Chemother. 50:3305-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merrill, M. R. 1993. NMR diffusion measurements using a composite gradient PGSE sequence. J. Magn. Reson. A 103:223-225. [Google Scholar]
- 55.Miyakawa, Y., P. Ratnakar, A. G. Rao, M. L. Costello, O. Mathieu-Costello, R. I. Lehrer, and A. Catanzaro. 1996. In vitro activity of the antimicrobial peptides human and rabbit defensins and porcine leukocyte protein against Mycobacterium tuberculosis. Infect. Immun. 64:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakayama, T., J. Shirane, K. Hieshima, M. Shibano, M. Watanabe, Z. Jin, D. Nagakubo, T. Saito, Y. Shimomura, and O. Yoshie. 2006. Novel antiviral activity of chemokines. Virology 350:484-492. [DOI] [PubMed] [Google Scholar]
- 57.Nilges, M., and S. I. O'Donoghue. 1998. Ambiguous NOEs and automated NOE assignment. Prog. Nucl. Magn. Reson. Spectrosc. 32:107-139. [Google Scholar]
- 58.Paavola, C. D., S. Hemmerich, D. Grunberger, I. Polsky, A. Bloom, R. Freedman, M. Mulkins, S. Bhakta, D. McCarley, L. Wiesent, B. Wong, K. Jarnagin, and T. M. Handel. 1998. Monomeric monocyte chemoattractant protein-1 (MCP-1) binds and activates the MCP-1 receptor CCR2B. J. Biol. Chem. 273:33157-33165. [DOI] [PubMed] [Google Scholar]
- 59.Pérez-Cañadillas, J. M., A. Zaballos, J. Gutiérrez, R. Varona, F. Roncal, J. P. Albar, G. Márquez, and M. Bruix. 2001. NMR solution structure of murine CCL20/MIP-3alpha, a chemokine that specifically chemoattracts immature dendritic cells and lymphocytes through its highly specific interaction with the beta-chemokine receptor CCR6. J. Biol. Chem. 276:28372-28379. [DOI] [PubMed] [Google Scholar]
- 60.Reference deleted.
- 61.Power, C. A., D. J. Church, A. Meyer, S. Alouani, A. E. I. Proudfoot, I. Clark-Lewis, S. Sozzani, A. Mantovani, and T. N. C. Wells. 1997. Cloning and characterization of a specific receptor for the novel CC chemokine MIP-3alpha from lung dendritic cells. J. Exp. Med. 186:825-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajarathnam, K., G. N. Prado, H. Fernando, I. Clark-Lewis, and J. Navarro. 2006. Probing receptor binding activity of interleukin-8 dimer using a disulfide trap. Biochemistry 45:7882-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Redfield, C., B. A. Schulman, M. A. Milhollen, P. S. Kim, and C. M. Dobson. 1999. Alpha-lactalbumin forms a compact molten globule in the absence of disulfide bonds. Nat. Struct. Biol. 6:948-952. [DOI] [PubMed] [Google Scholar]
- 64.Ruth, J. H., S. Shahrara, C. C. Park, J. C. M. Morel, P. Kumar, S. X. Qin, and A. E. Koch. 2003. Role of macrophage inflammatory protein-3 alpha and its ligand CCR6 in rheumatoid arthritis. Lab. Investig. 83:579-588. [DOI] [PubMed] [Google Scholar]
- 65.Schibli, D. J., H. N. Hunter, V. Aseyev, T. D. Starner, J. M. Wiencek, P. B. McCray, B. F. Tack, and H. J. Vogel. 2002. The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus. J. Biol. Chem. 277:8279-8289. [DOI] [PubMed] [Google Scholar]
- 66.Schibli, D. J., L. T. Nguyen, S. D. Kernaghan, Ø. Rekdal, and H. J. Vogel. 2006. Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures. Biophys. J. 91:4413-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scott, W. R. P., P. H. Hunenberger, I. G. Tironi, A. E. Mark, S. R. Billeter, J. Fennen, A. E. Torda, T. Huber, P. Krüger, and W. F. van Gunsteren. 1999. The GROMOS biomolecular simulation program package. J. Phys. Chem. A 103:3596-3607. [Google Scholar]
- 68.Strøm, M. B., Ø. Rekdal, and J. S. Svendsen. 2002. Antimicrobial activity of short arginine and tryptophan-rich peptides. J. Pept. Sci. 8:431-437. [DOI] [PubMed] [Google Scholar]
- 69.Strøm, M. B., Ø. Rekdal, and J. S. Svendsen. 2002. The effects of charge and lipophilicity on the antibacterial activity of undecapeptides derived from bovine lactoferricin. J. Pept. Sci. 8:36-43. [DOI] [PubMed] [Google Scholar]
- 70.Thomas, C. L., E. McKinnon, B. L. Granger, E. Harms, and R. L. Van Etten. 2002. Kinetic and spectroscopic studies of Tritrichomonas foetus low-molecular weight phosphotyrosyl phosphatase. Hydrogen bond networks and electrostatic effects. Biochemistry 41:15601-15609. [DOI] [PubMed] [Google Scholar]
- 71.Veldkamp, C. T., F. C. Peterson, A. J. Pelzek, and B. F. Volkman. 2005. The monomer-dimer equilibrium of stromal cell-derived factor-1 (CXCL 12) is altered by pH, phosphate, sulfate, and heparin. Protein Sci. 14:1071-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilkins, D. K., S. B. Grimshaw, V. Receveur, C. M. Dobson, J. A. Jones, and L. J. Smith. 1999. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry 38:16424-16431. [DOI] [PubMed] [Google Scholar]
- 73.Wishart, D. S., B. D. Sykes, and F. M. Richards. 1992. The chemical-shift index—a fast and simple method for the assignment of protein secondary structure through NMR-spectroscopy. Biochemistry 31:1647-1651. [DOI] [PubMed] [Google Scholar]
- 74.Wu, Z. B., D. M. Hoover, D. Yang, C. Boulègue, F. Santamaria, J. J. Oppenheim, J. Lubkowski, and W. Y. Lu. 2003. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human beta-defensin 3. Proc. Natl. Acad. Sci. USA 100:8880-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wüthrich, K. 1986. NMR of proteins and nucleic acids. John Wiley & Sons, Inc., New York, NY.
- 76.Yang, D., Q. Chen, D. M. Hoover, P. Staley, K. D. Tucker, J. Lubkowski, and J. J. Oppenheim. 2003. Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J. Leukoc. Biol. 74:448-455. [DOI] [PubMed] [Google Scholar]
- 77.Yang, D., O. Chertov, N. Bykovskaia, Q. Chen, M. J. Buffo, J. Shogan, M. Anderson, J. M. Schröder, J. M. Wang, O. M. Z. Howard, and J. J. Oppenheim. 1999. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-528. [DOI] [PubMed] [Google Scholar]
- 78.Zlotnik, A., O. Yoshie, and H. Nomiyama. 2006. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 7:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.