High-level resistance to aminoglycosides mediated by the production of 16S rRNA methylase among various gram-negative pathogens has been increasingly reported (4). Six 16S rRNA methylase enzymes have been identified: RmtA to RmtD, ArmA, and NpmA (4, 9). We recently described the emergence of ArmA among Acinetobacter baumannii strains in North America (3). Here, we report the isolation of an Escherichia coli strain coproducing 16S rRNA methylase RmtB and extended-spectrum β-lactamase (ESBL) CTX-M-65 from an ambulatory female with a history of sickle-cell anemia. E. coli grew in a urine culture ordered as part of her evaluation for chronic kidney disease. A review of the records indicated that she had been hospitalized briefly 4 months earlier, but the urine culture did not grow E. coli during or after that hospitalization. This is the first instance in which RmtB has been shown to be encoded on the same plasmid as an ESBL.
The isolate, E. coli ECRB1, was highly resistant to cefotaxime (MIC, 64 μg/ml) and gentamicin, tobramycin, and amikacin (MICs, >256 μg/ml) but was susceptible to trimethoprim-sulfamethoxazole and ciprofloxacin by Etest (AB Biodisk, Solna, Sweden). Transconjugants were obtained by the broth mating method on Luria-Bertani plates containing 50 μg of rifampin/ml and 50 μg of amikacin/ml by using E. coli XL1-Blue Rifr as the recipient. Transformants of E. coli DH10B were obtained on Luria-Bertani plates containing 50 μg of amikacin/ml following electroporation with plasmids purified from the parental strain. The MICs of cefotaxime (96 μg/ml) and the three aminoglycosides (≥128 μg/ml) were high for both the transconjugant and transformant strains. The results of PCR analysis of whole-cell lysates for CTX-M genes (6) were positive for both strains. Direct sequencing of the entire structural gene amplified from the transformant confirmed it to encode CTX-M-65, a variant of CTX-M-14. Multiplex PCR analysis of the transconjugant and transformant strains for 16S rRNA methylase genes (4) revealed the presence of rmtB. The results of PCR analysis for qepA, a quinolone efflux pump gene reported to be carried on the same plasmid as rmtB (10), were negative for the transconjugant and transformant strains.
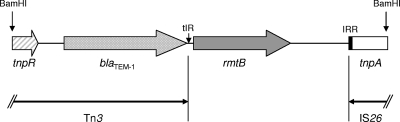
A 2.9-kb BamHI insert containing rmtB could be cloned from the transformant into vector pBCSK(−) (Stratagene, La Jolla, CA) with E. coli DH10B as the host. This plasmid conferred a high level of resistance to all three aminoglycosides tested (MICs, >256 μg/ml). Full sequencing of this insert revealed a genetic arrangement similar to those reported earlier (5, 10) in that the insert contained blaTEM-1 as part of Tn3 upstream from rmtB (Fig. 1). However, the putative transposase gene previously found downstream of rmtB was replaced by the 3′ end of another transposase gene comprising IS26.
FIG. 1.
Genetic support of rmtB. rmtB is flanked by Tn3 and IS26. tnpR, resolvase gene; tnpA, transposase gene; tIR, terminal inverted repeat of Tn3 (5′-CTTAACGTGAGTTTTCGTTCCACTGAGCGTCAGACCCC-3′); IRR, right inverted repeat of IS26 (5′-GGCACTGTTGCAAA-3′).
CTX-M-type ESBLs are increasingly associated with community-acquired E. coli infections throughout the world, including the United States (2, 8). rmtB has been found in strains carrying blaCTX-M but, unlike armA (1, 7, 11), on a separate plasmid from blaCTX-M. Our findings indicate, however, that rmtB may be captured by the same conjugative plasmid carrying blaCTX-M. The spread of such multidrug resistance plasmids among E. coli strains has a potential impact on the empirical management of complicated urinary tract infections that may be treated initially with cephalosporins and aminoglycosides.
Nucleotide sequence accession number.
The sequences determined in this work have been deposited in GenBank under accession no. EU213261 and EU213262.
Acknowledgments
Y.D. is supported by NIH training grant T32 AI007333. D.L.P. has received prior research funding from Pfizer, Elan, Merck, and AstraZeneca and is supported in part by NIH research grant R01 AI070896.
Footnotes
Published ahead of print on 14 January 2008.
REFERENCES
- 1.Bogaerts, P., M. Galimand, C. Bauraing, A. Deplano, R. Vanhoof, R. De Mendonca, H. Rodriguez-Villalobos, M. Struelens, and Y. Glupczynski. 2007. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J. Antimicrob. Chemother. 59:459-464. [DOI] [PubMed] [Google Scholar]
- 2.Doi, Y., J. Adams, A. O'Keefe, Z. Quereshi, L. Ewan, and D. L. Paterson. 2007. Community-acquired extended-spectrum β-lactamase producers, United States. Emerg. Infect. Dis. 13:1121-1123. [DOI] [PubMed] [Google Scholar]
- 3.Doi, Y., J. M. Adams, K. Yamane, and D. L. Paterson. 2007. Identification of 16S rRNA methylase-producing Acinetobacter baumanii clinical strains in North America. Antimicrob. Agents Chemother. 51:4209-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doi, Y., and Y. Arakawa. 2007. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 45:88-94. [DOI] [PubMed] [Google Scholar]
- 5.Doi, Y., K. Yokoyama, K. Yamane, J. Wachino, N. Shibata, T. Yagi, K. Shibayama, H. Kato, and Y. Arakawa. 2004. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob. Agents Chemother. 48:491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edelstein, M., M. Pimkin, I. Palagin, I. Edelstein, and L. Stratchounski. 2003. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob. Agents Chemother. 47:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galimand, M., S. Sabtcheva, P. Courvalin, and T. Lambert. 2005. Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob. Agents Chemother. 49:2949-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livermore, D. M., R. Canton, M. Gniadkowski, P. Nordmann, G. M. Rossolini, G. Arlet, J. Ayala, T. M. Coque, I. Kern-Zdanowicz, F. Luzzaro, L. Poirel, and N. Woodford. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165-174. [DOI] [PubMed] [Google Scholar]
- 9.Wachino, J. I., K. Shibayama, H. Kurokawa, K. Kimura, K. Yamane, S. Suzuki, N. Shibata, Y. Ike, and Y. Arakawa. 2007. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, NpmA, found in a clinically isolated Escherichia coli strain resistant to structurally diverse aminoglycosides. Antimicrob. Agents Chemother. 51:4401-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamane, K., J. Wachino, S. Suzuki, K. Kimura, N. Shibata, H. Kato, K. Shibayama, T. Konda, and Y. Arakawa. 2007. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 51:3354-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan, J. J., J. J. Wu, W. C. Ko, S. H. Tsai, C. L. Chuang, H. M. Wu, Y. J. Lu, and J. D. Li. 2004. Plasmid-mediated 16S rRNA methylases conferring high-level aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J. Antimicrob. Chemother. 54:1007-1012. [DOI] [PubMed] [Google Scholar]